Abstract
Bacteria use proteases to control three types of events temporally and spatially during processes of morphological development. These events are destruction of regulatory proteins, activation of regulatory proteins, and production of signals. While some of these events are entirely cytoplasmic, others involve intramembrane proteolysis of a substrate, trans-membrane signaling, or secretion. In some cases, multiple proteolytic events are organized into pathways, e.g., turnover of a regulatory protein activates a protease that generates a signal. We review well-studied and emerging examples, and identify recurring themes and important questions for future research. We focus primarily on paradigms learned from studies of model organisms, but we note connections to regulated proteolytic events that govern bacterial adaptation, biofilm formation and disassembly, and pathogenesis.
Keywords: Protease, regulatory proteolysis, signal, morphogenesis, differentiation, cell cycle
Introduction
Bacteria have evolved different regulatory strategies that allow them to adapt to changing conditions including changes in gene expression, cellular differentiation and changes in motility. In several of these strategies, proteolysis plays an essential role. Regulation by proteolysis is highly versatile and involved in diverse processes such as stress responses, growth, division, the cell cycle, development with cell differentiation, pathogenesis, biofilm formation and disassembly, and protein secretion [for recent, general reviews on regulated proteolysis, see (Gottesman, 2003, Jenal & Hengge-Aronis, 2003, Kirstein et al., 2009, Gur et al., 2011).
Proteolysis in bacteria comes in two forms, general proteolysis and regulated proteolysis (Schmidt et al., 2009). General proteolysis is important for protein homeostasis and the removal of misfolded or damaged proteins and is an essential part of the cellular protein quality control system. Regulated proteolysis is the specific removal or modification by proteolytic cleavage of proteins in response to specific signals. General as well as regulated proteolysis depends on a high degree of protease-substrate specificity to avoid haphazard degradation of proteins. In the case of regulated proteolysis, a substrate may contain one or more specific degradation signals, also referred to as degrons, which targets the protein to a protease (Kirstein et al., 2009). Alternatively, the substrate protein interacts with an adaptor protein that targets the substrate to the protease (Jenal & Hengge-Aronis, 2003, Kirstein et al., 2009).
Generally, regulated proteolysis may result in the complete degradation of a protein, also referred to as processive proteolysis, in that way effectively ridding a cell of that protein (Jenal & Hengge-Aronis, 2003). Alternatively, the protein substrate is not completely degraded but specifically cleaved, giving rise to a modified protein, which is the active form of the protein or has an altered activity compared to the uncleaved protein (Jenal & Hengge-Aronis, 2003). This type of proteolysis, which is sometimes referred to as non-processive proteolysis or processing, is typically important in the regulation of transcription factors as well as in the generation of intercellular signals.
The accumulation level of any cellular protein is the net result of the balance between synthesis and degradation. Because proteolysis is fast, it has been argued (Gottesman, 2003, Jenal & Hengge-Aronis, 2003) that regulation by proteolysis is advantageous in systems where a fast response is needed such as under stress conditions: (i) a protein can be quickly activated without the delay associated with transcriptional and translational control mechanisms; or, (ii) a protein can be efficiently removed when it is no longer needed on a much faster time-scale than the simple dilution by growth would allow. Because proteolysis is also irreversible, it is an especially effective regulatory mechanism in cases where a robust, irreversible commitment is required such as during cell cycle progression and cell differentiation. In addition to providing temporal control of regulatory proteins, proteolysis can be localized to a particular subcellular region, allowing spatial control of the accumulation of a regulatory protein that leads to the generation of cellular asymmetry, consequently dictating cell fate upon cell division.
Proteolysis is achieved by proteases, or peptidases, a group of enzymes that hydrolyze peptide bonds. They are catalogued on the basis of the active site residue or ion that carries out catalysis, i.e. serine, threonine, cysteine, glutamic, asparagine, aspartic, and metallo proteases (Rawlings et al., 2012). General as well as regulated processive proteolysis of cytoplasmic proteins is carried out by a set of related chaperone-protease complexes (Kirstein et al., 2009). The chaperones belong to the AAA+ protein family and use hydrolysis of ATP to unfold and then translocate a substrate protein into the proteolytic chamber of the associated protease. Examples of the chaperones include ClpA, ClpX, ClpC and HslU (also referred to as ClpY) and the associated proteases are ClpP and HslV (also referred to as ClpQ). Combining an ATPase with a protease gives rise to the so-called ATP-dependent proteases such as ClpAP, ClpXP, ClpCP and HslUV. The Lon and FtsH proteases are two exceptions to this general scheme and in these two proteins, the AAA+ ATPase and protease domains are located within the same polypeptide. The importance of regulating processive proteolysis is underscored by the recent finding that the antibiotic ADEP binds to ClpP and turns it into an uncontrolled protease (Kirstein et al., 2009, Lee et al., 2010). The proteases involved in non-processive proteolysis of substrate proteins are more diverse and often dedicated to cutting just one specific substrate. A theme that has emerged in regulated proteolysis of membrane-inserted protein substrates is the involvement of intramembrane proteases, which cleave substrates within a membrane or near its surface in a process called regulated intramembrane proteolysis (RIP).
Here, we review regulated proteolysis in bacterial development and differentiation. Bacteria use regulated proteolysis to control three types of events temporally and spatially during development. The events are destruction of regulatory proteins, activation of regulatory proteins, and production of intercellular signals (Fig. 1). These three types of events are reviewed in three major sections below. Of course, in some cases, multiple events are organized into pathways (e.g., turnover of a regulatory protein activates a protease that subsequently generates a signal). In these cases, we emphasize one event and review the pathway in the major section devoted to that type of event. We review well-studied and emerging examples, and we attempt to identify recurring themes and important questions for future research. We focus primarily on paradigms learned from studies of model organisms undergoing development and differentiation, but we note connections to regulated proteolytic events that govern bacterial adaptation, biofilm formation and disassembly, and pathogenesis.
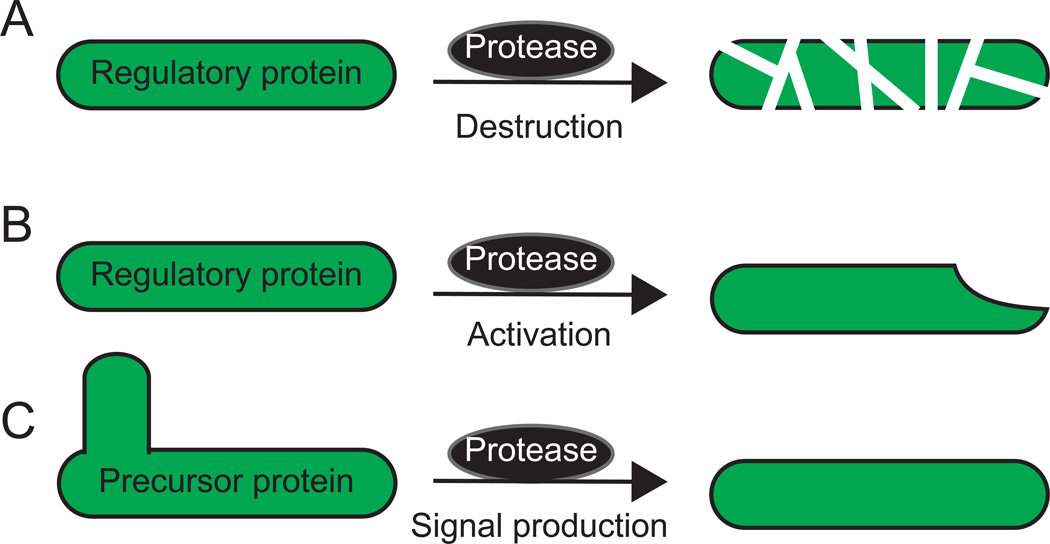
Figure 1. Events controlled by regulated proteolysis during bacterial development.
(A) Destruction of a regulatory protein by a protease, e.g., a Clp protease that processively degrades a regulatory protein.
(B) Activation of a regulatory protein by a protease, e.g., a protease (often dedicated to one specific substrate) that non-processively cleaves an inactive proprotein to produce the active regulatory protein.
(C) Production of an intercellular signal by a protease. In many cases, the protease(s) remains to be identified. A precursor protein is cleaved one or more times to produce a protein, peptide, or amino acid that acts as an intercellular signal.
Principles of regulated proteolysis from stress response studies
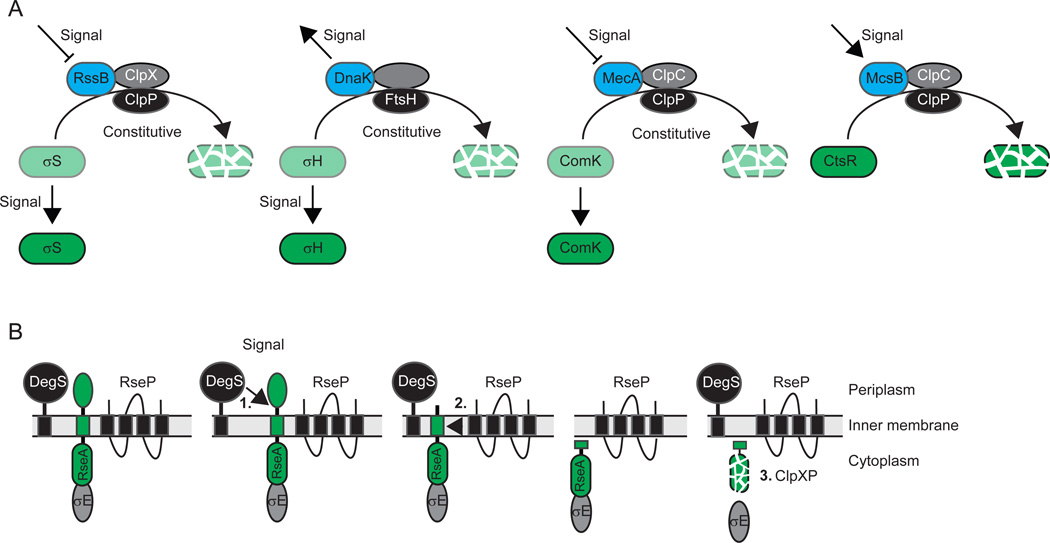
Regulated proteolysis is particularly well-studied in bacterial stress responses. Five examples are instructive to describe the circuit design for regulated proteolysis as well as mechanistic principles for regulation of proteolysis. Notably, regulated proteolysis in bacterial development and differentiation often involves variations over these circuit designs. Regulated proteolysis leading to the complete destruction of a protein substrate with a regulatory function is common in bacterial stress responses. In these responses, proteolysis of a transcription factor or of a protein that regulates the activity of a transcription factor is regulated in response to a particular stress cue or signal, resulting in the generation of a specific output response. In the cases of the alternative sigma factors σS and σH in Escherichia coli and the transcription activator ComK in Bacillus subtilis, proteolytic degradation is constitutive in the absence of the cognate signal. In response to the relevant signal, proteolytic degradation is inhibited and the transcription factor accumulates. Conversely, in the case of the transcriptional repressor CtsR in B. subtilis, proteolytic degradation is induced in response to the relevant signal, thus relieving repression. Similarly, in the case of the anti-sigma factor RseA in E. coli, the relevant signal results in the complete degradation of RseA so that σE is released from inhibition.
Briefly, σS is the master regulator of the general stress response in E. coli [for review, see (Battesti et al., 2011)]. σS is synthesized under non-stress conditions, however, it is immediately targeted for ClpXP-dependent proteolysis by the adaptor protein RssB also known as SprE (Fig. 2A). In response to different types of stresses including starvation, osmotic stress, temperature stress and pH stress, σS is stabilized. Two mechanisms involved in this stabilization are (i) regulation of RssB activity by anti-adaptor proteins such as IraP, IraM and IraD, which are synthesized in response to specific stresses and bind to RssB, interfering with RssB-dependent delivery of σS to ClpXP; and (ii) increased synthesis of σS, which out-titrates RssB. RssB activity has been suggested to be regulated by phosphorylation by the histidine protein kinase ArcB (Mika & Hengge, 2005), which is regulated by the cellular energy state; however, it remains controversial whether RssB phosphorylation is important for adaptor function (Peterson et al., 2004). Regardless, the net-accumulation of σS is the outcome of the balance between constitutive destruction by ClpXP on the one hand and regulation of adaptor activity and σS synthesis on the other hand.
Figure 2. Principles of regulated proteolysis from stress response studies.
(A) Representative examples of circuit design for the regulated destruction of regulatory cytoplasmic proteins. In all four examples, the level of green indicates the accumulation level of the substrate. Adaptors are indicated in blue, proteases in black and AAA+ proteins in grey. Note that in FtsH, the protease and AAA+ domains are part of the same polypeptide.
(B) Representative example for the regulated destruction of a regulatory inner membrane protein by RIP. The bitopic inner membrane protein substrate RseA is indicated in green. Cleavage by DegS, the site-1-protease, is immediately followed by cleavage by RseP, the site-2-protease, and degradation by ClpXP.
σH is the master regulator of the heat shock response in E. coli [for review, see (Narberhaus et al., 2009)]. σH is constitutively degraded by FtsH in the absence of heat shock and targeted to FtsH by DnaK (Fig. 2A). In response to heat shock, aggregated and unfolded proteins accumulate and titrate DnaK, causing stabilization of σH. Synthesis of σH is also increased [for review, see (Yura & Nakahigashi, 1999)]. Thus, in the case of σH, net-accumulation is determined by the balance between constitutive destruction by FtsH on the one hand and availability of the adaptor DnaK and σH synthesis on the other.
ComK is the master regulator of genetic competence in B. subtilis. ComK is targeted to the ClpCP protease by the adaptor protein MecA and is constitutively degraded [for review, see (Kirstein et al., 2009)] (Fig. 2A). At a high cell density, the intercellular signalling molecule ComX leads to synthesis of ComS, a 46-residue protein that binds to MecA and causes the release and stabilization of ComK. Here again, constitutive degradation is blocked in a specific manner, leading to the accumulation of a transcription factor.
The opposite type of regulation is observed for the regulator of the heat shock response CtsR in B. subtilis [for review, see (Kirstein et al., 2009)] (Fig. 2A). In the absence of heat shock, CtsR is stable and inhibits the expression of class III heat shock genes. In response to heat shock, the adaptor protein McsB binds CtsR, which also undergoes conformational changes in response to heat (Elsholz et al., 2010), and targets it for degradation by the ClpCP protease. As such, McsB is important for the heat-induced inactivation and subsequent degradation of CtsR. Interestingly, ClpC and ClpP localize dynamically to clusters in both polar regions of the cell (Kain et al., 2008, Kirstein et al., 2008, Simmons et al., 2008), and McsB and CtsR display similar localization patterns (Kirstein et al., 2008).
The fifth instructive example concerns RseA [for reviews, see (Ades, 2008, Clausen et al., 2011, Kroos & Akiyama, 2013)], which is a σE anti-sigma factor in E. coli (Fig. 2B). σE is held in an inactive state by its interaction with the cytoplasmic domain of the inner membrane protein RseA and is activated in response to unfolded outer membrane proteins in the periplasm. In response to the inducing signal, the inner membrane protease DegS is activated and cleaves RseA on the periplasmic side. This cleavage is immediately followed by cleavage of RseA in the trans-membrane segment by RseP (also known as YaeL) by RIP, causing the release of the cytoplasmic domain of RseA. This domain, in turn, is immediately degraded by ClpXP, freeing σE to direct transcription of its regulon. Thus, in this system, the accumulation of an anti-sigma factor is regulated and the presence of an inducing signal sets in motion a sequential and continuous proteolytic cascade that leads to the destruction of RseA and the release of σE. RIP by RseP homologs as part of a regulated proteolytic cascade has emerged as a common theme in regulated proteolysis not only in bacteria but in eukaryotes as well (Brown et al., 2000, Urban, 2009). Generally, in these cascades, the first proteolytic cleavage is the rate limiting step and is carried out by a protease often referred to as a site-1-protease; the second cleavage immediately follows and is carried out by an RseP homolog often referred to as the site-2-protease or intramembrane metalloprotease (IMMP). In the case of degradation of trans-membrane anti-sigma factors, the cytoplasmic domain is immediately degraded. However, in other cases, such as PodJ in Caulobacter crescentus (see details below), the product of the second cleavage accumulates. Moreover, in the case of Pro-σK in B. subtilis, substrate cleavage by the IMMP does not depend on prior cleavage of the substrate by a site-1-protease (see details below).
Destruction of regulatory proteins
Here, we review four examples of destruction of regulatory proteins (Fig. 1A) by ClpXP-dependent regulated proteolysis in bacterial development. Notably, these regulated proteolysis events are not induced by external cues or signals but are tied in with the cell cycle. Three of these, degradation of CtrA, PdeA and CpdR, are temporally regulated during the swarmer-to-stalk cell transition in Caulobacter crescentus and allow cell cycle progression to be coupled with pole development. In addition, degradation of CtrA is spatially regulated in predivisional cells and occurs specifically in the compartment destined to become the stalked cell, while CtrA is maintained in the compartment destined to become the swarmer cell, and in this case regulated proteolysis is important for cell fate determination. The fourth example involves the cell cycle-dependent destruction of Sda in B. subtilis during sporulation and allows chromosome status, i.e., DNA damage or ongoing replication, to be tied in with the initiation of sporulation. Finally in this section, we review two examples of destruction of regulatory proteins (Fig. 1A) by ClpCP-dependent proteolysis; compartment-specific degradation of SpoIIAB during B. subtilis sporulation and turnover of DegU and SlrR to regulate B. subtilis motility and biofilm disassembly, respectively.
Cell cycle-regulated degradation of CtrA, PdeA and CpdR in C. crescentus
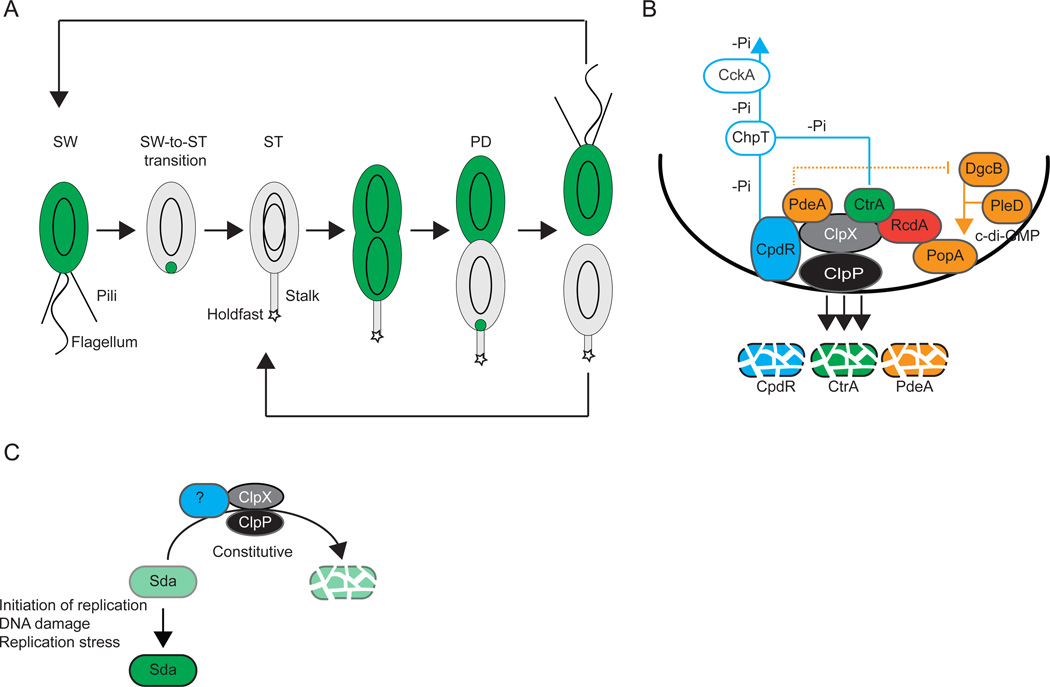
The C. crescentus cell cycle is characterized by an asymmetric cell division that results in the generation of two different cell types, the motile, flagellated and piliated swarmer (SW) cell and the sessile, stalked (ST) cell (Fig. 3A). After cell division, the ST cell immediately enters the S phase of a new cell cycle with initiation of replication and produces a new flagellum and pili at the pole opposite the stalk. In contrast, the SW cell does not initiate replication but remains in the G1 phase of the cell cycle. At some point, the SW cell differentiates into a ST cell with release of the flagellum and retraction of pili and their replacement with a stalk and its associated holdfast. In parallel, the cell enters the S phase of the cell cycle [for recent reviews on the C. crescentus cell cycle and its regulation, see (Laub et al., 2007, Jenal, 2009, Kirkpatrick & Viollier, 2012, Tsokos & Laub, 2012)]. Cell cycle progression in C. crescentus is controlled by four master regulators: DnaA, GcrA, CtrA and CcrM. The accumulation of all four regulators oscillates during the cell cycle and is controlled by a dynamic balance between temporally regulated synthesis and degradation by proteolysis (Jenal, 2009). The DNA replication initiator protein and transcriptional regulator DnaA is degraded in a cell cycle-dependent manner by ClpP and apparently independently of the ClpX and ClpA ATPases (Gorbatyuk & Marczynski, 2005); the protease involved in degradation of the global regulator GcrA is not known; and, CcrM, an adenine DNA methyltransferase, is constitutively degraded by Lon (Wright et al., 1996).
Figure 3. Destruction of regulatory proteins by regulated proteolysis.
(A) Schematic of the C. crescentus cell cycle including the presence and localization of CtrA (green). Polar appendages, swarmer (SW), stalked (ST) and predivisional (PD) cells are indicated. Black ovoids indicate chromosomes.
(B) Recruitment of ClpXP and CtrA to the incipient ST cell pole in C. crescentus. Polarly localized proteins are indicated in full colors. Blue lines indicate the reverse phosphate (-Pi) flow from CpdR and CtrA to ChpT and CckA. CpdR recruits ClpXP and functions as an adaptor for PdeA degradation by ClpXP. RcdA recruits CtrA but does not appear to function as an adaptor for ClpXP degradation of CtrA. Proteins involved in c-di-GMP metabolism and binding are shown in orange. The stippled orange line indicates the inhibition by PdeA of DgcB activity until PdeA degradation. The orange line indicates c-di-GMP synthesized by DgcB and PleD that binds to PopA.
(C) Regulation of Sda accumulation in response to chromosome status. Levels of green indicate levels of Sda accumulation. The putative adaptor (blue) required for Sda degradation by ClpXP is not known.
CtrA has two functions in C. crescentus cell cycle regulation: (i) inhibition of DNA replication in SW cells (Quon et al., 1998); and, (ii) regulation of expression of many cell-cycle dependent genes in predivisional (PD) cells (Laub et al., 2000). ctrA expression is initiated in late ST cells and peaks in late PD cells (Quon et al., 1996). CtrA is specifically degraded in the ST cell compartment of the late PD to allow the generation of the characteristic cellular asymmetry upon cell division (Fig. 3A). CtrA remains in SW cells until it is cleared by proteolytic degradation during the SW-to-ST cell transition to allow DNA replication to occur (Fig. 3A). CtrA is a response regulator and its activity is controlled at the levels of transcription, phosphorylation and protein stability (Domian et al., 1997). Thus, CtrA is degraded at two points during the cell cycle: degradation is temporally regulated during the SW-to-ST cell transition and spatially regulated in the ST compartment of the late PD cell.
CtrA is activated by phosphorylation by the CckA-ChpT phosphorelay (Biondi et al., 2006). (Domian et al., 1999)CtrA activity is eliminated during the SW-to-ST cell transition by two redundant mechanisms, dephosphorylation and proteolysis. During the SW-to-ST cell transition, the CckA-ChpT phosphorelay is inhibited and the phosphate flow is reversed, resulting in CtrA dephosphorylation (Biondi et al., 2006) (Fig. 3B). CtrA degradation depends on ClpXP (Jenal & Fuchs, 1998) and involves dynamic co-localization with ClpXP at the incipient ST cell pole (McGrath et al., 2006). Two signaling pathways converge to bring the protease and its substrate together at the same subcellular site: the CckA-ChpT phosphorelay together with the response regulator CpdR brings ClpXP to the incipient ST cell pole and the PleD/DgcB-PopA-RcdA pathway takes CtrA to the same pole (Fig. 3B).
CpdR is a single domain response regulator and is also phosphorylated by the CckA-ChpT phosphorelay (Biondi et al., 2006). As opposed to CtrA, which is active in the phosphorylated form, phosphorylation of CpdR keeps the protein inactive. As mentioned, during the SW-to-ST cell transition, the CckA-ChpT phosphorelay is inhibited; therefore, CpdR accumulates in its active unphosphorylated form at this stage of the cell cycle (Biondi et al., 2006, Iniesta et al., 2006). Unphosphorylated CpdR localizes to the incipient ST pole and recruits ClpXP to this pole (Iniesta et al., 2006) (Fig. 3B). Because CtrA and CpdR are both regulated by the CckA-ChpT phosphorelay, CtrA deactivation and degradation is synchronized in time.
At the SW-to-ST cell transition, polar localization of CtrA is accomplished by a cell cycle-dependent increase in the concentration of the second messenger c-di-GMP. This increase is a result of the activation of two diguanylate cyclases, PleD and DgcB (Duerig et al., 2009, Abel et al., 2011) (Fig. 3B). PleD activity is activated via its phosphorylation and localizes to the incipient ST cell pole resulting in local production of c-di-GMP (Paul et al., 2008). DgcB is present throughout the cell cycle and is activated by the proteolytic degradation of its antagonist, the phosphodiesterase PdeA, (Abel et al., 2011)by ClpXP at the SW-to-ST transition resulting in a boost in c-di-GMP accumulation (Abel et al., 2011) (Fig. 3B). c-di-GMP binds to and activates PopA. Activated PopA, in turn, localizes to the incipient ST cell pole (Duerig et al., 2009) and recruits RcdA, which subsequently serves as a polar targeting factor for CtrA (McGrath et al., 2006) (Fig. 3B). Once at the incipient ST pole, CtrA co-localizes with ClpXP and is destroyed (Jenal & Fuchs, 1998, McGrath et al., 2006). Importantly, to ensure spatial-temporal coordination of ClpXP and CtrA localization both signaling pathways are interconnected by (i) unphosphorylated CpdR not only serving as a polar recruitment factor for ClpXP but also as an adaptor for ClpXP-dependent degradation of PdeA (Abel et al., 2011) and (ii) RcdA being recruited to the pole not only by PopA (Duerig et al., 2009) but also by ClpX (McGrath et al., 2006) (Fig. 3B). Ultimately, CpdR is also degraded by ClpXP, causing the release of ClpXP from the pole and relieving the degradation of CtrA (Iniesta & Shapiro, 2008) (Fig. 3B).
CtrA is also specifically degraded in the ST compartment but not in the SW compartment of the late PD cell (Domian et al., 1997). The CckA-ChpT-CpdR phosphorelay is inactive in the ST and active in the SW compartment of the late PD cell [for reviews, see (Laub et al., 2007, Jenal, 2009, Tsokos & Laub, 2012)]. Therefore, CpdR is in its unphosphorylated form in the ST compartment allowing CpdR, ClpXP, RcdA and CtrA to localize to the ST pole where CtrA degradation occurs (Ryan et al., 2004, Iniesta et al., 2006, McGrath et al., 2006).
CpdR is not only a polar localization factor for ClpXP but also serves directly as an adaptor for PdeA degradation (Abel et al., 2011, Rood et al., 2012). In contrast, RcdA is not an adaptor for CtrA and is not required for CtrA proteolysis in vitro (Chien et al., 2007) suggesting that the primary function of RcdA is in polar positioning of CtrA. It, however, remains an open question whether co-localization of ClpXP and CtrA is essential for the regulated degradation of CtrA. On the one hand, ClpXP degradation of CtrA depends on positioning the protease at the cell pole because in a cpdR mutant ClpXP does not degrade CtrA (Iniesta et al., 2006). However, substitutions in RcdA that disrupt polar RcdA and CtrA localization do not affect CtrA proteolysis, suggesting that RcdA may not stimulate CtrA proteolysis by localizing CtrA at the cell pole (Taylor et al., 2009). Also, it is not known whether CpdR functions as an adaptor for CtrA delivery to ClpXP. Clearly, localized degradation of CtrA in the ST compartment of the late PD cells seems beneficial and could explain the spatially regulated destruction of CtrA in the ST compartment. The degradation of CtrA at the SW-to-ST cell transition could, at least in principle, also fulfill its function without localization of the involved proteins. However, the cell cycle-dependent localization of protease and substrate could be one way in which proteolytic activity is tied in with the cell cycle. Additional ClpXP substrates that are degraded in a cell cycle-dependent manner were recently identified (Bhat et al., 2013). It will be interesting to determine whether the activity of ClpXP in general depends on its localization.
Cell cycle-regulated degradation of structural proteins in C. crescentus
Proteolysis not only controls the accumulation of regulatory proteins in C. crescentus but also plays an important role in cell differentiation and maintenance of cellular asymmetry by ridding the SW cell of its polar flagellum, pili and chemotaxis machinery during the SW-to-ST cell transition (Jenal, 2009, Kirkpatrick & Viollier, 2012). Shedding of the polar flagellum is initiated by the degradation of FliF in the MS ring (Jenal & Shapiro, 1996). ClpAP is the protease responsible for FliF degradation in vivo (Grünenfelder et al., 2004); however, the molecular mechanism(s) responsible for activation of FliF proteolytic degradation by ClpAP remains unknown. The chemoreceptor proteins McpA and McpB are substrates of ClpXP and are also degraded during the SW-to-ST cell transition (Tsai & Alley, 2001, Potocka et al., 2002). Interestingly, degradation of McpA, similarly to CtrA, depends on CpdR (Iniesta et al., 2006).
Cell cycle-regulated degradation of Sda in B. subtilis
In response to nutrient limitations and at a high cell density, B. subtilis cells can initiate a developmental program that culminates in endospore formation (Fig. 4) [for review, see (Kroos, 2007)]. Initiation of spore formation is also cell cycle regulated and only occurs in cells containing two fully replicated, undamaged chromosomes. The coupling between the cell cycle, replication stress, DNA damage and initiation of sporulation depends on the Sda protein. Sda binds to and inhibits the histidine protein kinase KinA (Burkholder et al., 2001), which is a major kinase for the master regulator of sporulation, the response regulator Spo0A. The inhibition of KinA kinase activity results in reduced levels of phosphorylated Spo0A and, therefore, initiation of sporulation is blocked when Sda accumulates. The sda gene is transcribed in a cell cycle-dependent manner and activated by the DnaA protein at the onset of replication (Veening et al., 2009) (Fig. 3C). Moreover, sda transcription is activated by the DnaA protein in response to replication stress and DNA damage (Burkholder et al., 2001, Ruvolo et al., 2006) (Fig. 3C). Importantly, Sda is constitutively degraded (Ruvolo et al., 2006) (Fig. 3C). In vivo this degradation depends on ClpXP; however, in vitro ClpXP alone is insufficient to degrade Sda, suggesting that an adaptor protein may be involved in targeting Sda to ClpXP (Ruvolo et al., 2006). Replication stress and DNA damage does not affect Sda proteolysis (Ruvolo et al., 2006). Thus, in the case of the Sda protein, regulated activation of sda transcription in combination with constitutive proteolysis allows Sda to specifically accumulate in response to a cell cycle signal (initiation of replication), replication stress or DNA damage, in this way allowing the coupling between chromosome status and initiation of sporulation. Overall, the design of the regulatory circuit that allows accumulation of Sda is similar to the designs of the circuits governing σS, σH and ComK accumulation (Fig. 2A).
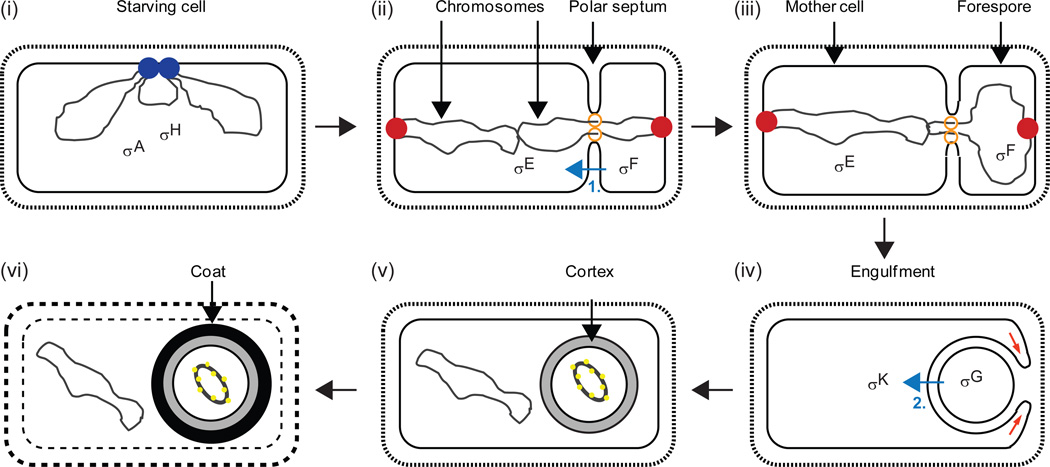
Figure 4. Morphological changes and sigma factors during B. subtilis endospore formation.
(i) The DNA replication machinery (blue) completes chromosome (black) duplication in a starving cell. The major vegetative sigma factor, σA, and the alternative sigma factor, σH, begin to direct transcription of sporulation genes. (ii) A region near each chromosome’s origin of replication is bound by a complex of proteins (red) attached to the membrane at opposite cell poles. The polar septum forms with a DNA translocase (orange) at the annulus. About one-third of the origin-proximal region of one chromosome is located in the smaller forespore compartment, creating genetic asymmetry for about 15 min. σF becomes active in the forespore and initiates a signaling pathway (blue arrow labelled “1”) that leads to proteolytic activation of σE in the mother cell as described in Figure 5. (iii) Most of one chromosome is translocated into the forespore. (iv) The mother cell membrane migrates around the forespore membrane (red arrows) as engulfment begins. σG becomes active in the forespore and initiates a signaling pathway (blue arrow labelled “2”) that leads to proteolytic activation of σK in the mother cell as described in Figures 6 and 7. (v) Cortex forms between the inner and outer forespore membranes. The forespore chromosome is condensed into a toroid by binding of small, acid-soluble spore proteins (yellow). (vi) Spore coat proteins complete their assembly on the forespore surface. The mother cell lyses. Adapted from (Kroos, 2007).
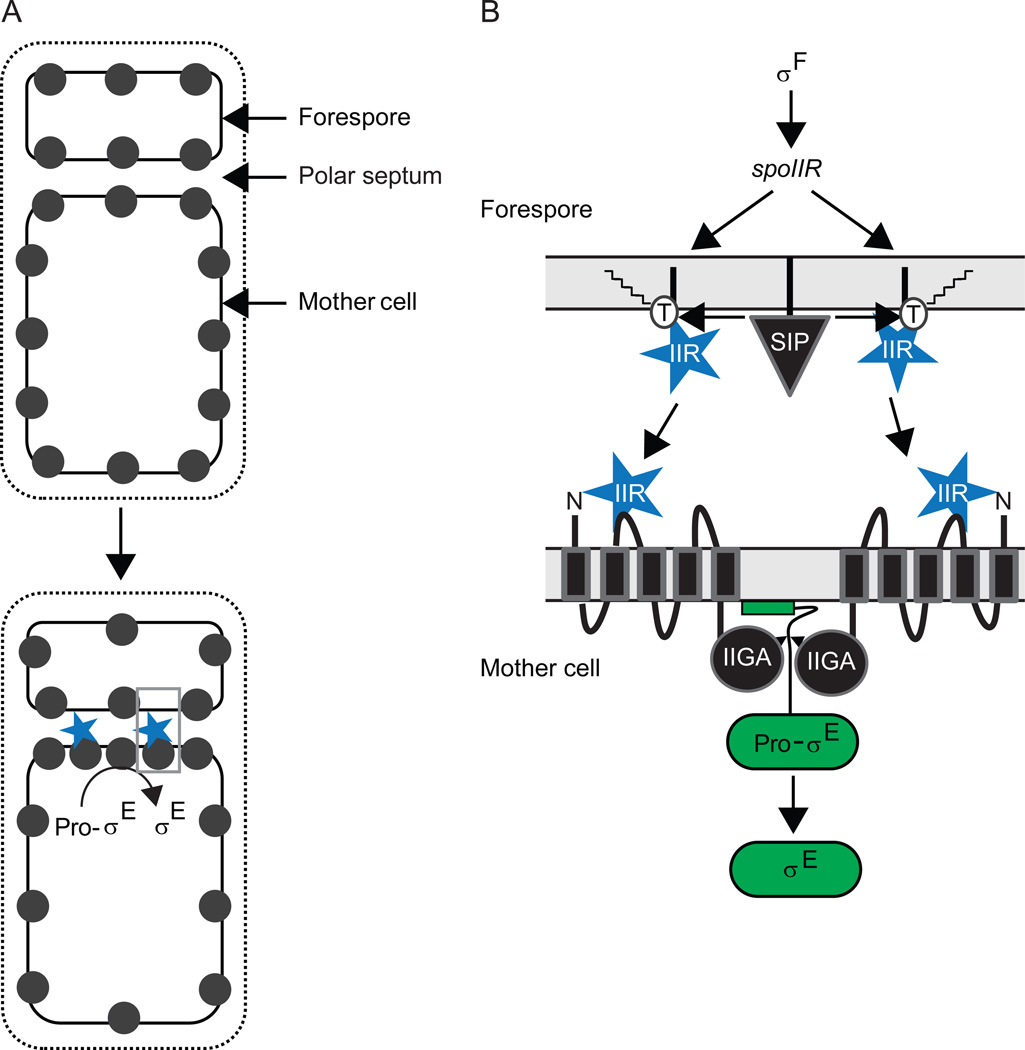
Compartment-specific degradation of regulators during B. subtilis sporulation
Sda is degraded as DNA replication proceeds in starving B. subtilis cells, allowing sporulation to initiate. Sporulation in B. subtilis depends on the creation of unequal cellular compartments by the asymmetrically-positioned septum (Fig. 4). Subsequent to septum formation, the smaller forespore is engulfed by the mother cell and then the spore cortex and coat are synthesized. Eventually, the mother cell lyses and the spore is released. The larger mother cell and the smaller forespore each receive a copy of the genome. Different genes are expressed in each compartment due to activation of different sigma factors [see details below; for review, see (Kroos, 2007)]. Briefly, σF becomes active in the forespore and this activity results in regulated proteolysis of Pro-σE in the mother cell in a process that depends on intercellular signaling. In the forespore, σF directs transcription of the gene encoding σG and σG is activated upon completion of forespore engulfment. In the mother cell, σE directs transcription of the gene for Pro-σK and regulated proteolysis results in active σK in the mother cell in a process that also depends on intercellular signaling since it is initiated by σG activity in the forespore.
σF and Pro-σE are synthesized before formation of the asymmetric septum. However, several mechanisms ensure that σF becomes active first, and only in the forespore. One of these mechanisms involves forespore-specific degradation of the anti-sigma factor SpoIIAB by ClpCP (Pan et al., 2001). As mentioned, ClpC and ClpP localize to clusters in the polar regions of growing cells (Kain et al., 2008, Kirstein et al., 2008, Simmons et al., 2008). During sporulation, ClpCP preferentially accumulates in the forespore, and this may contribute to forespore-specific degradation of SpoIIAB and activation of σF (Kain et al., 2008). σF activity in the forespore leads to proteolytic cleavage of Pro-σE and accumulation of active σE in the mother cell (see details below). Compartment-specific proteolysis appears to prevent Pro-σE and σE from accumulating in the forespore, but the protease(s) remains to be identified (Ju et al., 1998, Fujita & Losick, 2002). The polar localization of ClpCP during growth and its preferential forespore accumulation should be further investigated to see if themes similar to ST pole accumulation of ClpXP in C. crescentus emerge.
Destruction of regulatory proteins during biofilm disassembly, motility and pathogenesis
Key regulators of biofilm disassembly, motility, pathogenesis and many other bacterial processes undergo regulated proteolysis that is processive and causes their destruction. Regulation of motility and both biofilm formation and disassembly is intimately linked in a variety of bacteria [for review, see (Guttenplan & Kearns, 2013)]. In B. subtilis, ClpCP may degrade key regulators of both processes. Swimming motility requires flagella synthesized under control of σD, which is regulated by the anti-sigma factor FlgM, whose transcription is activated by phosphorylated DegU (Hsueh et al., 2011), a substrate of the MecA/ClpCP (Ogura & Tsukahara, 2010) adaptor/protease that also targets ComK as mentioned (Fig. 2A). Biofilm disassembly requires turnover of SlrR that appears to involve both autocleavage and ClpCP, although it remains to be seen whether SlrR is a direct substrate of ClpCP or whether the protease has multiple targets in biofilm formation and disassembly (Chai et al., 2010). The ability to form biofilms and move contributes to pathogenesis of many bacterial species, as does expression of other virulence factors. Clp, Lon, and FtsH proteases have all been implicated in bacterial pathogenesis, as has RIP of anti-sigma factors [for reviews, see (Ingmer & Brondsted, 2009, Urban, 2009)] (typically utilizing a circuit design like that in Fig. 2B). Studies of bacterial adaptation and development using model organisms continue to provide paradigms for discovery of regulated proteolysis that impacts human health.
Activation of regulatory proteins
Bacteria use regulated proteolysis during development not only to destroy regulatory proteins, as reviewed in the preceding section, but also to activate regulatory proteins (Fig. 1B). Activation involves precise proteolytic cleavage without immediate further degradation. In some cases, precise cleavage involves RIP by an IMMP similar to proteases that function in proteolytic cascades leading to destruction of anti-σ factors as in the case of σE in E. coli (Fig. 2B). In other cases, precise cleavage involves an aspartic protease, a type of protease that has not yet been implicated in regulatory protein destruction. As observed for some events that destroy regulatory proteins, activation events are temporally and spatially controlled, and occur at membranes. We review three well-studied activation events. Two of these, activation of σE and of σK, occur in the mother cell in response to signals from the forespore during B. subtilis sporulation (Fig. 4). Because these intercellular signals are produced by the activity of σF and σG in the forespore, and the activity of σF and σG is in turn coupled to formation of the asymmetric septum and completion of engulfment, respectively, the activation of σE and of σK by regulated proteolysis is indirectly coupled to morphogenesis. Both events are subject to multiple layers of control, as seen for the cell cycle-regulated degradation of key regulatory proteins in C. crescentus (Fig. 3B). The third activation event involves PodJ at the pole of C. crescentus SW cells and allows a shortened form of PodJ (PodJS) to serve as a scaffold for structural and signaling proteins that contribute to polar organelle development. We identify questions that remain about these three activation events. We note that Streptomyces coelicolor σBldN is made as a proprotein that is proteolytically processed during development of spore-bearing aerial hyphae, but the protease responsible has not been identified (Bibb & Buttner, 2003).
Activation of σE during B. subtilis sporulation
σE is initially made as inactive Pro-σE with an additional N-terminal 27 residues that are removed by regulated proteolysis (LaBell et al., 1987). Pro-σE begins to be synthesized before the polar septum forms, as Spo0A–P accumulates in the predivisional cell. Spo0A–P activates transcription of the spoIIG operon [for review, see (Losick & Stragier, 1992)], which codes for Pro-σE and the protease, SpoIIGA, that removes the pro-sequence from Pro-σE (Jonas et al., 1988, Stragier et al., 1988). Both Pro-σE and SpoIIGA localize to the polar septum when it forms (Peters & Haldenwang, 1991, Ju et al., 1997, Fawcett et al., 1998, Hofmeister, 1998), awaiting a signal from the forespore (Fig. 5B). The formation of the polar septum causes σF to be released from the SpoIIAB anti-σ in the forespore [see details above; for review, see (Kroos, 2007)], and σF RNA polymerase (RNAP) directs transcription of the spoIIR gene (Hofmeister et al., 1995, Karow et al., 1995, Londono-Vallejo & Stragier, 1995). The SpoIIR protein is believed to be secreted from the forespore into the septal space, where it signals SpoIIGA to cleave Pro-σE (Fig. 5B). In this way, activation of σE is linked temporally to activation of σF, which is in turn governed by formation of the polar septum (under control of Spo0A–P, although how this transcription factor redirects septum formation from midcell to a polar location remains a mystery).
Figure 5. Activation of σE during B. subtilis sporulation.
(A) Model for activation of σE primarily in the mother cell (Chary et al., 2010). SpoIIGA (black circles) is evenly distributed in the predivisional cell membrane, so shortly after the polar septum forms it is evenly distributed in membranes (top), but diffusion and capture at the septum is proposed to result in a higher concentration of SpoIIGA on the mother cell side (bottom), which would outcompete SpoIIGA on the forespore side for SpoIIR (blue) secreted into the septal space, resulting in cleavage of Pro-σE primarily in the mother cell. Grey box indicates region expanded in (B).
(B) Expanded view of a model for the activation of σE across the polar septum. σF directs spoIIR transcription in the forespore. SpoIIR is translocated across the forespore membrane of the polar septum, but remains associated with the membrane and becomes acylated on a threonine residue (T), which allows it to be cleaved by signal peptidase (SIP) and released into the septal space to interact with SpoIIGA in the mother cell membrane of the septum (Diez et al., 2012). Alternatively, SpoIIGA in the mother cell membrane of the septum might interact with SpoIIR prior to its release from the forespore membrane of the septum, but SpoIIR would still need to be released in order to activate SpoIIGA. Interaction of SpoIIR with the N-terminal domain of SpoIIGA is proposed to cause a conformational change in its C-terminal domain that allows the active aspartic protease dimer to cleave Pro-σE associated with the mother cell membrane of the septum, releasing σE into the mother cell (Imamura et al., 2008).
Several factors ensure that σE accumulates in the mother cell and not in the forespore. One of these was mentioned in the preceding section since it appears to involve destruction of Pro-σE in the forespore. A second factor contributing to the spatial regulation of σE is that the spoIIG operon is transcribed mainly in the mother cell after the polar septum forms (Fujita & Losick, 2003). This is due in part to the origin-distal location of the spoIIG operon since there is a short (about 15 minute) period of transient genetic asymmetry after polar septation during which both copies of spoIIG are present in the mother cell (Fig. 4ii). Also, the SpoIIAA protein appears to inhibit formation of Spo0A–P in the forespore, so the spoIIG operon is not transcribed as strongly as in the mother cell (Arabolaza et al., 2003). Circumventing some of these regulatory mechanisms by placing a second copy of the spoIIG operon near the origin of replication (so it is present in the forespore upon asymmetric septation) and driving its transcription in the forespore using a strong σF-directed promoter revealed a third factor that can contribute to spatial regulation of σE, but is not normally required for this purpose (Chary et al., 2010). CsfB, which inhibits premature σG activity in the forespore (Chary et al., 2007, Camp & Losick, 2008, Karmazyn-Campelli et al., 2008), can also inhibit σE activity in the forespore (Chary et al., 2010). The work of Chary et al. (Chary et al., 2010) also suggested that SpoIIGA and SpoIIR are limiting for σE accumulation in the forespore. This led them to propose that SpoIIGA evenly distributed in the membrane of the predivisional cell might diffuse and be captured at the polar septum when it forms (Fig. 5A). Owing to the larger size of the mother cell, more SpoIIGA would be captured on the mother cell side of the septum and it would outcompete SpoIIGA on the forespore side for SpoIIR secreted into the septal space, resulting in activation of SpoIIGA predominantly on the mother cell side and cleavage of Pro-σE there. An attractive feature of this model is that SpoIIGA would be poised to respond to SpoIIR upon septation, provided that SpoIIGA diffusion and capture at the septum is rapid. This could explain how σE can become active in the mother cell within 4 minutes of spoIIR being transcribed by σF RNAP in the forespore (Eldar et al., 2009). Rapid activation of σE in the mother cell is necessary since σE RNAP directs transcription of genes whose products prevent formation of a second polar septum in the mother cell (Eichenberger et al., 2001) and commit the mother cell to sporulation (Dworkin & Losick, 2005). It remains to be seen whether SpoIIGA is more abundant on the mother cell side of the septum than on the forespore side. Advances in total internal reflection fluorescence (TIRF) microscopy (Betzig et al., 2006) should allow this question to be addressed. Presumably, natural conditions have selected for the seemingly redundant mechanisms that ensure σE accumulates in the mother cell and not in the forespore.
How does the SpoIIR signal protein activate the SpoIIGA protease to cleave Pro-σE? Coexpression of SpoIIR with SpoIIGA and Pro-σE in E. coli resulted in accurate, rapid, and abundant cleavage of Pro-σE to σE (Imamura et al., 2008). Using this system, mutational analysis based on modeling of the SpoIIGA C-terminal domain provided evidence that it forms a dimeric aspartic protease similar to the human immunodeficiency virus type 1 (HIV-1) protease. This led to the simple notion that SpoIIR activates SpoIIGA by promoting dimer formation; however, in the absence of SpoIIR, SpoIIGA was found to be self-associated after detergent solubilization from E. coli membranes. Upon coexpression in E. coli, SpoIIR was found to interact with SpoIIGA. These findings suggest that SpoIIR secreted to the periplasm of E. coli can interact with extracellular parts of dimeric SpoIIGA, causing a conformational change that is transmitted across the membrane to activate the cytosolic C-terminal aspartic protease domain to cleave Pro-σE (Fig. 5B). A follow-up study investigated the specificity of B. subtilis SpoIIGA and its paralogs from several Bacilli for their Pro-σE substrates (Imamura et al., 2011). Insights to emerge are that residues distal from the cleavage site contribute to substrate specificity and that SpoIIGA paralogs exhibit a range in their breadth of ability to cleave Pro-σE paralogs. Much more work is needed to understand how SpoIIGA interacts with Pro-σE at the membrane surface. A recent study shed new light on the interaction between SpoIIGA and SpoIIR (Diez et al., 2012). Starting from a previous observation that processing of Pro-σE to σE requires fatty acid biosynthesis (Schujman et al., 1998), it was shown that SpoIIR signaling as well as cleavage of its N-terminal signal sequence probably depends on acylation of a conserved threonine residue (Diez et al., 2012). It was also shown that SpoIIGA recruits SpoIIR to the septum, but the results suggest that only acylated SpoIIR can be released from the forespore membrane into the septal space and activate SpoIIGA (Fig. 5B), which may be predominantly in the mother cell membrane of the septum (Fig. 5A). Acylation of SpoIIR is proposed to allow cleavage by an unidentified signal peptidase, perhaps in a membrane microdomain that harbors the signal peptidase (Diez et al., 2012). In this way, activation of σE and ensuing commitment to sporulation could be linked to fatty acid synthesis, coordinating metabolic conditions with developmental progression. Some aspects of the model need more work. Acylation of SpoIIR needs to be demonstrated and the acetyltransferase needs to be identified, as does the signal peptidase proposed to cleave acylated SpoIIR. It is worth noting that unacylated SpoIIR appears to be able to activate SpoIIGA made in the B. subtilis forespore (Diez et al., 2012). Whether SpoIIR remains anchored via its signal peptide in the forespore membrane of the polar septum and interacts with the N-terminal domain of SpoIIGA embedded in that membrane, or whether SpoIIR is released by a signal peptidase that does not require acylation of SpoIIR, is unknown. Likewise, it is unknown whether SpoIIR produced in E. coli is released from the inner membrane into the periplasm when it stimulates coexpressed SpoIIGA to cleave Pro-σE in the cytosol (Imamura et al., 2008). These instances of signaling by SpoIIR might not fully recapitulate intercellular signaling from the forespore to the mother cell, which requires acylation of SpoIIR and its release from the forespore membrane of the septum (Diez et al., 2012). Perhaps acylated SpoIIR is incapable of activating SpoIIGA located in the same membrane, making release of SpoIIR into the septal space necessary, thus promoting intercellular signaling due to the proposed abundance of SpoIIGA in the mother cell membrane of the polar septum (Fig. 5).
In summary, the protease(s) responsible for destruction of Pro-σE in the forespore has not been identified. It is but one of several mechanisms that ensure σE inactivity in the forespore. An unidentified signal peptidase is proposed to cleave acylated SpoIIR. If so, the pathway is a two-step proteolytic cascade involving production of an intercellular signal in the first step. In the second step, SpoIIGA activates a regulatory protein by cleaving Pro-σE to σE. SpoIIGA may accumulate more abundantly in the mother cell membrane of the polar septum and outcompete SpoIIGA in the forespore membrane of the septum for interaction with a limiting amount of the SpoIIR signal protein. SpoIIGA appears to be unique among aspartic proteases in terms of its structure and mechanism of activation, allowing it to assess protein (SpoIIR) and fatty acid (SpoIIR acylation) biosynthesis in the forespore and activate σE predominantly in the mother cell. Whether SpoIIR acylation reports energy status of the forespore to the mother cell or helps ensure predominantly intercellular signaling from the forespore to the mother cell, or both, remains to be seen.
Activation of σK during B. subtilis sporulation
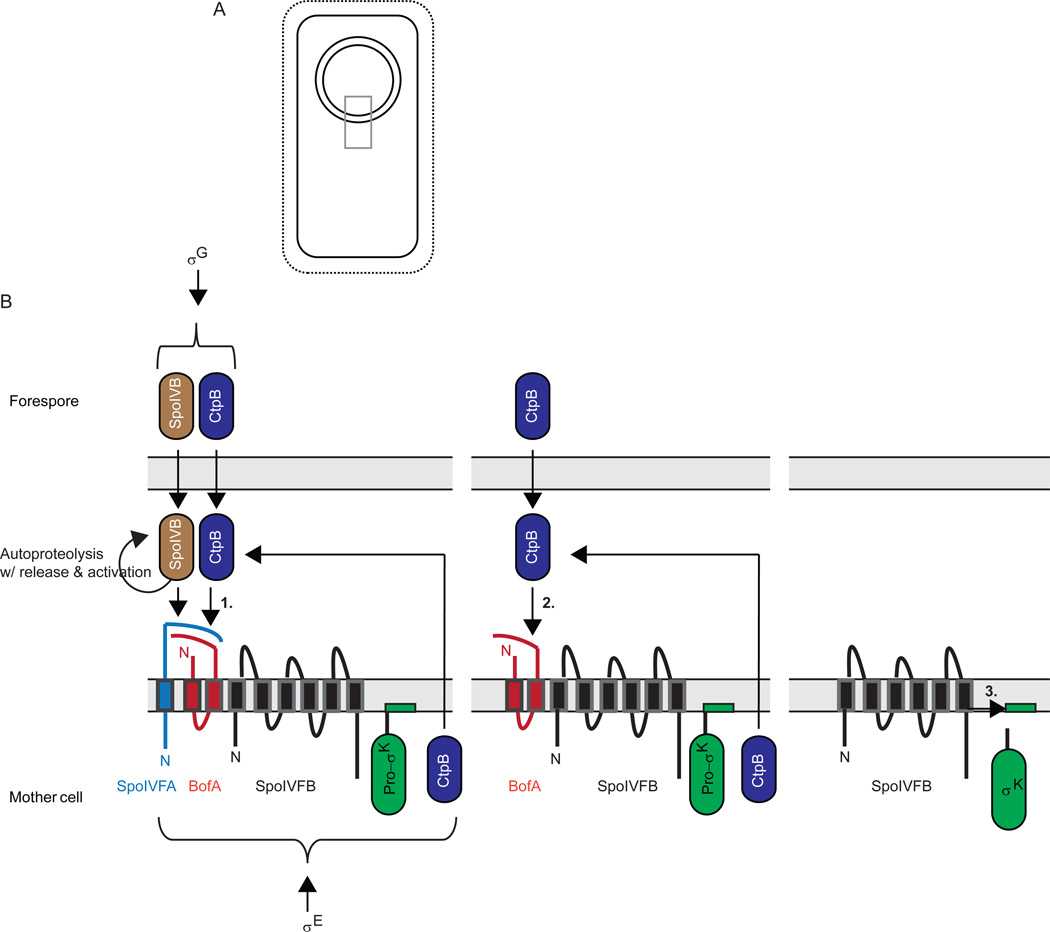
σK is initially made as inactive Pro-σK with an additional N-terminal 20 residues that are removed by regulated proteolysis (Kroos et al., 1989, Cutting et al., 1990, Lu et al., 1990). The gene coding for Pro-σK is created by a chromosomal rearrangement that occurs only in the mother cell due to σE-directed expression of a site-specific recombinase (Stragier et al., 1989, Kunkel et al., 1990, Sato et al., 1990, Sato et al., 1994). For this reason, and because the initial transcription of the rearranged sigK gene requires both σE RNAP and SpoIIID (Kunkel et al., 1988, Halberg & Kroos, 1994), a transcription factor that also depends on σE for its transcription (Kunkel et al., 1989, Stevens & Errington, 1990, Tatti et al., 1991, Jones & Moran, 1992), Pro-σK is made only in the mother cell (Zhang et al., 1998) and its appearance is delayed relative to most proteins in the σE regulon. Those proteins include SpoIVFB, an IMMP that removes the pro-sequence from Pro-σK (Cutting et al., 1990, Cutting et al., 1991, Lu et al., 1995, Rudner et al., 1999, Yu & Kroos, 2000, Zhou et al., 2009), and two proteins that inhibit the IMMP, SpoIVFA and BofA (Cutting et al., 1990, Cutting et al., 1991, Ricca et al., 1992, Resnekov & Losick, 1998, Rudner & Losick, 2002, Zhou & Kroos, 2004). The three proteins form a complex in the mother cell membrane of the polar septum (Figs. 6 and 7), which migrates around the forespore during the process of engulfment (Resnekov et al., 1996, Rudner & Losick, 2002, Rudner et al., 2002, Doan et al., 2005, Jiang et al., 2005). During engulfment, channels form, spanning the space between the two membranes surrounding the forespore and connecting it to the mother cell (Blaylock et al., 2004, Camp & Losick, 2008, Meisner et al., 2008, Camp & Losick, 2009, Doan et al., 2009). The channels are proposed to allow the mother cell to nurture the forespore by providing small molecules needed to maintain the integrity of the forespore and allow σG RNAP to transcribe genes (Camp & Losick, 2009, Doan et al., 2009). Among those genes is one that codes for SpoIVB (Cutting et al., 1991), a serine protease secreted from the forespore into the intermembrane space (Wakeley et al., 2000, Hoa et al., 2002), where it cleaves the extracellular domain of SpoIVFA (Fig. 6), a crucial first step toward activating the IMMP, SpoIVFB, to cleave Pro-σK (Dong & Cutting, 2003, Zhou & Kroos, 2005, Campo & Rudner, 2006). A second serine protease, CtpB, is secreted from both the forespore and the mother cell into the intermembrane space, where it can cleave both SpoIVFA and BofA, but this appears to be a fine-tuning mechanism since absence of CtpB only delays SpoIVFB cleavage of Pro-σK slightly (Pan et al., 2003, Zhou & Kroos, 2005, Campo & Rudner, 2006, Campo & Rudner, 2007). One or more additional proteases might participate in fully relieving SpoIVFB from inhibition by SpoIVFA and BofA. In any case, it is already clear that activation of σK in the mother cell involves at least two serine proteases secreted from the forespore, which destroy inhibitors of an IMMP located in the outermost membrane that surrounds the forespore after engulfment, allowing the IMMP to cleave Pro-σK and release active σK into the mother cell (Fig. 6). Hence, this proteolytic cascade involves both destruction of the regulatory proteins SpoIVFA and BofA, and activation of the regulatory protein σK via RIP. It is initiated by activation of σG in the forespore, which relies on channels formed during engulfment. Interestingly, channel proteins interact with SpoIVFA, which, in turn, interacts with engulfment proteins (Doan et al., 2005, Jiang et al., 2005), although the interactions might be indirect (Fig. 7). Nevertheless, the interactions likely promote close coupling between engulfment, channel formation, σG activity in the forespore, and activation of σK in the mother cell. Below, we discuss components of the σK activation pathway in more detail.
Figure 6. Activation of σK during B. subtilis sporulation.
(A) Upon completion of engulfment, the forespore is surrounded by two membranes. Grey box indicates region expanded in (B).
(B) Expanded view depicting a series of proteolytic cleavages (see text for references). First, σG in the forespore causes expression of serine proteases SpoIVB and CtpB (also expressed under σE control in the mother cell), which are translocated into the intermembrane space, where they cleave the C-terminal domain of SpoIVFA to initiate its degradation (1). SpoIVFA was in a complex with BofA and SpoIVFB in the outer membrane surrounding the forespore after these proteins were expressed in the mother cell under σE control. In a second step, CtpB and one or more other proteases (not shown) cleave BofA to initiate its degradation (2). Finally, SpoIVFB cleaves Pro-σK, releasing σK into the mother cell (3). The pro-sequence of Pro-σK is depicted to loop into the membrane based on findings that Pro-σK associates peripherally with membranes in B. subtilis (Zhang et al., 1998) or when expressed in E. coli (Zhou et al., 2013).
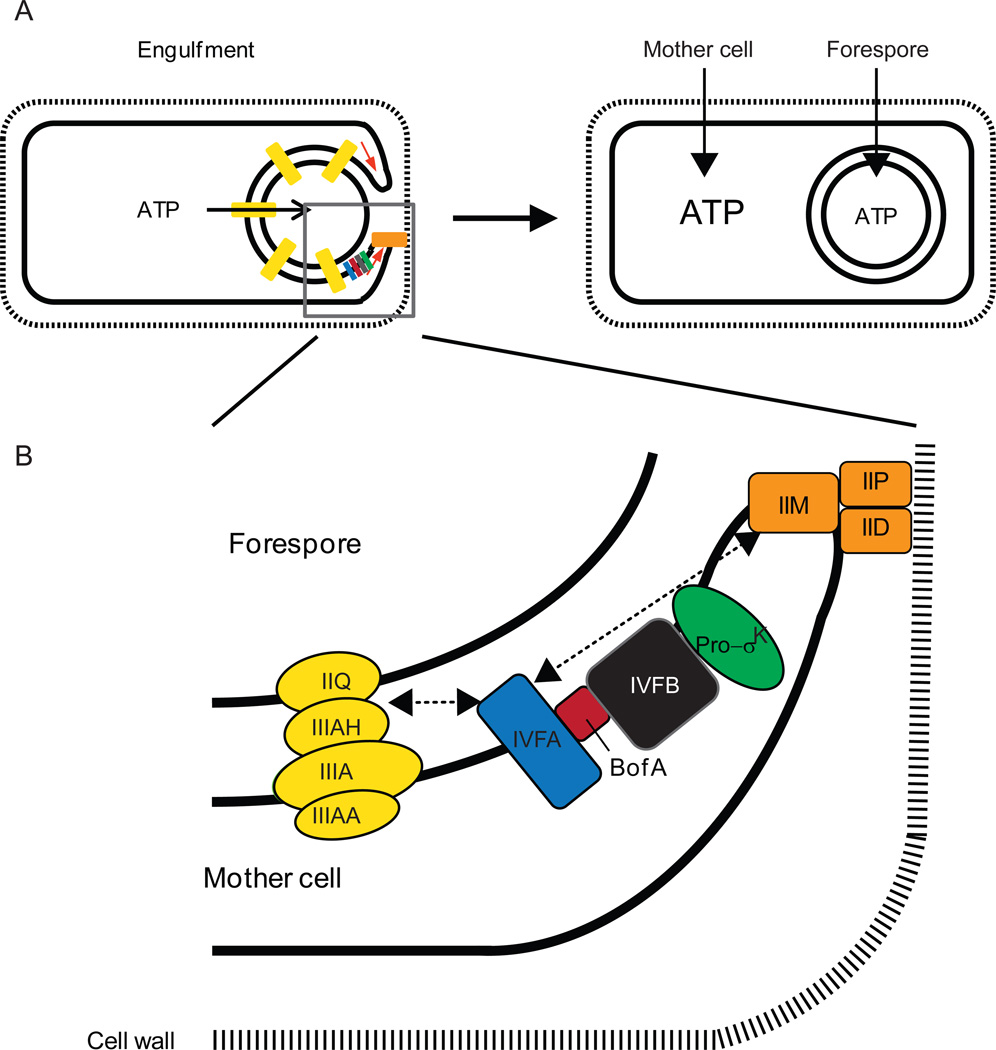
Figure 7. A model for ATP transport and accumulation during B. subtilis sporulation, and localization of the SpoIVFA-BofA-SpoIVFB complex with channel and engulfment complexes.
(A) A complex of proteins (orange) interacts with the cell wall and causes the mother cell membrane to engulf the forespore (left). During engulfment, channels (yellow) are formed that span the intermembrane space and have been proposed to allow small molecules like ATP to move from the mother cell into the forespore (see text for references). Upon completion of engulfment, the channels undergo reorganization and some components are degraded, perhaps allowing the ATP concentration to rise in the mother cell (right). ATP binding to the CBS domain of SpoIVFB would activate it to cleave Pro-σK, provided SpoIVFA and BofA have been degraded (Fig. 6). Grey box indicates region expanded in (B).
(B) Enlarged view of protein complexes during engulfment. SpoIVFA facilitates assembly of SpoIVFB with its inhibitor BofA and localizes the complex to foci that include the channel (yellow) and engulfment complexes (orange), although whether SpoIVFA interacts directly with a protein(s) in the other complexes or interacts indirectly is unknown (dashed arrows). Reprinted from (Kroos & Akiyama, 2013) with permission.
SpoIVB is the primary signal from the forespore (Fig. 6). (Gomez & Cutting, 1996, Gomez & Cutting, 1997, Wakeley et al., 2000)(Gomez & Cutting, 1996)After translocation across the innermost membrane that surrounds the forespore after engulfment, the PDZ domain of one SpoIVB molecule binds to the C-terminus of another SpoIVB molecule, facilitating autoproteolytic cleavage in trans near the N-terminus, which releases SpoIVB into the space between the two membranes surrounding the forespore (Dong & Cutting, 2004) (Fig. 6). SpoIVB then undergoes two autoproteolytic cleavages in cis near its new N-terminus, after which its PDZ domain appears to bind to the C-terminal region of BofA in the intermembrane space, allowing SpoIVB to cleave the C-terminal region of SpoIVFA at multiple sites in the intermembrane space (Dong & Cutting, 2003, Dong & Cutting, 2004, Campo & Rudner, 2006). A variant of SpoIVFA rendered uncleavable by SpoIVB due to multiple substitutions around the cleavage sites can be cleaved by CtpB, but this requires SpoIVB, perhaps to cleave another protein in complex with SpoIVFA (Campo & Rudner, 2006, Campo & Rudner, 2007). While CtpB may provide a backup mechanism in case spoIVFA has mutations, the finding that SpoIVFA decreased normally during development of a ctpB mutant, but not a spoIVB mutant, indicates SpoIVB is responsible for destruction of SpoIVFA (Zhou & Kroos, 2005), initiating a proteolytic cascade.
The primary role of CtpB may be to cleave BofA. Loss of BofA was delayed during development of a ctpB mutant in pulse-chase immunoprecipitation experiments, and cleavage of Pro-σK was similarly delayed (Zhou & Kroos, 2005). CtpB appeared to cleave BofA near its C-terminus upon coexpression in E. coli. Purified CtpB degraded purified BofA. Taken together, the results support a model in which SpoIVB first cleaves SpoIVFA, then CtpB cleaves BofA (Fig. 6). However, the finding that Pro-σK cleavage is delayed but not abolished in a ctpB null mutant suggests that an additional protease(s) might participate in the destruction of BofA (Pan et al., 2003, Zhou & Kroos, 2005).
BofA appears to be the primary inhibitor of the SpoIVFB IMMP. BofA, but not SpoIVFA, forms a complex with SpoIVFB and inhibits its activity upon coexpression in E. coli (Zhou & Kroos, 2004). A histidine residue (H57) in BofA was shown to be important for complex formation and inhibition, leading to the proposal that H57 provides a fourth zinc ligand that inhibits the SpoIVFB metalloprotease, analogous to the cysteine switch mechanism of matrix metalloprotease regulation (Van Wart & Birkedal-Hansen, 1990). The proposed mechanism of inhibition by BofA (Zhou & Kroos, 2004) remains to be tested further.
SpoIVFA functions primarily as an assembly and localization factor. It facilitates formation of a complex with BofA and SpoIVFB during B. subtilis sporulation (Rudner & Losick, 2002) and it enhances inhibition of SpoIVFB in combination with BofA upon coexpression in E. coli (Zhou & Kroos, 2005). SpoIVFA also localizes the complex to foci in the outermost membrane surrounding the forespore (Doan et al., 2005, Jiang et al., 2005). The foci contain proteins involved in engulfment and channel formation (Fig. 7). SpoIID, SpoIIM, and SpoIIP are normally required for engulfment (Abanes-De Mello et al., 2002, Aung et al., 2007). They form a complex that interacts with the cell wall and pulls the mother cell membrane around the forespore (Morlot et al., 2010). SpoIIQ and SpoIIIAH have extracellular domains that interact and zipper the mother cell membrane around the forespore during engulfment (Blaylock et al., 2004, Broder & Pogliano, 2006). The interaction between their extracellular domains allows SpoIIQ and SpoIIIAH to form channels that connect the mother cell and forespore cytoplasms (Blaylock et al., 2004, Broder & Pogliano, 2006). The channels likely include several other proteins encoded in the spoIIIA operon since these SpoIIIA proteins resemble components of secretion systems (Camp & Losick, 2008, Meisner et al., 2008). SpoIIIAA is similar to secretion ATPases and its ATPase motifs are important for sporulation (Doan et al., 2009). How SpoIVFA localizes the BofA- and SpoIVFB-containing complex to foci containing engulfment and channel proteins is unknown.
Why the SpoIVFA-BofA-SpoIVFB complex is localized to foci containing engulfment and channel proteins is also unknown. The finding that a bofA null mutation which bypasses the need for σG to activate σK (Ricca et al., 1992), does not bypass the need for engulfment and channel proteins to activate σK, suggested that engulfment and channel proteins govern σK activation in the mother cell independently of their role in activating σG in the forespore (Jiang et al., 2005). However, accumulation of SpoIVFB is diminished in the absence of BofA (Rudner & Losick, 2002) and it is further diminished in the absence of both BofA and an engulfment (SpoIID) or channel (SpoIIQ) protein (Doan & Rudner, 2007). A more stable form of SpoIVFB that accumulated in the absence of BofA, bypassed the need for engulfment and channel proteins to activate σK (Doan & Rudner, 2007). These investigators also found that the SpoIVB signal protein and other proteins secreted into the intermembrane space are degraded when engulfment is impaired, and this degradative response is turned off if engulfment is restored. The protease(s) mediating this reversible degradative response has not been identified, but it appears to couple the morphological process of engulfment to secretion-dependent SpoIVB and CtpB proteolysis of SpoIVFA and BofA, respectively, allowing SpoIVFB to cleave Pro-σK. Such coupling between morphogenesis, signaling, and gene expression is attractive, but a degradative response in the intermembrane space would not demand the observed co-localization of the SpoIVFA-BofA-SpoIVFB complex with engulfment and channel proteins. Rather, the co-localization could be related to the ATP dependence of SpoIVFB (Zhou et al., 2009). The channels have been proposed to secrete small molecules from the mother cell into the forespore in order to maintain its integrity and allow σG RNAP activity (Camp & Losick, 2009, Doan et al., 2009). As proposed recently, secretion of ATP through the channels and/or the ATPase activity of SpoIIIAA might result in a relatively low ATP concentration in the vicinity of channels, ensuring that channel-associated SpoIVFB remains inactive in case it escapes BofA inhibition (Kroos & Akiyama, 2013) (Fig. 7). The channels undergo reorganization and some components are degraded upon completion of engulfment (Chiba et al., 2007, Meisner et al., 2008). This would presumably allow the ATP concentration to rise in the mother cell, especially in the vicinity of the outermost membrane surrounding the forespore. Binding of ATP to the C-terminal CBS domain of SpoIVFB would activate the enzyme by changing its conformation or oligomeric state (Zhou et al., 2009), as observed for the CBS domains in its namesake cystathionine-β-synthase and in a variety of other proteins (Scott et al., 2004). This model is attractive since activation of σK would be coupled both to channel-dependent activation of σG in the forespore, leading to secretion of SpoIVB and CtpB proteases that target SpoIVFA and BofA (Fig. 6), and to engulfment completion and channel destruction, resulting in a rise in ATP that activates SpoIVFB to cleave Pro-σK (Fig. 7). A mechanism ensuring completion of engulfment and destruction of channels seems desirable since σK RNAP activity in the mother cell primarily leads to production of spore coat proteins that assemble on the forespore surface. It remains to be seen whether the ATP concentration rises in the mother cell as proposed and it remains to be understood how ATP activates the SpoIVFB IMMP.
In summary, activation of σK is the culmination of a proteolytic cascade that is highly integrated with morphogenesis and has multiple control points. (Zhang et al., 1998, Zhou et al., 2009, Zhou et al., 2013)The serine proteases SpoIVB and CtpB are secreted into the intermembrane space surrounding the forespore and target SpoIVFA and BofA for destruction, respectively, although another protease(s) that remains to be identified is implicated in BofA destruction, and one or more unknown proteases appear to ensure that proteins secreted into the intermembrane space, such as SpoIVB and CtpB, do not accumulate highly until engulfment is completed. Upon completion of engulfment, destruction of SpoIVFA, BofA, and channel proteins commences, liberating the IMMP SpoIVFB from inhibition and perhaps allowing its CBS domain to sense a rise in ATP in the mother cell, resulting in RIP of Pro-σK and release of active σK from the outermost membrane surrounding the forespore, into the mother cell. Identifying and characterizing other proteases involved in the pathway, and understanding exactly how and why the SpoIVFA·BofA·SpoIVFB complex interacts with engulfment and channel proteins, offer challenges that promise even deeper knowledge of temporally and spatially regulated proteolysis. Comparing the proteolytic cascade governing σK activation with that proposed to govern σE activation (Fig. 5B) and with that governing RseA destruction (releasing σE from inhibition) in E. coli (Fig. 2B), there are more proteases (SpoIVB, CtpB, SpoIVFB, and others that remain to be identified) and more substrates (SpoIVFA, BofA, and Pro-σK) in the σK activation pathway, providing more opportunities for regulated proteolytic events.
Modification of PodJ activity in C. crescentus by regulated proteolysis
A hallmark of the C. crescentus cell cycle is the close coupling between cell cycle progression and the morphogenesis of two distinct daughter cells with different polar structures. In the late PD cell, a flagellum is built and the bases of the type IV pili are assembled at the pole opposite to the stalked pole (Fig. 3A). After cell division, the type IV pili filaments are assembled. As described, at the SW-to-ST cell transition, the flagellum is shed and the pili are retracted, followed by formation of a stalk with a holdfast at its tip. Thus, the pole asymmetries are preserved by two mechanisms, (i) restriction of the assembly of the flagellum and the pili to the incipient SW pole; and, (ii) the removal of these structures at the SW-to-ST cell transition [for reviews, see (Jenal, 2009, Kirkpatrick & Viollier, 2012, Tsokos & Laub, 2012)]. The PodJ protein functions in this polar organelle development by serving as a polarly localized recruitment factor for other proteins including the pilus base protein CpaE (Viollier et al., 2002, Viollier et al., 2002), the holdfast protein HfaD (Hardy et al., 2010) and the regulatory proteins PleC, PopA and DivL (Viollier et al., 2002, Hinz et al., 2003, Lawler et al., 2006, Duerig et al., 2009, Curtis et al., 2012) involved in controlling cell cycle progression.
PodJ accumulation is cell cycle regulated by a dynamic balance between synthesis and proteolysis (Crymes et al., 1999, Viollier et al., 2002, Hinz et al., 2003). PodJ exists in two isoforms, the full-length PodJL and a shorter, truncated form (PodJS). PodJL consists of an N-terminal cytoplasmic domain, a trans-membrane domain and a C-terminal periplasmic domain (Viollier et al., 2002, Hinz et al., 2003, Lawler et al., 2006) (Fig. 8). PodJL is synthesized in the late ST cell/early PD cell and localizes to the incipient flagellated pole (Fig. 8). During cell division, PodJL is proteolytically processed to PodJS, which is also at the flagellated pole. Later, during the SW-to-ST cell transition, PodJS is completely degraded (Fig. 8). It has been suggested that the temporally regulated processing of PodJL to PodJS may serve to shut off the assembly of additional pilus bases and possibly also to allow PodJS to recruit other factors to the flagellated pole (Jenal, 2009, Curtis et al., 2012).
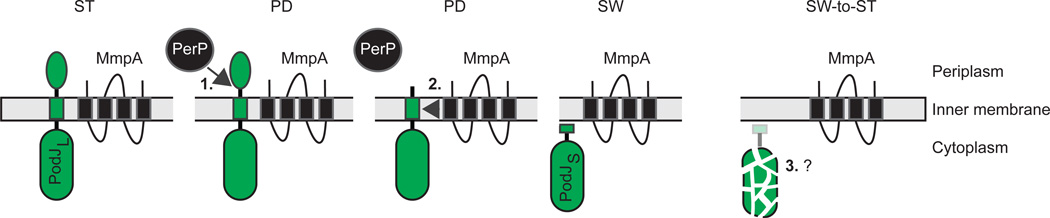
Figure 8. Regulation of PodJ in C. crescentus by sequential proteolysis.
The bitopic inner membrane protein substrate protein PodJL is shown in green. Cleavage of PodJL by PerP, the site-1-protease, is immediately followed by cleavage by the IMMP MmpA, the site-2-protease. Later, PodJS is cleaved at the SW-to-ST transition by an unknown protease. Swarmer (SW), stalked (ST) and predivisional (PD) cells are indicated. See also Figure 3A for details.
PodJ processing is an example of a proteolytic cascade with at least three proteases involved: PerP, which is a periplasmic aspartic protease of the retropepsin-like family, is a site-1-protease, makes the first cut and cleaves PodJL immediately after the trans-membrane domain to remove the periplasmic domain (Chen et al., 2006, Curtis et al., 2012) (Fig. 8). The cleavage by PerP is immediately followed by cleavage by MmpA, the site-2-protease, which is an IMMP similar to those involved in RseA degradation (Fig. 2B) and σK activation (Fig. 6). RIP by MmpA within the trans-membrane domain releases PodJ from the membrane giving rise to PodJS (Chen et al., 2005, Curtis et al., 2012). Finally, PodJS is degraded by a third protease during the SW-to-ST cell transition (Chen et al., 2005, Curtis et al., 2012). This third protease remains to be identified but it is likely that one of the ATP-dependent proteases is involved in this step (Chen et al., 2005).
PodJL processing to PodJS is synchronized with the cell cycle by controlling the accumulation of PerP (Chen et al., 2006, Curtis et al., 2012). perP transcription is CtrA-dependent, initiates in the late ST cell/early PD cell and reaches a maximum in the SW cell (Chen et al., 2006) (Chen et al., 2005). PerP is presumably constitutively active and cleaves PodJL as soon as it accumulates sufficiently. Because MmpA is present throughout the cell cycle, it has been suggested that PerP-dependent cleavage of PodJL is the rate-limiting step in PodJL processing to PodJS and that MmpA cleavage is “only waiting” for the cut by PerP (Curtis et al., 2012). The signal(s) and the protease(s) leading to PodJS degradation during the SW-to-ST cell transition are not known.
As pointed out (Curtis et al., 2012), PodJ processing shares many similarities with the RIP of the anti-σ factor RseA in E. coli (Fig. 2B) with some notable differences. In the case of RseA, the site-1-protease DegS is anchored in the inner membrane with its serine protease domain in the periplasm while PerP is apparently not anchored in the inner membrane and is an aspartic protease of the retropepsin-like family. Notably, DegS is present in an inactive form and is specifically activated in response to the accumulation of unfolded outer membrane proteins. Processing of RseA by DegS is immediately followed by RIP by RseP, causing the release of the cytoplasmic part of RseA. Subsequently, this domain is immediately degraded by ClpXP leading to the release of σE. Thus, in the case of RseA, the first protease needs to be activated and once the proteolytic cascade has been initiated it results in the complete degradation of the substrate RseA. In contrast, in the case of PodJ, PerP is apparently constitutively active and is regulated at the level of its synthesis, and proteolysis is arrested after RIP by MmpA, with PodJS only being completely degraded at the SW-to-ST transition. As mentioned, it is not known which protease(s) is involved in PodJS degradation and how this degradation is regulated. As for activation of σK by a proteolytic cascade involving RIP (Figs. 6 and 7), the themes of multilayer control and morphological coupling are present in the cell cycle-dependent and pole-localized regulated proteolysis of PodJ.
Production of intercellular signals
Bacteria use intercellular communication to coordinate the activities of groups of cells, via the synthesis and sensing of self-generated, secreted intercellular signalling molecules (Bassler & Losick, 2006). Quorum sensing is a major signalling modality that allows bacteria to assess cell density and to respond appropriately at a population-wide level (Bassler & Losick, 2006). Quorum sensing systems generate an output response once a threshold concentration of a diffusible intercellular signalling molecule has been reached. For Gram-negative and Gram-positive bacteria, common intercellular signalling molecules involved in quorum sensing are acyl-homoserine lactone derivatives and small peptides, respectively (Bassler & Losick, 2006). As described for the activation of σE and σK in B. subtilis, intercellular signalling may also involve events in which a producing cell (the forespore) communicates exclusively with a recipient (the mother cell) in close proximity.
Intercellular communication is essential for bacterial development as exemplified during fruiting body formation by Myxococcus xanthus [for review, see (Konovalova et al., 2010)], the development of genetic competence, biofilms, and spores by B. subtilis [for reviews, see (López & Kolter, 2010, Vlamakis et al., 2013)], and aerial hyphae formation by Streptomyces species [for reviews see (Flärdh & Buttner, 2009, Chater et al., 2010, Willey & Gaskell, 2011, McCormick & Flardh, 2012)]. Many of these signals are generated by the proteolytic processing of ribosomally-produced precursors using a multitude of different types of proteases that are subject to different types of regulation (Fig. 1C). In addition, these signals may contain post-translational modifications, e.g., lanthionine bridges. Many of the small peptide signals are either taken up by cells or function to regulate histidine protein kinases of two component systems. Larger precursors that are cleaved to generate relatively large signalling proteins is a recurring theme in bacterial development, but exactly how these signals are produced and how they function are open questions in many cases. Secreted peptides and proteins generated by proteolytic processing not only function as intercellular signals but can also function as toxins that kill siblings and delay sporulation as described for the SkfA peptide and the SdpC protein fragment of B. subtilis (Gonzalez-Pastor et al., 2003) [for review, see (Gonzalez-Pastor, 2011)], or they may function directly in morphogenesis as described for the SapB peptide of Streptomyces coelicolor [for review, see (Willey & Gaskell, 2011).
Here, we focus on production of intercellular signals that function during development of M. xanthus, B. subtilis, and Streptomyces species. Insights gained from studies of these model organisms provide paradigms for understanding how regulated proteolysis contributes to intercellular communication of other bacteria. In particular, peptide signaling occurs during Anabaena heterocyst formation [for review, see (Kumar et al., 2010); see also (Higa et al., 2012)] but the protease(s) involved has not been identified. The proteases involved in two other types of peptide signalling have been identified but will not be described in detail here since these systems have been reviewed recently. Hydrophobic peptide signals regulate mating and virulence of Enterococcus faecalis, and their production involves an initial cleavage of a lipoprotein by Type II signal peptidase followed by cleavage of the signal sequence by an IMMP called Eep [for reviews, see (Thoendel & Horswill, 2010, Dunny & Johnson, 2011)]. Cyclic peptide signals regulate pathogenesis and biofilm disassembly of Staphylococcus aureus, and the current model for their production involves cleavage, cyclization, and transport across the membrane by the cysteine protease AgrB, followed by cleavage by Type I signal peptidase to release the cyclic peptide from the cell [for review, see (Thoendel et al., 2011)]. Cyclic peptides produced by AgrB-like proteins are likely used for quorum sensing by diverse Gram-positive bacteria, and are implicated in E. faecalis, Listeria monocytogenes, and Clostridium perfringens pathogenesis [for review, see (Thoendel & Horswill, 2010)].
Production of intercellular signals by M. xanthus
M. xanthus is a social bacterium characterized by the ability to form spore-filled fruiting bodies in response to starvation. Fruiting bodies are macroscopic structures formed as a result of aggregation of thousands of cells. Those cells that have accumulated inside fruiting bodies undergo differentiation to spores. These spores are environmentally resistant, can survive extended periods of starvation, and germinate to produce vegetative cells when nutrients become available. Fruiting body formation is a multicellular developmental process and requires coordination of cell behaviour in time and space. This coordination is accomplished by means of extensive intercellular communication. Initial genetic screens suggested that there are at least five intercellular signals (referred to as the A- to E-signals) (Hagen et al., 1978, Downard et al., 1993); however, only two of these signals, the A- and the C-signal, have been characterized experimentally. Both of these signals are generated by proteolysis. Interestingly, one of the two Lon proteases in M. xanthus has been implicated in the generation of the B-signal (Gill et al., 1993, Tojo et al., 1993).
Production of A-signal
The A-signal accumulates early during development and has been suggested to be part of a quorum sensing-like system that measures the density of starving cells (Kuspa et al., 1986, Kuspa & Kaiser, 1989, Kuspa et al., 1992). Only if a threshold concentration is reached, development proceeds (Kuspa et al., 1992). The A-signal was purified from the supernatant of starving M. xanthus cells based on its ability to rescue development of and gene expression in A-signal deficient asg mutants (Kuspa et al., 1986, Kuspa et al., 1992, Kuspa et al., 1992). The A-signal consists of two fractions, a heat-stable and a heat-labile fraction (Kuspa et al., 1992, Plamann et al., 1992). The heat-labile fraction contains at least two secreted proteases, one of which has a substrate specificity similar to that of trypsin (Plamann et al., 1992). Interestingly, the separate addition of three different proteases, trypsin, pronase or proteinase K, which have different specificities and are unrelated to M. xanthus, to cells unable to synthesize the A-signal, rescues development of these mutants (Plamann et al., 1992). The heat-stable fraction consists of a mixture of amino acids and small peptides (Kuspa et al., 1992). Individual amino acids have A-signal activity and the A-signal activity of a peptide is equal to the sum of the A-signal activity of its constituent amino acids (Kuspa et al., 1992, Kuspa et al., 1992). Based on these findings it has been suggested that amino acids and a mixture of non-specific peptides likely constitute the A-signal (Kuspa et al., 1992, Plamann et al., 1992). Moreover, it has been suggested that secreted proteases early during starvation non-specifically degrade surface exposed proteins to generate the A-signal. In response to starvation, the RelA-dependent stringent response is initiated, resulting in an upshift in the concentration of the alarmone/second messenger (p)ppGpp (Manoil & Kaiser, 1980). The increase in (p)ppGpp as well as at least five Asg proteins [for review see (Kaiser, 2004)] and the alternative sigma factor, σD (Viswanathan et al., 2006) are involved in A-signal production.
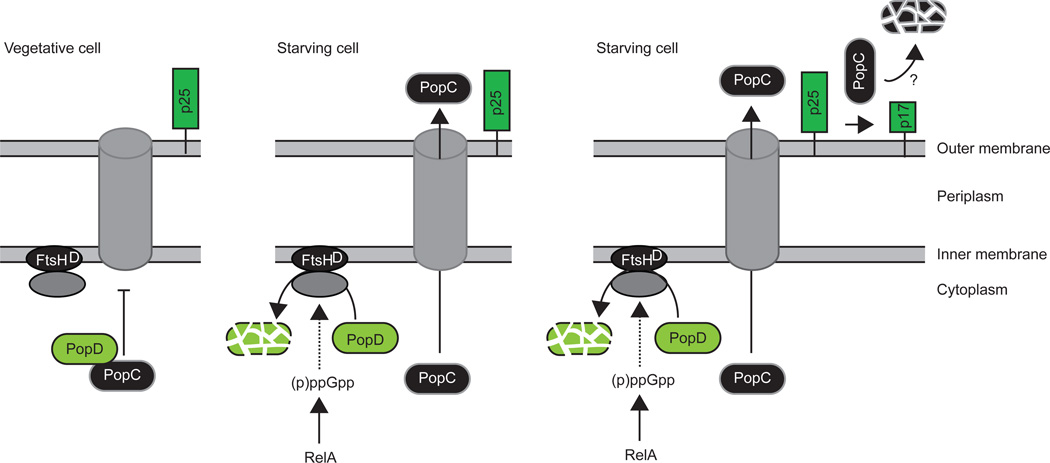
The identity of the A-signal generating proteases is currently unknown. However, recent global transcriptome analysis in two mutants unable to generate the A-signal (asgA and asgB) demonstrated that at least the AsgA and AsgB proteins are required for the expression of a large number of genes suggested to encode secreted proteins. Importantly, among these genes, 13 are predicted to encode secreted proteases including three trypsin-like proteases (Konovalova et al., 2012). These proteases are candidates for being involved in A-signal synthesis. A future challenge will be to identify the A-signalling proteases and to elucidate how their activity is restricted. The cellular response to A-signal also needs to be better-understood. Suppressor mutations that bypass the requirement for A-signalling for expression of certain genes appear to identify some components of the A-signal response pathway [for review, see (Kaplan, 2003)]. Interestingly, the asgA and asgB mutants have reduced expression of a secreted protease (PopC) necessary for C-signal production (see below), and restoring its expression in asg mutants rescued many of their developmental defects without restoring A-signalling (Konovalova et al., 2012).
Production of C-signal
The C-signal becomes important after 6 hrs of development (Kroos & Kaiser, 1987). The C-signal is essential for development and functions as a morphogen that induces the two morphogenetic events in fruiting body formation, aggregation of cells into fruiting bodies and their subsequent differentiation to spores (Shimkets et al., 1983). Moreover, available evidence suggests that the C-signal functions in a threshold dependent manner with a low level of signalling inducing aggregation and with sporulation being induced at a higher level of signalling (Kim & Kaiser, 1991, Li et al., 1992, Kruse et al., 2001).
The C-signal was initially purified from starving M. xanthus cells based on its ability to rescue development of a mutant, which is unable to synthesize the C-signal, and shown to be a 17 kDa protein (p17) encoded by the csgA gene (Kim & Kaiser, 1990, Kim & Kaiser, 1990). p17 is associated with the outer membrane (Lobedanz & Søgaard-Andersen, 2003) and likely exposed on the cell surface (Shimkets & Rafiee, 1990). In agreement with these observations, C-signal transmission is likely cell-cell contact-dependent (Kroos et al., 1988, Kim & Kaiser, 1990, Kim & Kaiser, 1990, Kim & Kaiser, 1990). p17 is generated by the specific proteolytic cleavage of the 25 kDa full-length CsgA protein (p25) and accumulates in response to starvation (Kruse et al., 2001, Lobedanz & Søgaard-Andersen, 2003). This cleavage results in the removal of the N-terminal 8 kDa of p25 although the precise cleavage site has yet to be mapped. Similarly to p17, p25 is associated with the outer membrane (Lobedanz & Søgaard-Andersen, 2003). p25 and p17 do not show similarity to outer membrane beta-barrel proteins (Lobedanz & Søgaard-Andersen, 2003) and, in fact, p25 shows sequence similarities to short-chain alcohol dehydrogenases (Lee et al., 1995). Also, p25 does not contain a lipoprotein signal peptide (Lobedanz & Søgaard-Andersen, 2003). Therefore, it is currently not known how p25 and p17 associate with the outer membrane but it has been speculated that both proteins may contain a hydrophobic modification that anchors them in the outer membrane (Lobedanz & Søgaard-Andersen, 2003).
p25 cleavage depends on a two-step proteolytic cascade that incorporates starvation sensing and regulation by compartmentalization (Rolbetzki et al., 2008, Konovalova et al., 2012). p25 as well as the subtilisin-like protease PopC that directly cleaves p25 to generate p17 accumulate in vegetative cells (Fig. 9). However, PopC is in the cytoplasm (Rolbetzki et al., 2008) whereas p25 is on the cell surface. PopC is held in the cytoplasm in a complex with the PopD protein (Konovalova et al., 2012) (Fig. 9). PopD is encoded in an operon with PopC, interacts directly with PopC and inhibits PopC secretion. Degradation of PopD is triggered in response to the starvation-induced, RelA-dependent upshift in (p)ppGpp (Konovalova et al., 2012) (Fig. 9) that also triggers A-signal production early in development(Konovalova et al., 2012). As a consequence of PopD degradation, PopC is released for secretion. PopD degradation depends on one of the two FtsH homologs in M. xanthus, FtsHD; however, it is not known whether FtsHD directly degrades PopD (Konovalova et al., 2012) (Fig. 9). Thus, PopD destruction is induced in response to a specific signal, starvation.
Figure 9. Generation of the C-signal (p17) in M. xanthus by a proteolytic cascade that incorporates destruction of a regulatory protein and regulation by compartmentalization.
The protease PopC and the substrate p25 are present in vegetative cells but PopC is held in a complex with PopD and not secreted. The PopC secretion system is not known and indicated by a grey barrel structure. In response to starvation, PopD is degraded in an FtsHD-dependent manner and PopC is secreted. Outside the cells, PopC cleaves p25 to generate p17 and is rapidly degraded. The question mark indicates that the protease(s) involved in PopC degradation is not known.
Once secreted, PopC gains access to the cell surface, where p25 is located and directly cleaves p25 (Rolbetzki et al., 2008). In total, this proteolytic cascade incorporates starvation sensing, which induces PopD degradation, and regulation of p25 cleavage by compartmentalization. (Rolbetzki et al., 2008) In vegetative cells, the protease and its substrate are in different cellular compartments (cytoplasm and cell surface, respectively) whereas in starving cells, protease and substrate are brought together in the same compartment. However, unlike in the case of CtrA, where protease and substrate are present within the same cellular compartment (the cytoplasm) and are brought together in a specific subcellular region within that compartment, regulation by compartmentalization for the p25/PopC system takes place across different cellular compartments. In total, the pathway that controls generation of the C-signal involves two different types of regulated proteolytic events: first, the degradation of PopD (destruction of a regulatory protein); and second, cleavage of p25 to generate active p17 (activation of a regulatory protein). In addition, p25 cleavage is regulated at the level of PopC stability (Rolbetzki et al., 2008). PopC is highly unstable after secretion independently of p17 cleavage and its own proteolytic activity (Fig. 9). In combination with the slow secretion of PopC, this may ensure the gradual accumulation of p17 that is important for its function as a temporal and spatial morphogen. How cells respond to p17 remains a challenging question. A receptor has not been identified. The response pathway involves the transcription factor FruA, which is similar to response regulators of two-component signal transduction systems (Ogawa et al., 1996, Ellehauge et al., 1998), but FruA lacks certain features typically found in response regulators and it has been proposed to function as a pseudoresponse regulator rather than being phosphorylated by a histidine kinase (Mittal & Kroos, 2009).
Production of intercellular signals and toxins by B. subtilis
Regulated proteolysis not only destroys and activates regulatory proteins that control B. subtilis genetic competence, heat shock response, sporulation, biofilm disassembly, and motility as described above, it also produces intercellular signals and toxins that govern some of these processes as well as biofilm formation and extracellular protease production. These signals are peptides cleaved from precursors ranging from about 40 to 200 residues in length. They appear to function in the context of other self-produced extracellular signals and environmental signals (both abiotic and ones produced by other organisms), which together allow subpopulations of cells in a community to differentiate [for review, see (López & Kolter, 2010)]. The peptide signals target key regulators of gene expression or they kill sensitive siblings. Many details of their production remain to be elucidated.
Production of the ComX signal
As B. subtilis grows to high density in culture, a 10-residue peptide accumulates in the culture media that is derived from the 55-codon comX gene (Magnuson et al., 1994). The peptide was called “competence pheromone” because it induced premature development of the ability to take up exogenous DNA in cells at low density. As noted above, intercellular signalling via ComX leads to synthesis of ComS and stabilization of ComK, the master regulator of genetic competence (Fig. 2A). The ComX peptide signal is sensed by ComP, a histidine protein kinase embedded in the cell membrane, that phosphorylates the ComA response regulator (Magnuson et al., 1994), and ComA-P activates the srf operon in which the 46-codon comS gene is embedded (D'Souza et al., 1994). Other genes in the srf operon code for proteins that synthesize surfactin, a non-ribosomally-produced lipopeptide signal that regulates biofilm formation [for review see (López & Kolter, 2010)]. Hence, ComX signalling governs biofilm and competence development.
The comX gene is in the comQXPA cluster, which has a similar organization as the S. aureus agrBDCA operon involved in cyclic peptide production and sensing (Tortosa et al., 2001, Thoendel & Horswill, 2010). ComQ does not exhibit sequence similarity to the AgrB cysteine protease but it may play similar roles in signal production. The ComX signal is not a cyclic peptide, but it is believed to be cleaved from the 55-residue translation product of comX and the resulting 10-residue linear peptide is isoprenylated on a tryptophan residue and secreted from the cell (Ansaldi et al., 2002). ComQ is similar to isoprenyl diphosphate synthases and substitutions in a putative isoprenoid binding domain of ComQ eliminated ComX signal production, suggesting that ComQ modifies the ComX peptide (Bacon Schneider et al., 2002). Expression of comQX in E. coli is sufficient for signal production (Tortosa et al., 2001). It is possible that an E. coli protease(s) cleaves the ComX precursor, and the peptide modified by ComQ is somehow released from the E. coli cells. Alternatively, analogous to the current model for AgrB function [for review, see (Thoendel et al., 2011)], ComQ might cleave the ComX precursor (but not cyclize the peptide), modify it, and transport it across the membrane. It is critical to determine whether ComQ or another B. subtilis protease(s) cleaves the ComX precursor, and whether this step is regulated, given the importance of ComX signalling in the regulation of genetic competence and biofilm formation.
Production of Phr signals
At least eight Phr peptides of 5–6 residues in length are believed to be derived from precursors that are 39–57 residues long, which are the translation products of phr genes that are genetically linked to rap genes [for review, see (Perego, 2013)]. Rap proteins regulate competence, sporulation, and extracellular protease production by dephosphorylating and/or binding to key response regulators. Whether Phr peptides function as intercellular quorum-sensing signals or as unshared extracellular timing devices has been debated (Perego & Brannigan, 2001, Pottathil & Lazazzera, 2003). The question is whether Phr peptides can leave the surface of the producing cell to signal another cell or whether they exclusively remain at the surface of the producing cell for import by that cell. In both cases, import is through an oligopeptide permease and the Phr peptide binds to its cognate Rap protein and inhibits its function [for review, see (Perego, 2013)].
Although Phr precursors have typical signal sequences, they do not appear to be cleaved by a Type I signal peptidase at the expected site (Stephenson et al., 2003). Nevertheless, using a substrate lacking the signal sequence, it was found that the extracellular serine proteases subtilisin and Vpr can produce CSF (PhrC) and PhrA, but not PhrE (Lanigan-Gerdes et al., 2007). Presumably, other extracellular proteases produce PhrE and perhaps other Phr peptides. Because B. subtilis secretes a large number of extracellular proteases, there is tremendous potential for regulation of Phr peptide production, but identifying the proteases involved and studying their regulation is very challenging.
Production of the SkfA and SdpC toxins
In a population of sporulating B. subtilis, some cells accumulate more Spo0A–P, triggering expression of the skf and sdp operons that lead to production of toxins that kill non-expressing siblings (Gonzalez-Pastor et al., 2003) [for review, see (Gonzalez-Pastor, 2011)]. Nutrients released from lysed cells delay sporulation of the cannibalistic cells, which go on to form spores if starvation persists.
Based on similarity to operons that produce peptide antibiotics, the skf operon was inferred to produce a toxic peptide, likely by cleaving and possibly modifying a predicted 55-residue SkfA translation product (Gonzalez-Pastor et al., 2003). Indeed, imaging mass spectrometry identified a candidate molecule and it was determined to be the 26 C-terminal residues of SkfA, cyclized and with two modifications, a disulfide bond and a novel thioether bridge (Liu et al., 2010). The product of skfC (formerly skfCD due to an erroneous stop codon) is similar to the CaaX family of proteases and is proposed to cleave and cyclize the SkfA precursor. SkfEF is similar to ABC transporters and was proposed to export the toxic peptide and confer resistance to it; in agreement, expression of skfEF was sufficient for immunity (Gonzalez-Pastor et al., 2003). Like ComQ involved in ComX signal production, SkfC is not similar in sequence to the AgrB cysteine protease involved in cyclic peptide production in S. aureus [for review, see (Thoendel et al., 2011)], but it may play similar roles (i.e., precursor cleavage and peptide cyclization, but not transport across the membrane). Biochemical work, perhaps modelled after that on AgrB (Qiu et al., 2005), can commence on SkfC to determine whether it functions as proposed.
The SpdC protein fragment was initially identified in conditioned medium as an extracellular factor that delays sporulation by inducing the yvbA yvaZ operon (Gonzalez-Pastor et al., 2003). The induced operon was subsequently renamed spdRI and SpdI was shown to confer immunity to the SpdC toxin (Ellermeier et al., 2006), which collapses the proton motive force of sensitive cells (Lamsa et al., 2012). The SpdC toxin was determined to be a 42-residue internal fragment from SpdC (residues 141–182) with one disulfide bond (Liu et al., 2010). Full-length SpdC appears to be secreted and cleaved by signal peptidase (after residue 32), but further cleavage requires SpdA and SpdB, although neither protein resembles known proteases so the requirement may be indirect (Perez Morales et al., 2013). Identifying the protease(s) that liberates SpdC toxin from its precursor might provide clues about regulation in addition to that provided by Spo0A–P at the level of transcription of the sdpABC operon.
Production of intercellular signals by Streptomyces species
The transition from growth of a vegetative (or substrate) mycelium to the formation of aerial hyphae by Streptomyces species involves intercellular signals, including some produced by regulated proteolysis [for reviews, see (Flärdh & Buttner, 2009, Chater et al., 2010, Willey & Gaskell, 2011, McCormick & Flardh, 2012)]. These signals include peptides that act in different ways. One or more short peptides is likely produced extracellularly and imported through an oligopeptide transporter, analogous to Phr peptides of B. subtilis, although the identity of the Streptomyces peptides and their intracellular targets are unknown. Modified peptides containing lanthionine bridges break surface tension to allow upward projection of aerial hyphae. Proteins anchored to the cell surface or secreted also participate in morphogenesis. Here, we focus on what is known about proteolytic events involved in signal production.
Extracellular proteases, a protease inhibitor, and an unidentified peptide
Streptomyces produce a large number of extracellular proteases that help acquire nutrients for growth, and they also use extracellular proteases, a protease inhibitor, and an unidentified short peptide to regulate the transition from growth to aerial hyphae formation [for review, see (Chater et al., 2010)]. For example, in Streptomyces griseus and in S. coelicolor the translation of a transcription factor, AdpA, is governed by the tRNA for the rare leucine codon UUA. The tRNA is specified by bldA, whose transcription is activated by AdpA, linking the two key regulators of early development in a positive feedback loop (Higo et al., 2011) (Fig. 10). AdpA also strongly activates transcription of a gene that codes for a secreted protease inhibitor in both species. The inhibitor protein, called STI in the case of S. coelicolor, inhibits at least one protease (SCO1355) that promotes aerial hyphae formation (Kim et al., 2008) (Fig. 10). Interestingly, inactivation of STI is partly due to another extracellular protease (SCO5913), which has a UUA codon needing the tRNA specified by bldA in order to be translated (Fig. 10). In the context of a general model, it has been speculated that induction of the inhibitor-inactivating enzyme (i.e., SCO5913) depends on a peptide signal imported by the oligopeptide transporter BldK (Chater et al., 2010) (Fig. 10). A short peptide signal (655 Da) was isolated that required BldK to restore aerial hyphae formation to a mutant blocked early in an extracellular signaling cascade, but the peptide sequence could not be determined (Nodwell & Losick, 1998). In addition to the identity of the peptide, its mode of production, and its intracellular target, a key question is how SCO1355 promotes aerial hyphae formation. SCO1355 is a putative serine protease with a P-domain that interacts with STI and is similar to P-domains of eukaryotic proprotein convertases, suggesting SCO1355 cleaves proproteins required for aerial hyphae formation (Fig. 10). Potential targets include SapB and secreted proteins described below. Despite the many questions that remain and the challenges inherent in studying proteases that function in the complex extracellular milieu, the lessons learned so far from investigations of M. xanthus A-signal, B. subtilis Phr peptides, and Streptomyces clearly indicate that extracellular proteases and peptide signals play important roles in bacterial development.
Figure 10. A model for regulation of aerial hyphae formation by extracellular proteases, a protease inhibitor, and a peptide in S. coelicolor (Chater et al., 2010).
The adpA and 5913 genes contain TTA DNA sequences that specify UUA codons, which require the tRNA product of the bldA gene in order to be translated. 5913 is an extracellular protease that negatively regulates the protease inhibitor STI (Streptomyces trypsin inhibitor), which binds to and inhibits 1355, an extracellular protease proposed to cleave proproteins required for aerial hyphae formation. An unidentified peptide signal imported by BldK may induce expression of 5913, although this is speculative, as indicated by the question mark.
Production of lanthionine bridge-containing peptides
Streptomyces produce modified peptides with surface-active properties that together with amphipathic chaplin proteins described below are believed to coat hyphae and allow them to project upward by breaking surface tension at the colony-air interface [for review, see (Willey & Gaskell, 2011). SapB of S. coelicolor is derived from the 42-codon ramS gene embedded in a locus with RamC, which is believed to catalyze formation of two lanthionine bridges, and with RamAB, an ABC transporter likely to export SapB. Recent work indicates that the 42-residue RamS translation product is produced constitutively and localizes to the membrane, where it awaits regulated expression of the other Ram proteins at the onset of aerial hyphae formation (Gaskell et al., 2012). A 21-residue leader peptide is removed during production of SapB, but the protease responsible and when it acts (i.e., before, during, or after transport) remain to be elucidated.
Production of anchored or secreted morphogenetic proteins
The chaplins are a family of cell wall-anchored or secreted proteins with surface-active properties that work with lanthionine-containing peptides described above to permit erection of aerial hyphae (Claessen et al., 2003, Elliot et al., 2003). The three long chaplins of S. coelicolor were predicted to have C-terminal sorting signals for cleavage and linkage to the cell wall by one or more sortase enzymes, and the five short amyloid-forming chaplins were predicted to be secreted and to self-assemble between the long chaplins, being organized into rodlets on the surfaces of aerial hyphae and the spores that eventually form at their tips, by another family of secreted proteins called rodlins (Claessen et al., 2004). Expression of the chaplins is regulated by σBldN (Elliot et al., 2003) and expression of rodlins depends on chaplins (Claessen et al., 2004), ensuring that the proteins are expressed at the time of aerial hyphae formation. Little is known about their secretion, but recent work on two sortases involved in cleavage and anchoring of the long chaplins yielded some surprises (Duong et al., 2012). A double mutant lacking both sortases was much more defective in aerial hyphae formation than a triple mutant lacking all three long chaplins. The double sortase mutant failed to transcribe aerial hyphae-specific genes, implicating the sortases in cleavage and anchoring of a protein other than chaplins that extracellularly regulates developmental progression. While the double sortase mutant lacked short chaplins on aerial surfaces as expected, the triple mutant lacking long chaplins unexpectedly had short chaplins on its surface, organized into rodlets with only slight defects. Therefore, the secreted short chaplins and the secreted rodlins assemble rodlets without anchoring to the cell wall by long chaplins. Nevertheless, the long chaplin mutant was significantly delayed for aerial hyphae formation, indicating that the long chaplins do play a role in morphogenesis.
It is worth noting that the S. coelicolor peptides and the short chaplins described above qualify as intercellular signals in the sense that producing cells can complement mutants extracellularly and/or the peptide or protein accumulates extracellularly and rescues aerial hyphae formation when added to mutants (Willey et al., 1991, Willey et al., 1993, Nodwell & Losick, 1998, Claessen et al., 2003). Whether the short peptide normally leaves the surface of the producing cell before being imported by BldK is a question analogous to that debated for B. subtilis Phr peptides as described above. Similarly, the extent to which SapB or short chaplins normally leave the surface of the producing cell is unknown. In any case, SapB and the short chaplins are not believed to be taken up by recipient cells, nor are they believed to interact with a cell surface receptor. Rather, they are believed to coat hyphae and allow them to project upward. Interestingly, the coating has been proposed to form a cylindrical compartment that facilitates diffusion of nutrients and metabolites (perhaps signals too?) from the substrate to the growing tips of the aerial hyphae (Chater et al., 2010).
A protein that may function more like the M. xanthus C-signal has been described in S. griseus. The M. xanthus C-signal (p17) is believed to remain at the surface of the producing cell and interact with a receptor on the surface of a recipient cell as described above. Ironically, the S. griseus protein is named Factor C [for reviews, see (Chater et al., 2010, Willey & Gaskell, 2011)]. It is a 31-kDa protein with a 38-residue N-terminal TAT secretion signal that is cleaved upon export, but whether the protein remains membrane-associated via an N-terminal potential membrane spanning segment and whether the protein undergoes additional cleavage are unclear, although Factor C does not appear to be a diffusible signal.
Concluding remarks
We have reviewed examples of regulated proteolysis that involve destruction or activation of a regulatory protein, or production of a signal during bacterial development (Fig. 1). A major theme in the section on destruction of regulatory proteins is that the same Clp proteases mediating adaptive responses can be temporally and spatially controlled to destroy key regulatory proteins during the cell cycle, sporulation, or biofilm disassembly and transition to motility. These proteases, as well as IMMPs that target membrane-associated substrates, make pathogens more deadly, so further studies of regulated proteolysis in model organisms will continue to positively impact efforts to improve human health. IMMPs and other types of membrane-embedded or secreted proteases cleave membrane-associated substrates to activate them or change their activity, as reviewed in the second section. In the examples described, themes of multilayer control, morphological coupling, and proteolytic cascades emerged more prominently, adding complexity to the temporal and spatial control of the regulated proteolytic events. In the last section, perhaps the most important theme is that much remains to be learned about the production of intercellular signals by regulated proteolysis. With a few notable exceptions, the proteases involved in signal production remain to be identified. The few that have been identified are only beginning to be understood in terms of their regulation. Studying regulated proteolytic events that occur at membranes or in the extracellular milieu poses many challenges. It is our hope that the themes, connections, and questions identified in this review will stimulate further progress on the most important challenges facing a better understanding of regulated proteolysis in bacterial development and related areas.
Acknowledgements
We thank Urs Jenal for helpful discussions. Research in the authors’ laboratories is supported by National Institutes of Health Grant GM43585 and National Science Foundation Grant MCB-0744343 (to L.K.), by Michigan State University AgBioResearch, and by research grants from the German Research Council within the frameworks of the Collaborative Research Center 987 "Microbial Diversity in Environmental Signal Response" and the graduate programme “Intra- and Inter-Cellular Transport and Communication”, and the LOEWE Research Center for Synthetic Microbiology, and intramural funds of the Max Planck Society (to L.S.-A.).
References
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA, Jenal U. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell. 2011;43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ades SE. Regulation by destruction: design of the σE envelope stress response. Current Opinion in Microbiology. 2008;11:535–540. doi: 10.1016/j.mib.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Ansaldi M, Marolt D, Stebe T, Mandic-Mulec I, Dubnau D. Specific activation of the Bacillus quorum-sensing systems by isoprenylated pheromone variants. Mol Microbiol. 2002;44:1561–1573. doi: 10.1046/j.1365-2958.2002.02977.x. [DOI] [PubMed] [Google Scholar]
- Arabolaza AL, Nakamura A, Pedrido ME, Martelotto L, Orsaria L, Grau RR. Characterization of a novel inhibitory feedback of the anti-anti-sigma SpoIIAA on Spo0A activation during development in Bacillus subtilis . Mol Microbiol. 2003;47:1251–1263. doi: 10.1046/j.1365-2958.2003.03376.x. [DOI] [PubMed] [Google Scholar]
- Aung S, Shum J, Abanes-De Mello A, Broder DH, Fredlund-Gutierrez J, Chiba S, Pogliano K. Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation. Mol Microbiol. 2007;65:1534–1546. doi: 10.1111/j.1365-2958.2007.05887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon Schneider K, Palmer TM, Grossman AD. Characterization of comQ and comX, two genes required for production of ComX pheromone in Bacillus subtilis . J Bacteriol. 2002;184:410–419. doi: 10.1128/JB.184.2.410-419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli . Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Bhat NH, Vass RH, Stoddard PR, Shin DK, Chien P. Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development. Mol Microbiol. 2013;88:1083–1092. doi: 10.1111/mmi.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb MJ, Buttner MJ. The Streptomyces coelicolor developmental transcription factor σBldN is synthesized as a proprotein. J Bacteriol. 2003;185:2338–2345. doi: 10.1128/JB.185.7.2338-2345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- Blaylock B, Jiang X, Rubio A, Moran CP, Jr, Pogliano K. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 2004;18:2916–2928. doi: 10.1101/gad.1252704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder DH, Pogliano K. Forespore engulfment mediated by a ratchet-like mechanism. Cell. 2006;126:917–928. doi: 10.1016/j.cell.2006.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis . Cell. 2001;104:269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Camp AH, Losick R. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol. 2008;69:402–417. doi: 10.1111/j.1365-2958.2008.06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis . Genes Dev. 2009;23:1014–1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo N, Rudner DZ. A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Mol Cell. 2006;23:25–35. doi: 10.1016/j.molcel.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Campo N, Rudner DZ. SpoIVB and CtpB are both forespore signals in the activation of the sporulation transcription factor sigmaK in Bacillus subtilis . J Bacteriol. 2007;189:6021–6027. doi: 10.1128/JB.00399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Kolter R, Losick R. Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability. Mol Microbiol. 2010;78:218–229. doi: 10.1111/j.1365-2958.2010.07335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chary VK, Xenopoulos P, Piggot PJ. Expression of the σF-directed csfB locus prevents premature appearance of σG activity during sporulation of Bacillus subtilis . J Bacteriol. 2007;189:8754–8757. doi: 10.1128/JB.01265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chary VK, Xenopoulos P, Eldar A, Piggot PJ. Loss of compartmentalization of σE activity need not prevent formation of spores by Bacillus subtilis . J Bacteriol. 2010;192:5616–5624. doi: 10.1128/JB.00572-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater KF, Biro S, Lee KJ, Palmer T, Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiol Rev. 2010;34:171–198. doi: 10.1111/j.1574-6976.2009.00206.x. [DOI] [PubMed] [Google Scholar]
- Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- Chen JC, Hottes AK, McAdams HH, McGrath PT, Viollier PH, Shapiro L. Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. EMBO J. 2006;25:377–386. doi: 10.1038/sj.emboj.7600935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Coleman K, Pogliano K. Impact of membrane fusion and proteolysis on SpoIIQ dynamics and interaction with SpoIIIAH. J Biol Chem. 2007;282:2576–2586. doi: 10.1074/jbc.M606056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, Perchuk BS, Laub MT, Sauer RT, Baker TA. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc Natl Acad Sci USA. 2007;104:6590–6595. doi: 10.1073/pnas.0701776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, Rink R, de Jong W, Siebring J, de Vreugd P, Boersma FG, Dijkhuizen L, Wosten HA. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 2003;17:1714–1726. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, Stokroos I, Deelstra HJ, Penninga NA, Bormann C, Salas JA, Dijkhuizen L, Wosten HA. The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol Microbiol. 2004;53:433–443. doi: 10.1111/j.1365-2958.2004.04143.x. [DOI] [PubMed] [Google Scholar]
- Clausen T, Kaiser M, Huber R, Ehrmann M. HTRA proteases: regulated proteolysis in protein quality control. Nat Rev Mol Cell Biol. 2011;12:152–162. doi: 10.1038/nrm3065. [DOI] [PubMed] [Google Scholar]
- Crymes WB, Jr, Zhang D, Ely B. Regulation of podJ expression during the Caulobacter crescentus cell cycle. J Bacteriol. 1999;181:3967–3973. doi: 10.1128/jb.181.13.3967-3973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis PD, Quardokus EM, Lawler ML, Guo X, Klein D, Chen JC, Arnold RJ, Brun YV. The scaffolding and signalling functions of a localization factor impact polar development. Mol Microbiol. 2012;84:712–735. doi: 10.1111/j.1365-2958.2012.08055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting S, Roels S, Losick R. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis . J Mol Biol. 1991;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-σK processing in Bacillus subtilis . Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother-cell gene expression during development in Bacillus subtilis . Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- D’Souza C, Nakano MM, Zuber P. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis . Proc Natl Acad Sci USA. 1994;91:9397–9401. doi: 10.1073/pnas.91.20.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez V, Schujman GE, Gueiros-Filho FJ, de Mendoza D. Vectorial signalling mechanism required for cell-cell communication during sporulation in Bacillus subtilis . Mol Microbiol. 2012;83:261–274. doi: 10.1111/j.1365-2958.2011.07929.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Rudner DZ. Perturbations to engulfment trigger a degradative response that prevents cell-cell signalling during sporulation in Bacillus subtilis . Mol Microbiol. 2007;64:500–511. doi: 10.1111/j.1365-2958.2007.05677.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, Jr, Rudner DZ. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis . PLoS Genet. 2009;5:e1000566. doi: 10.1371/journal.pgen.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Domian IJ, Reisenauer A, Shapiro L. Feedback control of a master bacterial cell-cycle regulator. Proc Natl Acad Sci U S A. 1999;96:6648–6653. doi: 10.1073/pnas.96.12.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong TC, Cutting SM. SpoIVB-mediated cleavage of SpoIVFA could provide the intercellular signal to activate processing of Pro-σK in Bacillus subtilis . Mol Microbiol. 2003;49:1425–1434. doi: 10.1046/j.1365-2958.2003.03651.x. [DOI] [PubMed] [Google Scholar]
- Dong TC, Cutting SM. The PDZ domain of the SpoIVB transmembrane signalling protein enables cis-trans interactions involving multiple partners leading to the activation of the pro-σK processing complex in Bacillus subtilis . J Biol Chem. 2004;279:43468–43478. doi: 10.1074/jbc.M407048200. [DOI] [PubMed] [Google Scholar]
- Downard J, Ramaswamy SV, Kil KS. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus xanthus development. J Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM, Johnson CM. Regulatory circuits controlling enterococcal conjugation: lessons for functional genomics. Curr Opin Microbiol. 2011;14:174–180. doi: 10.1016/j.mib.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong A, Capstick DS, Di Berardo C, Findlay KC, Hesketh A, Hong HJ, Elliot MA. Aerial development in Streptomyces coelicolor requires sortase activity. Mol Microbiol. 2012;83:992–1005. doi: 10.1111/j.1365-2958.2012.07983.x. [DOI] [PubMed] [Google Scholar]
- Dworkin J, Losick R. Developmental commitment in a bacterium. Cell. 2005;121:401–409. doi: 10.1016/j.cell.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Eichenberger P, Fawcett P, Losick R. A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis . Mol Microbiol. 2001;42:1147–1162. doi: 10.1046/j.1365-2958.2001.02660.x. [DOI] [PubMed] [Google Scholar]
- Eldar A, Chary VK, Xenopoulos P, Fontes ME, Loson OC, Dworkin J, Piggot PJ, Elowitz MB. Partial penetrance facilitates developmental evolution in bacteria. Nature. 2009;460:510–514. doi: 10.1038/nature08150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellehauge E, Norregaard-Madsen M, Sogaard-Andersen L. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol Microbiol. 1998;30:807–817. doi: 10.1046/j.1365-2958.1998.01113.x. [DOI] [PubMed] [Google Scholar]
- Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Elliot MA, Karoonuthaisiri N, Huang J, Bibb MJ, Cohen SN, Kao CM, Buttner MJ. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor . Genes Dev. 2003;17:1727–1740. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsholz AK, Michalik S, Zuhlke D, Hecker M, Gerth U. CtsR, the Gram-positive master regulator of protein quality control, feels the heat. EMBO J. 2010;29:3621–3629. doi: 10.1038/emboj.2010.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett P, Melnikov A, Youngman P. The Bacillus SpoIIGA protein is targeted to sites of spore septum formation in a SpoIIE-independent manner. Mol Microbiol. 1998;28:931–943. doi: 10.1046/j.1365-2958.1998.00849.x. [DOI] [PubMed] [Google Scholar]
- Flärdh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Micro. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- Fujita M, Losick R. An investigation into the compartmentalization of the sporulation transcription factor σE in Bacillus subtilis . Mol Microbiol. 2002;43:27–38. doi: 10.1046/j.1365-2958.2002.02732.x. [DOI] [PubMed] [Google Scholar]
- Fujita M, Losick R. The master regulator for entry into sporulation in Bacillus subtilis becomes a cell-specific transcription factor after asymmetric division. Genes Dev. 2003;17:1166–1174. doi: 10.1101/gad.1078303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskell AA, Giovinazzo JA, Fonte V, Willey JM. Multi-tier regulation of the streptomycete morphogenetic peptide SapB. Mol Microbiol. 2012;84:501–515. doi: 10.1111/j.1365-2958.2012.08041.x. [DOI] [PubMed] [Google Scholar]
- Gill RE, Karlok M, Benton D. Myxococcus xanthus encodes an ATP-dependent protease which is required for developmental gene transcription and intercellular signaling. J Bacteriol. 1993;175:4538–4544. doi: 10.1128/jb.175.14.4538-4544.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez M, Cutting SM. Expression of the Bacillus subtilis spoIVBgene is under dual σF/σG control. Microbiol. 1996;142:3453–3457. doi: 10.1099/13500872-142-12-3453. [DOI] [PubMed] [Google Scholar]
- Gomez M, Cutting SM. bofCencodes a putative forespore regulator of the Bacillus subtilis σK checkpoint. Microbiol. 1997;143:157–170. doi: 10.1099/00221287-143-1-157. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Pastor JE. Cannibalism: a social behavior in sporulating Bacillus subtilis . FEMS Microbiol Rev. 2011;35:415–424. doi: 10.1111/j.1574-6976.2010.00253.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk B, Marczynski GT. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus . Mol Microbiol. 2005;55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Grünenfelder B, Tawfilis S, Gehrig S, M OS, Eglin D, Jenal U. Identification of the protease and the turnover signal responsible for cell cycle-dependent degradation of the Caulobacter FliF motor protein. J Bacteriol. 2004;186:4960–4971. doi: 10.1128/JB.186.15.4960-4971.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur E, Biran D, Ron EZ. Regulated proteolysis in Gram-negative bacteria — how and when? Nat Rev Micro. 2011;9:839–848. doi: 10.1038/nrmicro2669. [DOI] [PubMed] [Google Scholar]
- Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev. 2013 doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen DC, Bretscher AP, Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus . Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Halberg R, Kroos L. Sporulation regulatory protein SpoIIID from Bacillus subtilisactivates and represses transcription by both mother-cell-specific forms of RNA polymerase. J Mol Biol. 1994;243:425–436. doi: 10.1006/jmbi.1994.1670. [DOI] [PubMed] [Google Scholar]
- Hardy GG, Allen RC, Toh E, Long M, Brown PJ, Cole-Tobian JL, Brun YV. A localized multimeric anchor attaches the Caulobacter holdfast to the cell pole. Mol Microbiol. 2010;76:409–427. doi: 10.1111/j.1365-2958.2010.07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa KC, Rajagopalan R, Risser DD, Rivers OS, Tom SK, Videau P, Callahan SM. The RGSGR amino acid motif of the intercellular signalling protein, HetN, is required for patterning of heterocysts in Anabaena sp. strain PCC 7120. Mol Microbiol. 2012;83:682–693. doi: 10.1111/j.1365-2958.2011.07949.x. [DOI] [PubMed] [Google Scholar]
- Higo A, Horinouchi S, Ohnishi Y. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus . Mol Microbiol. 2011;81:1607–1622. doi: 10.1111/j.1365-2958.2011.07795.x. [DOI] [PubMed] [Google Scholar]
- Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- Hoa NT, Brannigan JA, Cutting SM. The Bacillus subtilis signaling protein SpoIVB defines a new family of serine peptidases. J Bacteriol. 2002;184:191–199. doi: 10.1128/JB.184.1.191-199.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeister A. Activation of the proprotein transcription factor Pro-σE is associated with three changes in its subcellular localization during sporulation in Bacillus subtilis . J Bacteriol. 1998;180:2426–2433. doi: 10.1128/jb.180.9.2426-2433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeister AEM, Londono-Vallejo A, Harry E, Stragier P, Losick R. Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis . Cell. 1995;83:219–226. doi: 10.1016/0092-8674(95)90163-9. [DOI] [PubMed] [Google Scholar]
- Hsueh YH, Cozy LM, Sham LT, Calvo RA, Gutu AD, Winkler ME, Kearns DB. DegU-phosphate activates expression of the anti-sigma factor FlgM in Bacillus subtilis . Mol Microbiol. 2011;81:1092–1108. doi: 10.1111/j.1365-2958.2011.07755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura D, Zhou R, Feig M, Kroos L. Evidence that the Bacillus subtilisSpoIIGA protein is a novel type of signal-transducing aspartic protease. J Biol Chem. 2008;283:15287–15299. doi: 10.1074/jbc.M708962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura D, Kuwana R, Kroos L, Feig M, Takamatsu H, Watabe K. Substrate Specificity of SpoIIGA, a Signal-transducing Aspartic Protease in Bacilli . J Biochem. 2011;149:665–671. doi: 10.1093/jb/mvr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmer H, Brondsted L. Proteases in bacterial pathogenesis. Res Microbiol. 2009;160:704–710. doi: 10.1016/j.resmic.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Iniesta AA, Shapiro L. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc Natl Acad Sci USA. 2008;105:16602–16607. doi: 10.1073/pnas.0808807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U. The role of proteolysis in the Caulobacter crescentus cell cycle and development. Res Microbiol. 2009;160:687–695. doi: 10.1016/j.resmic.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Jenal U, Shapiro L. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 1996;15:2393–2406. [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Hengge-Aronis R. Regulation by proteolysis in bacterial cells. Curr Opin Microbiol. 2003;6:163–172. doi: 10.1016/s1369-5274(03)00029-8. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rubio A, Chiba S, Pogliano K. Engulfment-regulated proteolysis of SpoIIQ: evidence that dual checkpoints control sigma activity. Mol Microbiol. 2005;58:102–115. doi: 10.1111/j.1365-2958.2005.04811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas RM, Weaver EA, Kenney TJ, Moran CP, Jr, Haldenwang WG. The Bacillus subtilis spoIIG operon encodes both σE and a gene necessary for σE activation. J Bacteriol. 1988;170:507–511. doi: 10.1128/jb.170.2.507-511.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CH, Moran CP. Mutant σ factor blocks transition between promoter binding and initiation of transcription. Proc Natl Acad Sci USA. 1992;89:1958–1962. doi: 10.1073/pnas.89.5.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju J, Luo T, Haldenwang W. Bacillus subtilispro-σE fusion protein localizes to the forespore septum and fails to be processed when synthesized in the forespore. J Bacteriol. 1997;179:4888–4893. doi: 10.1128/jb.179.15.4888-4893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju J, Luo T, Haldenwang W. Forespore expression and processing of the SigE transcription factor in wild-type and mutant Bacillus subtilis . J Bacteriol. 1998;180:1673–1681. doi: 10.1128/jb.180.7.1673-1681.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kain J, He GG, Losick R. Polar localization and compartmentalization of ClpP proteases during growth and sporulation in Bacillus subtilis. J Bacteriol. 2008;190:6749–6757. doi: 10.1128/JB.00589-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D. Signaling in Myxobacteria. Annu Rev Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- Kaplan H. Multicellular development and gliding motility in Myxococcus xanthus . Curr Opin Microbiol. 2003;6:572–577. doi: 10.1016/j.mib.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Karmazyn-Campelli C, Rhayat L, Carballido-Lopez R, Duperrier S, Frandsen N, Stragier P. How the early sporulation sigma factor sigmaF delays the switch to late development in Bacillus subtilis. Mol Microbiol. 2008;67:1169–1180. doi: 10.1111/j.1365-2958.2008.06121.x. [DOI] [PubMed] [Google Scholar]
- Karow ML, Glaser P, Piggot PJ. Identification of a gene, spoIIR, that links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis . Proc Natl Acad Sci USA. 1995;92:2012–2016. doi: 10.1073/pnas.92.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Hesketh A, Kim ES, Song JY, Lee DH, Kim IS, Chater KF, Lee KJ. Complex extracellular interactions of proteases and a protease inhibitor influence multicellular development of Streptomyces coelicolor . Mol Microbiol. 2008;70:1180–1193. doi: 10.1111/j.1365-2958.2008.06471.x. [DOI] [PubMed] [Google Scholar]
- Kim SK, Kaiser D. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus . Cell. 1990;61:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- Kim SK, Kaiser D. Purification and properties of Myxococcus xanthus C-factor, an intercellular signaling protein. Proc Natl Acad Sci USA. 1990;87:3635–3639. doi: 10.1073/pnas.87.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Kaiser D. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates fruiting body morphogenesis of Myxococcus xanthus . Genes Dev. 1990;4:896–904. doi: 10.1101/gad.4.6.896. [DOI] [PubMed] [Google Scholar]
- Kim SK, Kaiser D. Cell alignment required in differentiation of Myxococcus xanthus . Science. 1990;249:926–928. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- Kim SK, Kaiser D. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J Bacteriol. 1991;173:1722–1728. doi: 10.1128/jb.173.5.1722-1728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick CL, Viollier PH. Decoding Caulobacter development. FEMS Microbiol Rev. 2012;36:193–205. doi: 10.1111/j.1574-6976.2011.00309.x. [DOI] [PubMed] [Google Scholar]
- Kirstein J, Moliere N, Dougan DA, Turgay K. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol. 2009;7:589–599. doi: 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- Kirstein J, Strahl H, Molière N, Hamoen LW, Turgay K. Localization of general and regulatory proteolysis in Bacillus subtilis cells. Mol Microbiol. 2008;70:682–694. doi: 10.1111/j.1365-2958.2008.06438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J, Hoffmann A, Lilie H, Schmidt R, Rübsamen-Waigmann H, Brötz-Oesterhelt H, Mogk A, Turgay K. The antibiotic ADEP reprogrammes ClpP, switching it from a regulated to an uncontrolled protease. EMBO Mol Med. 2009;1:37–49. doi: 10.1002/emmm.200900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova A, Petters T, Søgaard-Andersen L. Extracellular biology of Myxococcus xanthus . FEMS Microbiol Rev. 2010;34:89–106. doi: 10.1111/j.1574-6976.2009.00194.x. [DOI] [PubMed] [Google Scholar]
- Konovalova A, Lobach S, Søgaard-Andersen L. A RelA-dependent two-tiered regulated proteolysis cascade controls synthesis of a contact-dependent intercellular signal in Myxococcus xanthus . Mol Microbiol. 2012;84:260–275. doi: 10.1111/j.1365-2958.2012.08020.x. [DOI] [PubMed] [Google Scholar]
- Konovalova A, Wegener-Feldbrugge S, Søgaard-Andersen L. Two intercellular signals required for fruiting body formation in Myxococcus xanthus act sequentially but non-hierarchically. Mol Microbiol. 2012;86:65–81. doi: 10.1111/j.1365-2958.2012.08173.x. [DOI] [PubMed] [Google Scholar]
- Kroos L. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu Rev Genet. 2007;41:13–39. doi: 10.1146/annurev.genet.41.110306.130400. [DOI] [PubMed] [Google Scholar]
- Kroos L, Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- Kroos L, Akiyama Y. Biochemical and structural insights into intramembrane metalloprotease mechanisms. BBA Biomemb. 2013 doi: 10.1016/j.bbamem.2013.03.032. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L, Kunkel B, Losick R. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science. 1989;243:526–529. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- Kroos L, Hartzell P, Stephens K, Kaiser D. A link between cell movement and gene expression argues that motility is required for cell-cell signaling during fruiting body development. Genes Dev. 1988;2:1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- Kruse T, Lobedanz S, Berthelsen NM, Søgaard-Andersen L. C-signal: a cell surface-associated morphogen that induces and co-ordinates multicellular fruiting body morphogenesis and sporulation in Myxococcus xanthus . Mol Microbiol. 2001;40:156–168. doi: 10.1046/j.1365-2958.2001.02365.x. [DOI] [PubMed] [Google Scholar]
- Kumar K, Mella-Herrera RA, Golden JW. Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol. 2010;2:a000315. doi: 10.1101/cshperspect.a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel B, Losick R, Stragier P. The Bacillus subtilis gene for the developmental transcription factor σK is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev. 1990;4:525–535. doi: 10.1101/gad.4.4.525. [DOI] [PubMed] [Google Scholar]
- Kunkel B, Sandman K, Panzer S, Youngman P, Losick R. The promoter for a sporulation gene in the spoIVC locus of Bacillus subtilis and its use in studies of temporal and spatial control of gene expression. J Bacteriol. 1988;170:3513–3522. doi: 10.1128/jb.170.8.3513-3522.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel B, Kroos L, Poth H, Youngman P, Losick R. Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis . Genes Dev. 1989;3:1735–1744. doi: 10.1101/gad.3.11.1735. [DOI] [PubMed] [Google Scholar]
- Kuspa A, Kaiser D. Genes required for developmental signalling in Myxococcus xanthus: three asg loci. J Bacteriol. 1989;171:2762–2772. doi: 10.1128/jb.171.5.2762-2772.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A, Kroos L, Kaiser D. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus . Dev Biol. 1986;117:267–276. doi: 10.1016/0012-1606(86)90369-6. [DOI] [PubMed] [Google Scholar]
- Kuspa A, Plamann L, Kaiser D. A-signalling and the cell density requirement for Myxococcus xanthus development. J Bacteriol. 1992;174:7360–7369. doi: 10.1128/jb.174.22.7360-7369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A, Plamann L, Kaiser D. Identification of heat-stable A-factor from Myxococcus xanthus . J Bacteriol. 1992;174:3319–3326. doi: 10.1128/jb.174.10.3319-3326.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBell TL, Trempy JE, Haldenwang WG. Sporulation-specific sigma factor, σ29, of Bacillus subtilis is synthesized from a precursor protein, P31 . Proc Nat Acad Sci USA. 1987;84:1784–1788. doi: 10.1073/pnas.84.7.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamsa A, Liu WT, Dorrestein PC, Pogliano K. The Bacillus subtilis cannibalism toxin SDP collapses the proton motive force and induces autolysis. Mol Microbiol. 2012;84:486–500. doi: 10.1111/j.1365-2958.2012.08038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanigan-Gerdes S, Dooley AN, Faull KF, Lazazzera BA. Identification of subtilisin, Epr and Vpr as enzymes that produce CSF, an extracellular signalling peptide of Bacillus subtilis . Mol Microbiol. 2007;65:1321–1333. doi: 10.1111/j.1365-2958.2007.05869.x. [DOI] [PubMed] [Google Scholar]
- Laub MT, Shapiro L, McAdams HH. Systems biology of Caulobacter . Ann Rev Genet. 2007;41:429–441. doi: 10.1146/annurev.genet.41.110306.130346. [DOI] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- Lawler ML, Larson DE, Hinz AJ, Klein D, Brun YV. Dissection of functional domains of the polar localization factor PodJ in Caulobacter crescentus . Mol Microbiol. 2006;59:301–316. doi: 10.1111/j.1365-2958.2005.04935.x. [DOI] [PubMed] [Google Scholar]
- Lee B-G, Park EY, Lee K-E, Jeon H, Sung KH, Paulsen H, Rubsamen-Schaeff H, Brotz-Oesterhelt H, Song HK. Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat Struct Mol Biol. 2010;17:471–478. doi: 10.1038/nsmb.1787. [DOI] [PubMed] [Google Scholar]
- Lee B-U, Lee K, Mendez J, Shimkets LJ. A tactile sensory system of Myxococcus xanthus involves an extracellular NAD(P)+-containing protein. Genes Dev. 1995;9:2964–2973. doi: 10.1101/gad.9.23.2964. [DOI] [PubMed] [Google Scholar]
- Li S, Lee B-U, Shimkets LJ. csgAexpression entrainsMyxococcus xanthus development. Genes Dev. 1992;6:401–410. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- Liu WT, Yang YL, Xu Y, et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis . Proc Natl Acad Sci USA. 2010;107:16286–16290. doi: 10.1073/pnas.1008368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobedanz S, Søgaard-Andersen L. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus . Genes Dev. 2003;17:2151–2161. doi: 10.1101/gad.274203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Stragier P. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis . Genes Dev. 1995;9:503–508. doi: 10.1101/gad.9.4.503. [DOI] [PubMed] [Google Scholar]
- López D, Kolter R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis . FEMS Microbiol Rev. 2010;34:134–149. doi: 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis . Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- Lu S, Halberg R, Kroos L. Processing of the mother-cell σfactor, σK, may depend on events occurring in the forespore during Bacillus subtilis development. Proc Natl Acad Sci USA. 1990;87:9722–9726. doi: 10.1073/pnas.87.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cutting S, Kroos L. Sporulation protein SpoIVFB from Bacillus subtilis enhances processing of the sigma factor precursor pro-σK in the absence of other sporulation gene products. J Bacteriol. 1995;177:1082–1085. doi: 10.1128/jb.177.4.1082-1085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis . Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- Manoil C, Kaiser D. Accumulation of guanosine tetraphosphate and guanosine pentaphosphate in Myxococcus xanthus during starvation and myxospore formation. J Bacteriol. 1980;141:297–304. doi: 10.1128/jb.141.1.297-304.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JR, Flardh K. Signals and regulators that govern Streptomyces development. FEMS Microbiol Rev. 2012;36:206–231. doi: 10.1111/j.1574-6976.2011.00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Meisner J, Wang X, Serrano M, Henriques AO, Moran CP., Jr A channel connecting the mother cell and forespore during bacterial endospore formation. Proc Natl Acad Sci USA. 2008;105:15100–15105. doi: 10.1073/pnas.0806301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Hengge R. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of σS (RpoS) in E. coli . Genes Dev. 2005;19:2770–2781. doi: 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal S, Kroos L. A combination of unusual transcription factors binds cooperatively to control Myxococcus xanthus developmental gene expression. Proc Natl Acad Sci U S A. 2009;106:1965–1970. doi: 10.1073/pnas.0808516106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot C, Uehara T, Marquis KA, Bernhardt TG, Rudner DZ. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis . Genes Dev. 2010;24:411–422. doi: 10.1101/gad.1878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus F, Obrist M, Führer F, Langklotz S. Degradation of cytoplasmic substrates by FtsH, a membrane-anchored protease with many talents. Res Microbiol. 2009;160:652–659. doi: 10.1016/j.resmic.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Nodwell JR, Losick R. Purification of an extracellular signaling molecule involved in production of aerial mycelium by Streptomyces coelicolor . J Bacteriol. 1998;180:1334–1337. doi: 10.1128/jb.180.5.1334-1337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Fujitani S, Mao X, Inouye S, Komano T. FruA, a putative transcription factor essential for the development of Myxococcus xanthus . Mol Microbiol. 1996;22:757–767. doi: 10.1046/j.1365-2958.1996.d01-1725.x. [DOI] [PubMed] [Google Scholar]
- Ogura M, Tsukahara K. Autoregulation of the Bacillus subtilis response regulator gene degU is coupled with the proteolysis of DegU-P by ClpCP. Mol Microbiol. 2010;75:1244–1259. doi: 10.1111/j.1365-2958.2010.07047.x. [DOI] [PubMed] [Google Scholar]
- Pan Q, Garsin DA, Losick R. Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-sigma factor in B. subtilis . Mol Cell. 2001;8:873–883. doi: 10.1016/s1097-2765(01)00362-8. [DOI] [PubMed] [Google Scholar]
- Pan Q, Losick R, Rudner DZ. A second PDZ-containing serine protease contributes to activation of the sporulation transcription factor σK in Bacillus subtilis . J Bacteriol. 2003;185:6051–6056. doi: 10.1128/JB.185.20.6051-6056.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Jaeger T, Abel S, Wiederkehr I, Folcher M, Biondi EG, Laub MT, Jenal U. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell. 2008;133:452–461. doi: 10.1016/j.cell.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M. Forty years in the making: understanding the molecular mechanism of peptide regulation in bacterial development. PLoS Biol. 2013;11:e1001516. doi: 10.1371/journal.pbio.1001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M, Brannigan JA. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides. 2001;22:1541–1547. doi: 10.1016/s0196-9781(01)00490-9. [DOI] [PubMed] [Google Scholar]
- Perez Morales TG, Ho TD, Liu WT, Dorrestein PC, Ellermeier CD. Production of the cannibalism toxin SDP is a multi-step process that requires SdpA and SdpB. J Bacteriol. 2013;195:3244–3251. doi: 10.1128/JB.00407-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters HK, Haldenwang WG. Synthesis and fractionation properties of SpoIIGA, a protein essential for Pro-σE processing in Bacillus subtilis . J Bacteriol. 1991;173:7821–7827. doi: 10.1128/jb.173.24.7821-7827.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CN, Ruiz N, Silhavy TJ. RpoS proteolysis is regulated by a mechanism that does not require the SprE (RssB) response regulator phosphorylation site. J Bacteriol. 2004;186:7403–7410. doi: 10.1128/JB.186.21.7403-7410.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann L, Kuspa A, Kaiser D. Proteins that rescue A-signal-defective mutants of Myxococcus xanthus . J Bacteriol. 1992;174:3311–3318. doi: 10.1128/jb.174.10.3311-3318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocka I, Thein M, M OS, Jenal U, Alley MR. Degradation of a Caulobacter soluble cytoplasmic chemoreceptor is ClpX dependent. J Bacteriol. 2002;184:6635–6641. doi: 10.1128/JB.184.23.6635-6641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottathil M, Lazazzera BA. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis . Front Biosci. 2003;8:d32–d45. doi: 10.2741/913. [DOI] [PubMed] [Google Scholar]
- Qiu R, Pei W, Zhang L, Lin J, Ji G. Identification of the putative staphylococcal AgrB catalytic residues involving the proteolytic cleavage of AgrD to generate autoinducing peptide. J Biol Chem. 2005;280:16695–16704. doi: 10.1074/jbc.M411372200. [DOI] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci U S A. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnekov O, Losick R. Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis . Proc Natl Acad Sci USA. 1998;95:3162–3167. doi: 10.1073/pnas.95.6.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnekov O, Alper S, Losick R. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis . Genes Cells. 1996;1:529–542. doi: 10.1046/j.1365-2443.1996.d01-262.x. [DOI] [PubMed] [Google Scholar]
- Ricca E, Cutting S, Losick R. Characterization of bofA, a gene involved in intercompartmental regulation of pro-σK processing during sporulation in Bacillus subtilis . J Bacteriol. 1992;174:3177–3184. doi: 10.1128/jb.174.10.3177-3184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolbetzki A, Ammon M, Jakovljevic V, Konovalova A, Søgaard-Andersen L. Regulated secretion of a protease activates intercellular signaling during fruiting body formation in M. xanthus . Dev cell. 2008;15:627–634. doi: 10.1016/j.devcel.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Rood KL, Clark NE, Stoddard PR, Garman SC, Chien P. Adaptor-dependent degradation of a cell-cycle regulator uses a unique substrate architecture. Structure. 2012;20:1223–1232. doi: 10.1016/j.str.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner D, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Losick R. A sporulation membrane protein tethers the pro-σK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 2002;16:1007–1018. doi: 10.1101/gad.977702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Pan Q, Losick RM. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci U S A. 2002;99:8701–8706. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo MV, Mach KE, Burkholder WF. Proteolysis of the replication checkpoint protein Sda is necessary for the efficient initiation of sporulation after transient replication stress in Bacillus subtilis . Mol Microbiol. 2006;60:1490–1508. doi: 10.1111/j.1365-2958.2006.05167.x. [DOI] [PubMed] [Google Scholar]
- Ryan KR, Huntwork S, Shapiro L. Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc Natl Acad Sci USA. 2004;101:7415–7420. doi: 10.1073/pnas.0402153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Samori Y, Kobayashi Y. The cisA cistron of Bacillus subtilis sporulation gene spoIVC encodes a protein homologous to a site-specific recombinase. J Bacteriol. 1990;172:1092–1098. doi: 10.1128/jb.172.2.1092-1098.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Harada K, Ohta Y, Kobayashi Y. Expression of the Bacillus subtilis spoIVCA gene, which encodes a site-specific recombinase, depends on the spoIIGB product. J Bacteriol. 1994;176:935–937. doi: 10.1128/jb.176.3.935-937.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Bukau B, Mogk A. Principles of general and regulatory proteolysis by AAA+ proteases in Escherichia coli. Res Microbiol. 2009;160:629–636. doi: 10.1016/j.resmic.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Schujman GE, Grau R, Gramajo HC, Ornella L, de Mendoza D. De novo fatty acid synthesis is required for establishment of cell type-specific gene transcription during sporulation in Bacillus subtilis . Mol Microbiol. 1998;29:1215–1224. doi: 10.1046/j.1365-2958.1998.01004.x. [DOI] [PubMed] [Google Scholar]
- Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets LJ, Rafiee H. CsgA, an extracellular protein essential for Myxococcus xanthus development. J Bacteriol. 1990;172:5299–5306. doi: 10.1128/jb.172.9.5299-5306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets LJ, Gill RE, Kaiser D. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc Natl Acad Sci USA. 1983;80:1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Grossman AD, Walker GC. Clp and Lon proteases occupy distinct subcellular positions in Bacillus subtilis . J Bacteriol. 2008;190:6758–6768. doi: 10.1128/JB.00590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson S, Mueller C, Jiang M, Perego M. Molecular analysis of Phr peptide processing in Bacillus subtilis . J Bacteriol. 2003;185:4861–4871. doi: 10.1128/JB.185.16.4861-4871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CM, Errington J. Differential gene expression during sporulation in Bacillus subtilis: structure and regulation of the spoIIID gene. Mol Microbiol. 1990;4:543–552. doi: 10.1111/j.1365-2958.1990.tb00622.x. [DOI] [PubMed] [Google Scholar]
- Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: How morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- Stragier P, Kunkel B, Kroos L, Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989;243:507–512. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- Tatti KM, Jones CH, Moran CP. Genetic evidence for interaction of σE with the spoIIID promoter in Bacillus subtilis . J Bacteriol. 1991;173:7828–7833. doi: 10.1128/jb.173.24.7828-7833.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JA, Wilbur JD, Smith SC, Ryan KR. Mutations that alter RcdA surface residues decouple protein localization and CtrA proteolysis in Caulobacter crescentus . J Mol Biol. 2009;394:46–60. doi: 10.1016/j.jmb.2009.08.076. [DOI] [PubMed] [Google Scholar]
- Thoendel M, Horswill AR. Biosynthesis of peptide signals in gram-positive bacteria. Adv Appl Microbiol. 2010;71:91–112. doi: 10.1016/S0065-2164(10)71004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojo N, Inouye S, Komano T. The lonD gene is homologous to the lon gene encoding an ATP-dependent protease and is essential for the development of Myxococcus xanthus . J Bacteriol. 1993;175:4545–4549. doi: 10.1128/jb.175.14.4545-4549.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortosa P, Logsdon L, Kraigher B, Itoh Y, Mandic-Mulec I, Dubnau D. Specificity and genetic polymorphism of the Bacillus competence quorum-sensing system. J Bacteriol. 2001;183:451–460. doi: 10.1128/JB.183.2.451-460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Alley MR. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J Bacteriol. 2001;183:5001–5007. doi: 10.1128/JB.183.17.5001-5007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokos CG, Laub MT. Polarity and cell fate asymmetry in Caulobacter crescentus. Curr Opin Microbiol. 2012;15:744–750. doi: 10.1016/j.mib.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S. Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nat Rev Microbiol. 2009;7:411–423. doi: 10.1038/nrmicro2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wart HE, Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc Natl Acad Sci USA. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Murray H, Errington J. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis . Genes Dev. 2009;23:1959–1970. doi: 10.1101/gad.528209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J. 2002;21:4420–4428. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci USA. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan P, Singer M, Kroos L. Role of σD in regulating genes and signals during Myxococcus xanthus development. J Bacteriol. 2006;188:3246–3256. doi: 10.1128/JB.188.9.3246-3256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 2013;11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley P, Hoa NT, Cutting S. BofC negatively regulates SpoIVB-mediated signalling in the Bacillus subtilis σK-checkpoint. Mol Microbiol. 2000;36:1415–1424. doi: 10.1046/j.1365-2958.2000.01962.x. [DOI] [PubMed] [Google Scholar]
- Wakeley PR, Dorazi R, Hoa NT, Bowyer JR, Cutting SM. Proteolysis of SpoIVB is a critical determinant in signalling of Pro-σK processing in Bacillus subtilis . Mol Microbiol. 2000;36:1336–1348. doi: 10.1046/j.1365-2958.2000.01946.x. [DOI] [PubMed] [Google Scholar]
- Willey J, Schwedock J, Losick R. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor . Genes Dev. 1993;7:895–903. doi: 10.1101/gad.7.5.895. [DOI] [PubMed] [Google Scholar]
- Willey J, Santamaria R, Guijarro J, Geistlich M, Losick R. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor . Cell. 1991;65:641–650. doi: 10.1016/0092-8674(91)90096-h. [DOI] [PubMed] [Google Scholar]
- Willey JM, Gaskell AA. Morphogenetic signaling molecules of the streptomycetes. Chem Rev. 2011;111:174–187. doi: 10.1021/cr1000404. [DOI] [PubMed] [Google Scholar]
- Wright R, Stephens C, Zweiger G, Shapiro L, Alley MR. CaulobacterLon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 1996;15:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- Yu Y-TN, Kroos L. Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. J Bacteriol. 2000;182:3305–3309. doi: 10.1128/jb.182.11.3305-3309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T, Nakahigashi K. Regulation of the heat-shock response. Curr Opin Microbiol. 1999;2:153–158. doi: 10.1016/S1369-5274(99)80027-7. [DOI] [PubMed] [Google Scholar]
- Zhang B, Hofmeister A, Kroos L. The pro-sequence of pro-σK promotes membrane association and inhibits RNA polymerase core binding. J Bacteriol. 1998;180:2434–2441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Kroos L. BofA protein inhibits intramembrane proteolysis of pro-σK in an intercompartmental signaling pathway during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 2004;101:6385–6390. doi: 10.1073/pnas.0307709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Kroos L. Serine proteases from two cell types target different components of a complex that governs regulated intramembrane proteolysis of pro-σK during Bacillus subtilis development. Mol Microbiol. 2005;58:835–846. doi: 10.1111/j.1365-2958.2005.04870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Cusumano C, Sui D, Garavito RM, Kroos L. Intramembrane proteolytic cleavage of a membrane-tethered transcription factor by a metalloprotease depends on ATP. Proc Natl Acad Sci USA. 2009;106:16174–16179. doi: 10.1073/pnas.0901455106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Chen K, Xiang X, Gu L, Kroos L. Features of Pro-σK Important for cleavage by SpoIVFB, an intramembrane metalloprotease. J Bacteriol. 2013;195:2793–2806. doi: 10.1128/JB.00229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]