Abstract
Transcription potential is determined by the accessibility of DNA sequences within the context of chromatin, which is coordinately controlled by various epigenetic modifications. Chemical inhibition of epigenetic regulators provides a quick and effective approach to investigate the roles of epigenetic modifications in controlling many biological processes, especially for species in which genetic information is limited. This mini-review provides a brief overview of epigenetic regulators in the model organism Arabidopsis thaliana and summarizes compounds that have been applied in plant epigenetics studies, with highlights in the applications of these chemical probes in mechanistic and functional investigations.
Keywords: plant epigenetics, chemical inhibitors, DNA methylation, histone modification, methyltransferase inhibitors
Transcription of genetic information is a principle biological process. Plant genomic DNA is hierarchically organized into complex chromatin structures with histones and non-histone architectural proteins. The status of chromatin is oftentimes termed as euchromatic or heterochromatic, indicating the accessibility of DNA to the transcriptional machinery being easy or difficult, respectively. While DNA can be methylated at the 5th position of the pyrimidine ring of cytosine, histones are subject to various modifications on different amino acid residues. These epigenetic marks, in combination with other epigenetic regulators such as chromatin-remodeling proteins, determine the accessibility of chromatin to transcriptional activities, and can be faithfully inherited through cell divisions. Compounds that alter the establishment or removal of certain epigenetic marks have been useful tools in elucidating mechanisms underlying epigenetic regulation in Arabidopsis, in which many epigenetic regulators have been identified through genetic studies. In other plant species where genetic mutants are less available, biological significance of epigenetic modifications are commonly demonstrated by applying chemical molecules that are known to interfere with epigenetic modifications in Arabidopsis or mammalian cells.
Epigenetic Regulators —Potential Targets of Chemical Inhibitors/Activators
Epigenetic regulators in plants can be roughly categorized based on their functions in controlling DNA methylation and/or histone modifications. In addition, certain epigenetic regulators such as MORC family ATPases and MOM1 do not fall into these categories, in that their mutations release heterochromatic transcriptional silencing largely independently of changes in the levels of DNA methylation or the repressive histone H3 lysine-9 dimethylation.1-4
DNA methylation
In Arabidopsis, establishment of DNA methylation can be mediated by DRM2, which catalyzes methylation in all cytosine contexts, namely CG, CHG, and CHH (H represents either A, T, or G). Once established, DNA methylation at the symmetric CG and CHG contexts is maintained by MET1 and CMT3, respectively, whereas cytosines in CHH context need de novo methylation during every cell cycle due to its asymmetric nature.5 In addition to DNA methyltransferases, non-coding RNAs (ncRNAs) play important roles in establishing DNA methylation. In the RNA-directed DNA methylation (RdDM) pathway, complementary pairing between 24 nt small interfering RNA and nascent scaffold RNA guides DRM2 to its target loci, with the aid of protein-protein interactions among the components within the silencing complex. The list of RdDM components continues to expand and have been reviewed elsewhere in detail.5-10
Active DNA dememthylation
DNA methylation can be lost as a result of passive or active demethylation processes. The former refers to the failure in maintaining DNA methylation due to DNA methyltransferase dysfunction or shortage of methyl-group supplies, whereas the latter is an outcome of enzymatic action resulting in the replacement of 5-methylcytosine with cytosine. Plant cytosine methylation can be actively removed by a subfamily of bi-functional DNA glycosylases represented by ROS1 and DME. These DNA demethylases directly excise the 5-methylcytosine base and then cleave the DNA backbone at the abasic site. The resultant single-nucleotide gap is subsequently filled with an unmodified cytosine through the DNA base excision repair pathway.11,12 Recruitment of ROS1 to its target loci has been suggested to be assisted by ROS3, an RNA-binding protein.13 The zinc finger DNA 3′phosphoesterase ZDP removes the 3′-phosphate from ROS1-nicked DNA, allowing subsequent base excision repair that can be mediated by XRCC1 in Arabidopsis.14,15 DME-mediated demethylation involves the DNA LIGASE 1 (LIG1), as indicated by genetic interaction analyses of maternal effects on seed development.16 Mutation of SSRP1, a non-histone chromosomal protein, leads to DNA hypermethylation as well as additional repressive chromatin modifications, thereby disrupting regulation of many paternally imprinted genes in the central cell.17
Histone modifications
Proteins involved in histone post-translational modification are potential epigenetic regulators, since histone proteins are core components of chromatin structures. Cytosine methylation and demethylation are both tightly linked with histone modifications, of which methylation and acetylation are known to be important for epigenetic regulation of gene expression. Arabidospis possesses protein families that catalyze either acetylation, deacetylation, methylation, or demethylation at various histone lysine residues. This review focuses on histone modifications that are known to be closely related to DNA methylation.
Histone acetylation is an active epigenetic mark that is conserved in eukaryotes.18,19 Increased DNA methylation was observed in Arabidopsis mutant defective in IDM1, an acetyltransferase that catalyzes acetylation of histone H3 lysine 18 (H3K18) and lysine 23 (H3K23) for subsequent DNA demethylation.20 The histone deacetylase HDA6 is required for DNA methylation at some loci, and its mutation significantly increases all lysine acetylation on the N-termini of histone H3 and H4, except H4K16, at its target loci.21 In Arabidopsis, mono- and dimethylation of H3K9 (H3K9me and H2K9me2, respectively), H3K27me, H3K27me2, and H4K20me are repressive histone marks associated with heterochromatin, while H3K4me3 is an active histone mark permissive to RNA transcription.22,23 SUVH2 maintains all types of repressive histone methylation,22 while SUVH4/KYP is the major histone methyltransferase responsible for H3K9me2, which can also be catalyzed by SUVH5 and SUVH6.24 SUVH4 and CMT3 bind to CHG methylation and H3K9me2, respectively, thereby forming a reinforcing loop of DNA methyaltion and H3K9me2.5,25 Indeed, maintenance of DNA methylation in the CHG context can be impaired by IBM1, which is a histone demethylase that prevents H3K9 methylation.26,27
Histone H2B ubiquitination is another known epigenetic modification. Mutation of SUP32/UBP26, an Arabidopsis deubiquitination enzyme, increases histone H2B mono-ubiquitinatioin at lysine 143 and H3K4me3 levels, accompanied by reduced H3K9me2 and suppression of RdDM-mediated heterochromatic silencing.28 In addition, concomitant elevation in H2B ubiquitination and H3K4 methylation were observed in Arabidopsis mutants defective in OTLD1 or KDM1C, which are histone deubiquitinase and histone demethylase, respectively.29 In mediating transcriptional regulation, histone modification can also involve ncRNAs such as COLDAIR, a cold-inducible long ncRNA that increases H3K27me3 marks at chromatin of the floral repressor FLC through recruitment of polycomb repressive complex 2.30 Moreover, DNA methylation shows a global anti-correlation with deposition of the histone variant H2A.Z, loss of which leads misregulation of many genes that are disproportionately associated with response to environmental and developmental stimuli.31,32
Compounds for Epigenetic Regulation in Plants
Based on their modes of action, chemicals that have been applied in plant epigenetics studies to date can be categorized into 3 groups: DNA methyltransferase inhibitors, histone deacetylase inhibitors, and molecules that disrupt normal methyl supplies for methyl-transferring reactions (Table 1).
Table 1. Compounds applied in plant epigenetics studies.
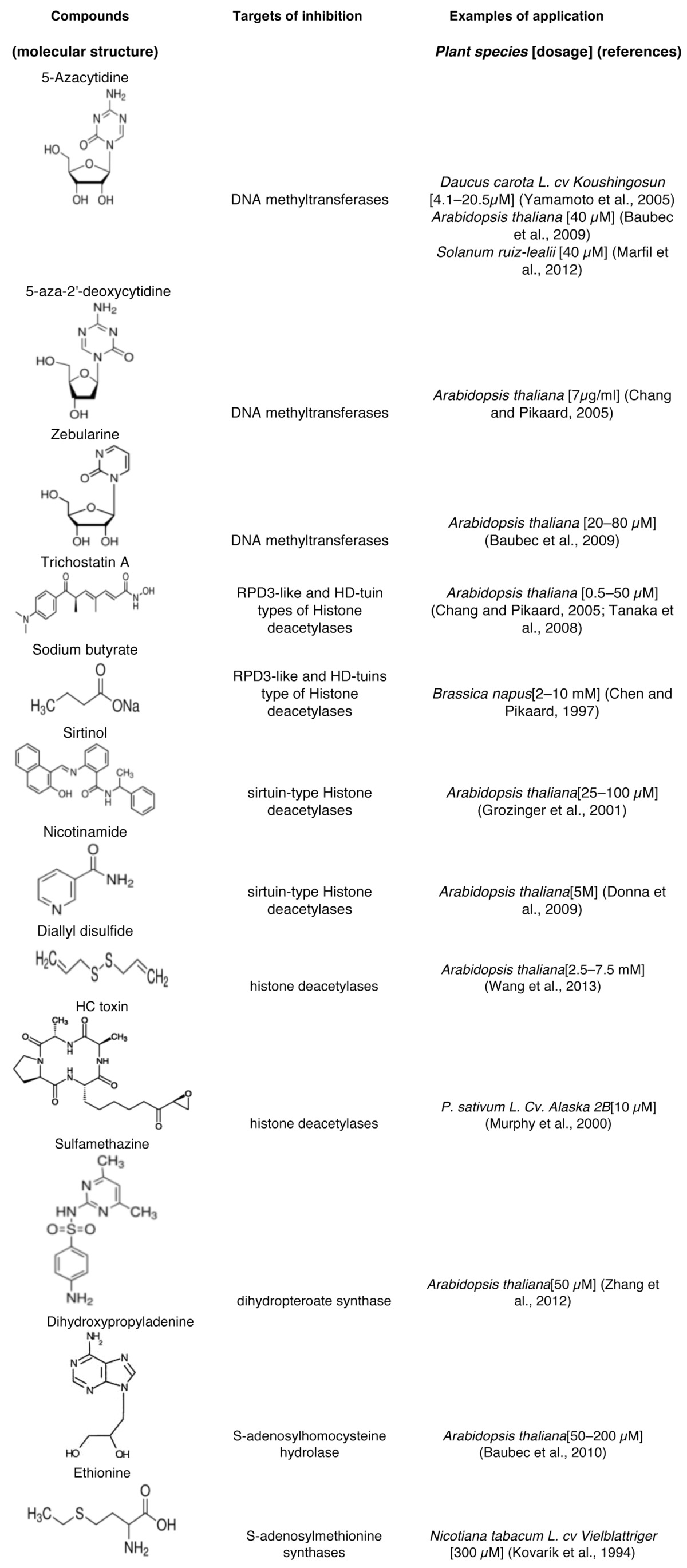
DNA methyltransferase inhibitors
During the catalysis of methyl transferring, the DNA methylatransferase forms a covalent bond with the cytosine carbon at the 6th position, and then transfers the methyl group from S-adenosylmethionine (SAM) to the 5th position of the cytosine ring. Following the methyl transferring, DNA methyltransferase will be released from its covalent bond with cytosine and move on to catalyze the next methylation reaction. Chemical analogs of cytosine that are incorporated in the DNA can also form covalent adducts with DNA methylatransferases, but a lack of methyl transferring reaction will then trap the enzymes on the DNA, thereby causing reduction in genome-wide DNA methylation.33
The two cytosine analogs, 5-Azacytidine (5-aza) and 5-aza-2'-deoxycytidine (aza-dC), are DNA methyltransferases inhibitors that are commonly applied in epigenetics studies. Global changes in the transcriptome have been shown in Arabidopsis treated with aza-dC.34 Consistent with the genome-wide DNA hypomethylation, genes responsive to aza-dC are distributed throughout the five chromosomes of Arabidopsis. Meanwhile, genes upregulated by aza-dC treatment were also shown to be induced in Arabidopsis mutants that are defective in cytosine methylation. Interestingly, genes that were downregulated by aza-dC treatment were enriched in stress response genes, though the functional significance of such enrichment remains unknown.34 Besides Arabidopsis, many other plant species have been studied with 5-aza or aza-dC to explore the biological significance of DNA methylation. In a carrot somatic embryogenesis system, treatment with 5-aza suppressed the formation of embryogenic cell clumps from epidermal cells.35 In the wild potato Solanum ruiz-lealii, application of 5-aza induced early flowering and changes in leaf morphology, which were possibly linked with transcriptional induction of four miRNAs including miR172.36 Flowering induced by 5-aza was also observed in Silene armeria, Pharbitis nil, and Perilla frutescens, while such induction seemed to be independent of the stability of the photoperiodically induced flowering state.37 In the plant Secale cereale, 5-aza treatment increased nuclear and nucleolar sizes and reallocated most of the rDNA from perinucleolar heterochromatin into the nucleolus, accompanied with an increase in rRNA gene transcription.38
Zebularine is another cytosine analog that is commonly used as a DNA methylatransferase inhibitor. Compared with 5-aza and aza-dC that are unstable in aqueous solution,39,40 zebularine is much more stable under physiological conditions and shows fewer side-effects.41 Zebularine treatments in Arabidopsis decreased DNA methylation levels in a dose-dependent and transient manner independent of cytosine contexts, demonstrating the effectiveness of zebularine in plant epigenetics studies.42
Histone deacetylase inhibitors
Trichostatin A (TSA) is a histone deacetylase inhibitor that has been applied in plant epigenetics studies. Crystallographic studies of a bacterial HDAC revealed that TSA binds to HDAC by inserting its long aliphatic chain into the catalytically active pocket, where it makes multiple contacts with the enzyme and thereby inhibits the enzymatic activity of HDAC.43 TSA and other HDAC inhibitors have been applied in plant epigenetics studies, often in combination with mutagenetic studies, to investigate the importance of histone acetylation to various biological processes at phenotypic and molecular levels. In an Arabidopsis HDA6 repression line, TSA treatment caused growth arrest due to releasing of the suppressed gene expression of LEAFY COTYLEDON1 (LEC1), FUSCA3 (FUS3), and ABSCISIC ACID INSENSITIVE3 (ABI3), all of which are embryonic transcription factors functioning in the maintenance of embryonic properties.44 An HDA6/HDA19 double-repression line similarly displayed growth arrest and embryo-like structures on mature leaves in the absence of HDAC inhibitor,44 suggesting that inhibition of HDA19, which functions redundantly with HDA6 in promoting normal post-germination growth, mediates TSA-induced growth arrest. TSA also alters Arabidopsis root hair cell development, likely through its inhibition of HDA18, since mutation of HDA18 resulted in a similar phenotype as was observed in TSA-treated wild type plants.45
Either TSA or 5-aza treatment can de-repress silenced rRNA genes in plants, while simultaneous treatment of both chemicals did not further elevate rRNA transcript levels,46 indicating that histone deacetylation and DNA methylation function in the same pathway to regulate rRNA transcriptional silencing. This is consistent with the general notion that histone deacetylation confers transcriptional gene silencing and is correlated with repressive epigenetic marks such as DNA methylation. On the other hand, studies with chemical probes also revealed unrelated- or even antagonistic gene regulation by histone acetylation and DNA methylation. TSA has been shown to have no effects on transcriptional silencing of some transgenes that can be induced by 5-aza treatments.47,48 Meanwhile, comparative transcriptional profiling of Arabidopsis treated with TSA, aza-dC, or both, disclosed little overlap between genes that were differentially regulated by these two chemicals, as well as the fact that TSA and aza-dC are often antagonistic in regulating gene expression.34
Similar to TSA, sodium butyrate activates silent rRNA genes.46 Arabidopsis treated with sodium butyrate exhibits increased acetylation of histone H4 at lysines 5, 8, 12, and 16, all of which are found at the promoters of active rRNA genes.49 As a result of the enrichment in the pool of acetylated histones, sodium butyrate treatment also rendered subsequent mass spectrometric analyses to discover the processive nature of histone H4 hyperacetylation.49 Activities of sirtuin-type HDACs are unaffected by TSA or sodium butyrate, but can be inhibited by sirtinol, which inhibits proper vascularization as well as hypocotyl and root development in Arabidopsis.50 In addition, several other histone deacetylation inhibitors including nicotinamide, diallyl disulfide, and HC toxin have also been applied for studying the importance of histone deacetylation in plants.51-53
Compounds that disrupt normal methyl supplies
Folate-dependent C1 metabolism produces S-adenosylmethionine (SAM), which is the universal methyl donor utilized by most methyltranferases to methylate DNA, RNA, and histones and other proteins.54 In a chemical genetic screening, sulfamethazine (SMZ) was identified as a chemical suppressor of epigenetic gene silencing in plants.55 SMZ treatment releases transcriptional silencing of transgenes as well as endogenous transposons and other repetitive elements. Arabidopsis treated with SMZ exhibited substantially reduced levels of DNA methylation and histone H3K9 dimethylation but did not show changes in the levels of heterochromatic siRNAs.55 SMZ belongs to the family of sulfonamides, which are known antibacterial compounds functioning through impairment of folate synthesis. As structural analogs and competitive antagonists to p-aminobenzoic acid (PABA), sulfonamides competitively inhibit dihydropteroate synthase-catalyzed biosynthesis of dihydropteroic acid, the precursor of tetrahydrofolic acid in the folate biosynthesis pathway.56 SMZ treatments decreased plant folate pool size and caused methyl deficiency as demonstrated by reductions in SAM levels and in global DNA methylation,55 indicating that SMZ confers epigenetic regulation via impairment of folate-dependent methyl supplies. Consistently, exogenous application of PABA or its downstream compounds in the folate biosynthesis pathway restored transcriptional silencing in SMZ-treated plants, whereas release of epigenetic gene silencing can also be observed in plants treated with methotrexate,55 which is an antifolate compound that causes folate depletion in Arabidopsis.57
Removal of the methyl group from SAM generates S-adenosylhomocysteine (SAH). The enzyme SAHH1 (SAH hydrolase 1) breaks SAH down to homocysteine and adenosine, a process that is essential to regenerate SAM and to maintain SAM-dependent methylation. Inhibition of Arabidopsis SAHH1 by dihydroxypropyladenine (DHPA) releases epigenetic silencing of a transgene, of which transcription requires simultaneous reduction in the levels of DNA methylation and histone H3K9 dimethylation.58 In another study, DHPA treatments of germinating tobacco seeds resulted in dosage-dependent global DNA hypomethylation and pleiotropic developmental defects.59 Similar to DHPA, ethionine treatment caused DNA hypomethylation in tobacco cells, possibly due to its function as an antagonist of methionine,60 which is the immediate precursor of SAM.
Perspective
In parallel to mutagenesis, chemical inhibition of epigenetic regulators provides a quick and effective approach to investigate the roles of epigenetic modifications in controlling various biological processes, especially for plant species of which genetic information is limited. In the model organism Arabidopsis thaliana, multi-gene families are commonly present for epigenetic regulators, though proteins in a gene family may individually have specific functions. In the cases where chemical inhibitors target the conserved functional domain of an enzyme family, the strategy of chemical biology can bypass the problem of gene redundancy. While chemical inhibitors are useful tools in studying plant epigenetics, it is worth to note that they should be used with care since most of them are also effective, and actually were first discovered, in mammalian studies. SMZ is one exception because it targets dihydropteroate synthase that is absent in humans, which do not synthesize folate de novo.61 In comparison to the various types of epigenetic modifications discussed herein and beyond, to date only a small number of compounds have been applied in plant epigenetics studies. Future research efforts in chemical biology, such as by means of chemical genetics screens, will expand the list of small molecule probes and consequently provide further mechanistic and functional insights into epigenetic regulations in plants.
Acknowledgments
We apologize to those authors whose work could not be cited due to space limitations. This work is supported by National Institutes of Health grants R01GM070795 and R01GM059138 to J-K Zhu.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/25364
References
- 1.Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature. 2000;405:203–6. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- 2.Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, Stroud H, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–51. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorković ZJ, Naumann U, Matzke AJ, Matzke M. Involvement of a GHKL ATPase in RNA-directed DNA methylation in Arabidopsis thaliana. Curr Biol. 2012;22:933–8. doi: 10.1016/j.cub.2012.03.061. [DOI] [PubMed] [Google Scholar]
- 4.Nishimura T, Molinard G, Petty TJ, Broger L, Gabus C, Halazonetis TD, et al. Structural basis of transcriptional gene silencing mediated by Arabidopsis MOM1. PLoS Genet. 2012;8:e1002484. doi: 10.1371/journal.pgen.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–76. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 7.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12:483–92. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Zhu JK. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14:142–7. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wierzbicki AT. The role of long non-coding RNA in transcriptional gene silencing. Curr Opin Plant Biol. 2012;15:517–22. doi: 10.1016/j.pbi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Pikaard CS, Haag JR, Pontes OM, Blevins T, Cocklin R. A Transcription Fork Model for Pol IV and Pol V-dependent RNA-Directed DNA Methylation. Cold Spring Harb Symp Quant Biol 2013; PMID:23567894. [DOI] [PubMed] [Google Scholar]
- 11.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–66. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Zhu JK. Active DNA Demethylation in Plants and Animals. Cold Spring Harb Symp Quant Biol 2012; PMID:23197304. [DOI] [PMC free article] [PubMed]
- 13.Zheng X, Pontes O, Zhu J, Miki D, Zhang F, Li WX, et al. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–62. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez-Macías MI, Qian W, Miki D, Pontes O, Liu Y, Tang K, et al. A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol Cell. 2012;45:357–70. doi: 10.1016/j.molcel.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez-Macías MI, Córdoba-Cañero D, Ariza RR, Roldán-Arjona T. The DNA repair protein XRCC1 functions in the plant DNA demethylation pathway by stimulating cytosine methylation (5-meC) excision, gap tailoring, and DNA ligation. J Biol Chem. 2013;288:5496–505. doi: 10.1074/jbc.M112.427617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreuzza S, Li J, Guitton AE, Faure JE, Casanova S, Park JS, et al. DNA LIGASE I exerts a maternal effect on seed development in Arabidopsis thaliana. Development. 2010;137:73–81. doi: 10.1242/dev.041020. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda Y, Kinoshita Y, Susaki D, Ikeda Y, Iwano M, Takayama S, et al. HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev Cell. 2011;21:589–96. doi: 10.1016/j.devcel.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Wolffe AP. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–2. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 19.Horn D. Histone deacetylases. Adv Exp Med Biol. 2008;625:81–6. doi: 10.1007/978-0-387-77570-8_7. [DOI] [PubMed] [Google Scholar]
- 20.Qian W, Miki D, Zhang H, Liu Y, Zhang X, Tang K, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336:1445–8. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.To TK, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, et al. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 2011;7:e1002055. doi: 10.1371/journal.pgen.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer A, Hofmann I, Naumann K, Reuter G. Heterochromatin proteins and the control of heterochromatic gene silencing in Arabidopsis. J Plant Physiol. 2006;163:358–68. doi: 10.1016/j.jplph.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 23.Latham JA, Dent SY. Cross-regulation of histone modifications. Nat Struct Mol Biol. 2007;14:1017–24. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- 24.Liu C, Lu F, Cui X, Cao X. Histone methylation in higher plants. Annu Rev Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
- 25.Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–80. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saze H, Shiraishi A, Miura A, Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319:462–5. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- 27.Miura A, Nakamura M, Inagaki S, Kobayashi A, Saze H, Kakutani T. An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J. 2009;28:1078–86. doi: 10.1038/emboj.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, Hasegawa PM, et al. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447:735–8. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- 29.Krichevsky A, Zaltsman A, Lacroix B, Citovsky V. Involvement of KDM1C histone demethylase-OTLD1 otubain-like histone deubiquitinase complexes in plant gene repression. Proc Natl Acad Sci USA. 2011;108:11157–62. doi: 10.1073/pnas.1014030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–9. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 31.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–9. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coleman-Derr D, Zilberman D. DNA Methylation, H2A.Z, and the Regulation of Constitutive Expression. Cold Spring Harb Symp Quant Biol 2012; PMID: 23250988. [DOI] [PubMed]
- 33.Wu JC, Santi DV. On the mechanism and inhibition of DNA cytosine methyltransferases. Prog Clin Biol Res. 1985;198:119–29. [PubMed] [Google Scholar]
- 34.Chang S, Pikaard CS. Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. J Biol Chem. 2005;280:796–804. doi: 10.1074/jbc.M409053200. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto N, Kobayashi H, Togashi T, Mori Y, Kikuchi K, Kuriyama K, et al. Formation of embryogenic cell clumps from carrot epidermal cells is suppressed by 5-azacytidine, a DNA methylation inhibitor. J Plant Physiol. 2005;162:47–54. doi: 10.1016/j.jplph.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 36.Marfil CF, Asurmendi S, Masuelli RW. Changes in micro RNA expression in a wild tuber-bearing Solanum species induced by 5-Azacytidine treatment. Plant Cell Rep. 2012;31:1449–61. doi: 10.1007/s00299-012-1260-x. [DOI] [PubMed] [Google Scholar]
- 37.Kondo H, Miura T, Wada KC, Takeno K. Induction of flowering by 5-azacytidine in some plant species: relationship between the stability of photoperiodically induced flowering and flower-inducing effect of DNA demethylation. Physiol Plant. 2007;131:462–9. doi: 10.1111/j.1399-3054.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 38.Caperta AD, Neves N, Viegas W, Pikaard CS, Preuss S. Relationships between transcription, silver staining, and chromatin organization of nucleolar organizers in Secale cereale. Protoplasma. 2007;232:55–9. doi: 10.1007/s00709-007-0277-4. [DOI] [PubMed] [Google Scholar]
- 39.Beisler JA. Isolation, characterization, and properties of a labile hydrolysis product of the antitumor nucleoside, 5-azacytidine. J Med Chem. 1978;21:204–8. doi: 10.1021/jm00200a012. [DOI] [PubMed] [Google Scholar]
- 40.Constantinides PG, Jones PA, Gevers W. Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature. 1977;267:364–6. doi: 10.1038/267364a0. [DOI] [PubMed] [Google Scholar]
- 41.Cheng JC, Matsen CB, Gonzales FA, Ye W, Greer S, Marquez VE, et al. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst. 2003;95:399–409. doi: 10.1093/jnci/95.5.399. [DOI] [PubMed] [Google Scholar]
- 42.Baubec T, Pecinka A, Rozhon W, Mittelsten Scheid O. Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J. 2009;57:542–54. doi: 10.1111/j.1365-313X.2008.03699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–93. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka M, Kikuchi A, Kamada H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2007;146:149–61. doi: 10.1104/pp.107.111674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu CR, Liu C, Wang YL, Li LC, Chen WQ, Xu ZH, et al. Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proc Natl Acad Sci USA. 2005;102:14469–74. doi: 10.1073/pnas.0503143102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen ZJ, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 1997;11:2124–36. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murfett J, Wang XJ, Hagen G, Guilfoyle TJ. Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell. 2001;13:1047–61. doi: 10.1105/tpc.13.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sako K, Maki Y, Kanai T, Kato E, Maekawa S, Yasuda S, et al. Arabidopsis RPT2a, 19S proteasome subunit, regulates gene silencing via DNA methylation. PLoS ONE. 2012;7:e37086. doi: 10.1371/journal.pone.0037086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS. In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J. 2007;52:615–26. doi: 10.1111/j.1365-313X.2007.03264.x. [DOI] [PubMed] [Google Scholar]
- 50.Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J Biol Chem. 2001;276:38837–43. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- 51.Bond DM, Dennis ES, Pogson BJ, Finnegan EJ. Histone acetylation, VERNALIZATION INSENSITIVE 3, FLOWERING LOCUS C, and the vernalization response. Mol Plant. 2009;2:724–37. doi: 10.1093/mp/ssp021. [DOI] [PubMed] [Google Scholar]
- 52.Murphy JP, McAleer JP, Uglialoro A, Papile J, Weniger J, Bethelmie F, et al. Histone deacetylase inhibitors and cell proliferation in pea root meristems. Phytochemistry. 2000;55:11–8. doi: 10.1016/S0031-9422(00)00195-3. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z, Cao H, Sun Y, Li X, Chen F, Carles A, et al. Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell. 2013;25:149–66. doi: 10.1105/tpc.112.108191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loenen WA. S-adenosylmethionine: jack of all trades and master of everything? Biochem Soc Trans. 2006;34:330–3. doi: 10.1042/BST20060330. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H, Deng X, Miki D, Cutler S, La H, Hou YJ, et al. x Sulfamethazine suppresses epigenetic silencing in Arabidopsis by impairing folate synthesis. Plant Cell. 2012;24:1230–41. doi: 10.1105/tpc.112.096149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ortelli F, Maxwell CA, Curtis J, Watkins WM. Studies on anti-folate antimalarials in east Africa. Parassitologia. 1999;41:313–4. [PubMed] [Google Scholar]
- 57.Loizeau K, De Brouwer V, Gambonnet B, Yu A, Renou JP, Van Der Straeten D, et al. A genome-wide and metabolic analysis determined the adaptive response of Arabidopsis cells to folate depletion induced by methotrexate. Plant Physiol. 2008;148:2083–95. doi: 10.1104/pp.108.130336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baubec T, Dinh HQ, Pecinka A, Rakic B, Rozhon W, Wohlrab B, et al. Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic States in Arabidopsis. Plant Cell. 2010;22:34–47. doi: 10.1105/tpc.109.072819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fulneček J, Matyášek R, Votruba I, Holý A, Křížová K, Kovařík A. Inhibition of SAH-hydrolase activity during seed germination leads to deregulation of flowering genes and altered flower morphology in tobacco. Mol Genet Genomics. 2011;285:225–36. doi: 10.1007/s00438-011-0601-8. [DOI] [PubMed] [Google Scholar]
- 60.Kovarík A, Koukalová B, Holý A, Bezdĕk M. Sequence-specific hypomethylation of the tobacco genome induced with dihydroxypropyladenine, ethionine and 5-azacytidine. FEBS Lett. 1994;353:309–11. doi: 10.1016/0014-5793(94)01048-X. [DOI] [PubMed] [Google Scholar]
- 61.Hanson AD, Gregory JF., 3rd Folate biosynthesis, turnover, and transport in plants. Annu Rev Plant Biol. 2011;62:105–25. doi: 10.1146/annurev-arplant-042110-103819. [DOI] [PubMed] [Google Scholar]


