Abstract
Aim:
To establish and characterize primary lung cancer cell lines from Chinese population.
Methods:
Lung cancer specimens or pleural effusions were collected from Chinese lung cancer patients and cultured in vitro with ACL4 medium (for non-small cell lung carcinomas (NSCLC)) or HITES medium (for small cell lung carcinomas (SCLC)) supplemented with 5% FBS. All cell lines were maintained in culture for more than 25 passages. Most of these cell lines were further analyzed for oncogenic mutations, karyotype, cell growth kinetics, and tumorigenicity in nude mice.
Results:
Eight primary cell lines from Chinese lung cancer patients were established and characterized, including seven NSCLC cell lines and one SCLC cell line. Five NSCLC cell lines were found to harbor epidermal growth factor receptor (EGFR) kinase domain mutations.
Conclusion:
These well-characterized primary lung cancer cell lines from Chinese population provide a unique platform for future studies of the ethnic differences in lung cancer biology and drug response.
Keywords: lung cancer, primary cell lines, epidermal growth factor receptor, small cell lung carcinomas, non-small cell lung carcinomas
Introduction
Lung cancer is one of the most common causes of cancer-related death worldwide1, 2, with a five year survival rate of approximately 15%. In China, the situation is even worse. The vast majority of lung cancers are carcinomas derived from epithelial cells and are pathologically divided into two major types: non-small cell lung carcinomas (NSCLC) (∼80%) and small cell lung carcinomas (SCLC) (∼20%).
Lung cancer cell lines are crucial for translational and biomedical research. Lung cancer cells were successfully cultured approximately 25 years after the establishment of a human cervical carcinoma (HeLa) cell line3, 4. Since then, many attempts have been made to establish permanent lung cancer cell lines5, 6, 7, 8, 9, 10. The emergence of lung cancer cell lines reached a peak after serum-free chemically defined media (for example, ACL4, C-based medium, and HITES) were introduced11, 12. Since then, more than 200 lung cancer cell lines have been successfully established. These permanent cell lines, derived from either primary or metastatic cancers, provide important experimental systems for studying the genetic changes and biology associated with tumor initiation and progression. As a pure cancer cell population, the global using of in vitro permanent cell lines over the past several decades has led to great achievements, including elucidating the molecular and translational biology of cancer and further drug screening3, 13. To date, more than 9000 citations, including several important biomedical discoveries, have resulted from the usage of these lines3. However, it is noteworthy that among all of the widely used cell lines, almost none have a Chinese genetic background.
Studies demonstrate that ethnic differences in genetic background are important in defining cancer biology as well as in drug toxicity14, 15, 16, 17, 18, 19, 20, 21. For example, the epidermal growth factor receptor (EGFR) kinase domain mutations occur in approximately 10% of NSCLC patients in the Caucasian population22, but occur in 30%–50% of NSCLC patients from East Asian populations15, 17, 18, 21, 23. Ethnic differences in the expression of allelic variants may produce altered pharmacokinetics and result in differential toxicity for the same anticancer treatments19, 20. Understanding the causes of ethnic differences in cytotoxic metabolism may help to improve cancer treatment in the clinic. From this standpoint, the establishment of lung cancer cell lines from Chinese genetic background is urgently needed.
We have worked to establish Chinese lung cancer cell lines. A total of eight primary Chinese lung cancer cell lines were successfully established, including seven NSCLC cell lines and one SCLC cell line. We characterized most of these cell lines for oncogenic mutations, growth kinetics, karyotype, and tumorigenicity in nude mice. These cell lines provide a very useful platform for studying the ethnic differences in cancer biology and drug response in the future.
Materials and methods
Collection of clinical specimens
All patients were from the Chinese population and underwent surgery for potentially curative resections. The lung cancer specimens or pleural effusions were collected with patient consent. Solid tumors were immediately immersed in ice-cold RPMI-1640 supplemented with P/S (1000 U/mL penicillin G and 1000 mg/L streptomycin), while pleural effusions were kept on ice. Samples were transported to the laboratory for primary cell culturing within one hour of collection.
Preparation of lung cancer cells from resected samples
Solid tumor specimens were rinsed twice with PBS supplemented with P/S and finely minced with scissors. Both necrotic tissue and apparently normal tissue were discarded. Tumor fragments were then immersed into ACL4 medium (for NSCLC)24, 25 or HITES medium (for SCLC)11, 12 supplemented with 5% fetal bovine serum (FBS) and P/S, and they were pipetted more than 50 times. The cell suspension was then transferred to a collagen-coated flask. The pleural effusions were subjected to red blood cell lysis and then to Ficoll gradient separation to remove lymphocytes. The interface was collected and washed three times with PBS plus P/S. Cells were suspended in ACL4 medium with 5% FBS and P/S and then cultured in a collagen-coated flask. The medium was changed every 3 d. A cell scraper was used to remove visible fibroblast growth whenever it occurred. Once the cells were confluent, they were digested with 0.05% EDTA-trypsin for passage. All established cell lines were maintained in culture for more than 25 passages.
Karyotype analysis
Cells were seeded and cultured for 70 h or until confluence. Colchicine was added to cells with a final concentration of 0.2 μg/mL. Cells were incubated for another 2 h and then digested using 0.05% EDTA-trypsin and resuspended in 0.075 mol/L KCl. After fixation in 3:1 methanol/glacial acetic acid, cell suspensions were dropped onto ice-cold slides, which were then stained by Giemsa for 8 min. Chromosome ploidy was estimated from at least 29 metaphases per cell line under a microscope.
EGFR and KRAS mutation detection
Detection of EGFR and KRAS mutations was previously described18, 21. Briefly, genomic DNA and RNA were extracted from Chinese lung cancer cell lines per standard protocol (RNeasy Mini Kit, and QIAamp DNA Mini Kit, Qiagen, Hilden, Germany). Total RNA was reverse transcribed into single-stranded cDNA using a RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, EU, USA) and sequenced. The cDNA regions of EGFR (exons 18–22) and KRAS (exons 2–3) were then PCR amplified and sequenced. All mutations found from the cDNA sequencing were further confirmed by genomic DNA sequencing.
Fluorescence in situ hybridization (FISH)
Cells were applied onto slides as described in Karyotype analyses. Slides were then incubated in 0.01% pepsin solution (pH 2.0) for 10 min at 37 °C and then fixed in 1% formaldehyde for another 10 min at room temperature. Vysis EGFR probe (Abbott Laboratories, Des Plaines, IL, USA) was applied to the targeted area, and slides were sealed with rubber cement. Slides were then incubated at 83 °C for 5 min, followed by another 16 h at 37 °C. Slides were washed with 0.4×SSC/0.3% NP-40 at 73 °C for 4 min and then 2×SSC/0.1% NP-40 at ambient temperature for 2 min. Chromatin was counterstained with 4′,6-diamidino-2-phenylindole. Fluorescence was analyzed in more than 50 cells per cell line for both EGFR and chromosome 7 centromere signals. We divided the EGFR copy number variation into 6 categories (disomy, low trisomy, high trisomy, low polysomy, high polysomy, gene amplification) as previously described26.
Cell growth assay
Cells were seeded in 96-well plates with ACL4 plus 5% FBS (5000 cells per well for cell lines SH-437, SH-450; 7000 cells per well for cell lines SH-416, SH-289). Cell growth kinetics were assessed by MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay daily as previously described27.
Soft agar assay
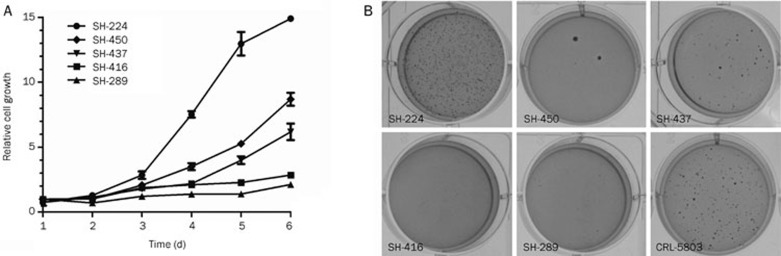
To assess soft agar colony formation, each of the cell lines (SH-224, SH-289, SH-416, SH-437, and SH-450. CRL-5803 serves as a control, which was obtained from ATCC) was suspended in a top layer of RPMI 1640 containing 10% FBS and 0.4% Select agar at 5000 cells per well in 6-well plates and plated on a bottom layer of RPMI-1640 containing 10% FBS and 1% Select agar. Cells were stained with 0.5 mL crystal violet for 1 h.
Tumor formation in nude mice
Cells (5×106 to 1×107) were mixed with matrigel and then transplanted subcutaneously in nude mice. The animals were killed after 2 weeks, and the tumors were fixed in formalin for hematoxylin-eosin staining and pathological analysis.
Results
Establishment of the primary Chinese lung cancer cell lines
From 2008 to 2010, we collected and used 179 fresh lung cancer specimens for the establishment of primary Chinese lung cancer cell lines. Eventually, eight cell lines were successfully established and maintained in cell culture for more than 25 passages. Some cell lines even exceed 40 passages. Among these cell lines, seven are NSCLC cell lines, including three from adenocarcinomas, one from squamous cell carcinoma, one from adeno-squamous carcinoma, one from large cell carcinoma and one from poorly differentiated carcinoma (Figure 1, Table 1). All NSCLC cell lines were cultured using ACL4 medium with 5% FBS. Three cell lines including SH-543, SH-450, and SH-437 can also be maintained in RPMI-1640 with 10% FBS. Because SCLC patients seldom go through surgery and very few tumor specimens are available, we have established only one SCLC cell line.
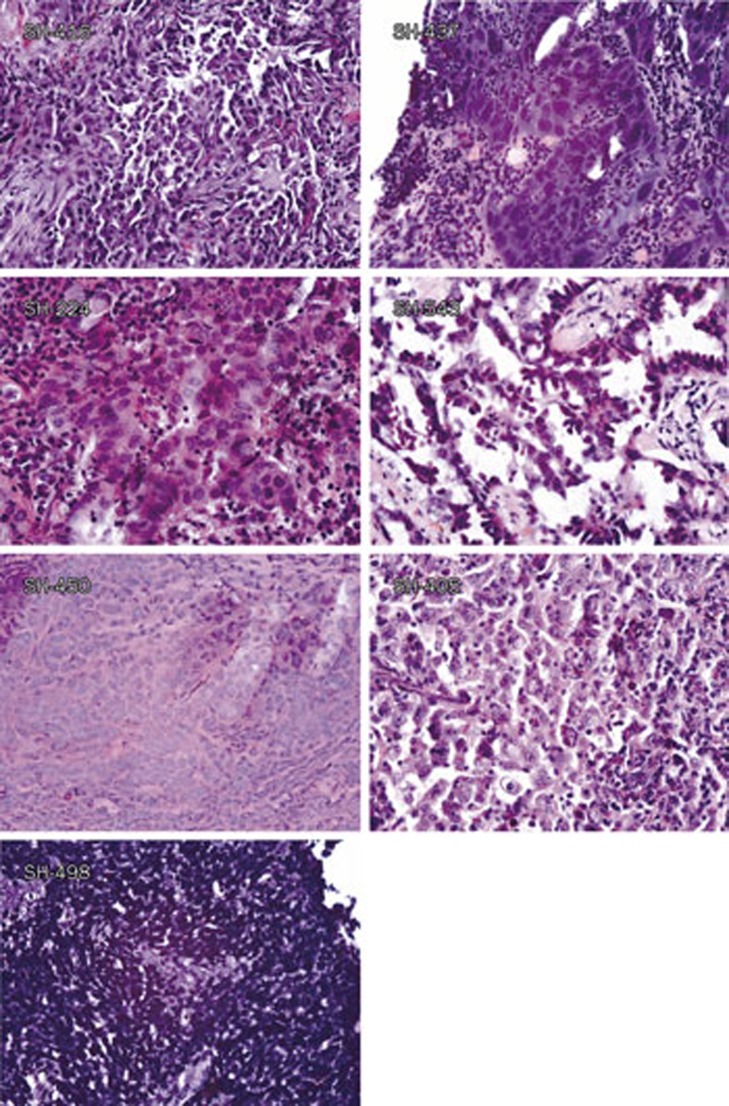
Figure 1.
Typical pathological images of the tumor tissues used to establish primary Chinese lung cancer cell lines. Typical pathological images of the original tumors of primary Chinese lung cancer cell lines are shown, including SH-416 (SCC), SH-437 (LC), SH-224 (Ad), SH-543 (Ad), SH-450 (AS), SH-405 (PDE), SH-498 (SCLC).
Table 1. Clinical and pathologic summary of the established primary Chinese lung cancer cell lines.
| Cell lines |
Gender |
Age (Year-old) |
Pathology |
Staging |
Origin |
EGFR mutations |
EGFR CNV |
|
|---|---|---|---|---|---|---|---|---|
| Tumor | Cell line | |||||||
| SH-224 | Female | 71 | Ad | III | Primary tumor | Exon 19 deletion | Exon 19 deletion | High polysomy |
| SH-289 | Female | 58 | Ad | IV | Pleural fluid | Exon 19 deletion | Exon 19 deletion | Amplification |
| SH-543 | Female | 62 | Ad | I | Primary tumor | WT | WT | Disomy |
| SH-416 | Female | 57 | SCC | III | Primary tumor | Exon 19 deletion | Exon 19 deletion | Amplification |
| SH-450 | Male | 59 | AS | III | Primary tumor | Exon 19 deletion | Exon 19 deletion | High polysomy |
| SH-437 | Male | 49 | LC | II | Primary tumor | Exon 21 P848L | Exon 21 P848L | High polysomy |
| SH-405 | Male | 63 | PDE | IIIa | Primary tumor | WT | WT | NA |
| SH-498 | Male | 53 | SCLC | LS | Primary tumor | WT | WT | NA |
Ad: Adenocarcinoma; SCC: Squamous cell carcinoma; LC: Large cell carcinoma; AS: Adeno-squamous carcinoma; PDE: Poorly differentiated carcinoma; WT: wild type; SCLC: Small cell lung carcinoma; LS: Limited stage; NA: not available; CNV: Copy number variation.
The clinical and pathologic details for all the patients from whom the cell lines were derived are summarized in Table 1. Most lines were established from primary solid tumors, with typical pathological images shown in Figure 1, except SH-289, which was derived from pleural effusions. All of the patients were from the Chinese population with an age range from 49 to 71 years old and with different stages of disease.
Morphological appearance of primary Chinese lung cancer cell lines
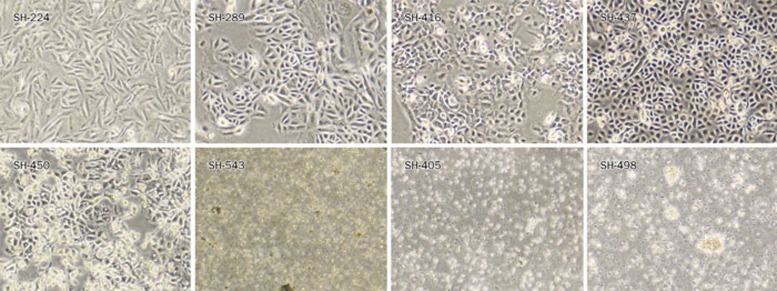
Five NSCLC cell lines, including SH-224, SH-289, SH-416, SH-437, and SH-450, grow in cell culture as an attached monolayer (Figure 2). Adherent cells were, in general, large, and they exhibited characteristic epithelioid 'cobble-stone' morphology and were occasionally multi-nucleated or vacuolated. SH-450 cells were slightly unique due to their unconstrained growth and foci formation even after they reach 100% confluence. Another two NSCLC cell lines, SH-405 and SH-543, grew in suspension like SCLC cell lines. This is not common because the majority of NSCLC cell lines are adherent cells. Only 5% (8 of 154) of NSCLC cell lines grow as suspension cells, and none has been established before in a Chinese genetic background. Typical suspension cell growth was seen in the SCLC cell line SH-498, which grows as either single cell or cell clusters.
Figure 2.
Representative morphology of primary Chinese lung cancer cell lines under light microscope. Typical morphology of cell lines is shown, including SH-224 (Ad), SH-289 (Ad), SH-416 (SCC), SH-437 (LC), SH-450 (AS), SH-543 (Ad), SH-405 (PDE), and SH-498 (SCLC).
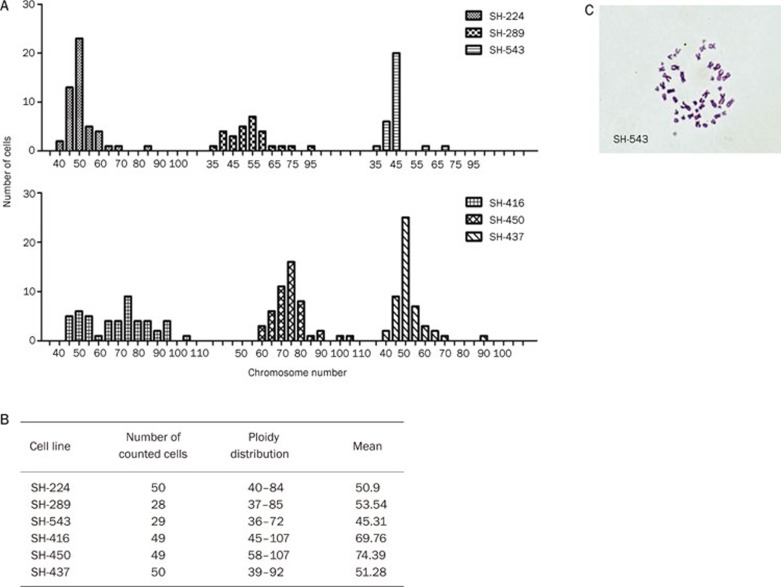
Karyotype of the primary Chinese lung cancer cell lines
We have performed karyotype for the cell lines, except for SH-498 and SH-405, which grow very slow in suspension. The human origin of these cell lines is confirmed by a peak on nearly 46 chromosomes (Figure 3A). Most of the cell lines are aneuploid, consistent with their tumor origin. Significant subpopulations with both lower and higher chromosome ploidy were shown in all analyzed cell lines, with their modal chromosome numbers ranging from 36 to 107 (Figure 3B). SH-416 and SH-289 cells harbor a wide range of chromosomes number, while the suspension cell line SH-543 displayed almost uniformed diploidy (Figure 3A, 3C).
Figure 3.
Ploidy and cytogenetic analyses of primary Chinese lung cancer cell lines. (A) Ploidy distribution of six Chinese NSCLC cell lines. (B) The summary of chromosome counting in the six Chinese NSCLC cell lines. (C) The karyotype of SH-543 is shown as one example.
Status of EGFR mutations and gene amplification in the primary Chinese lung cancer cell lines
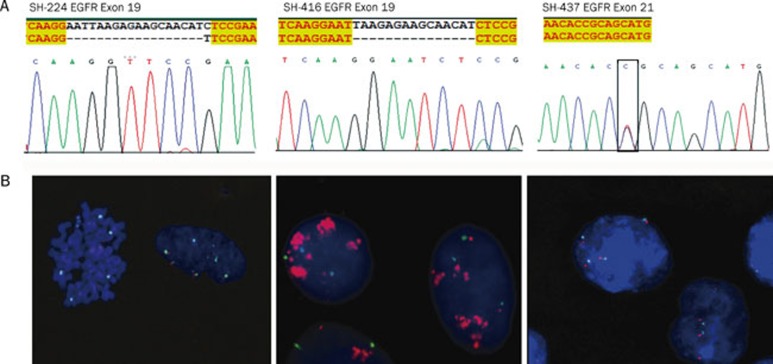
The EGFR kinase domain mutations, which are frequently associated with clinical tyrosine kinase inhibitor response, often occur in NSCLC patients from East Asia including China. We then asked if any of the above established cell lines harbor EGFR mutations, which may be useful for drug response studies. Using cDNA sequencing of the EGFR tyrosine kinase domain region, we found that five lines harbor EGFR mutations — four among them have exon 19 deletions (SH-224, SH-289, SH-416, and SH-450) and one has an exon 21 point mutation (SH-437) (Figure 4A). We further checked the gene amplification of EGFR allele, and the results from FISH analyses showed that all 5 cell lines displayed various degree of gene copy number increase (Table 1). Three cell lines (SH-224, SH-437, and SH-450) showed high polysomy, while another two (SH-289 and SH-416) showed gene amplification (Figure 4B).
Figure 4.
Status of EGFR mutation and gene amplification in primary Chinese lung cancer cell lines. (A) DNA sequencing results showed EGFR mutations in the indicated cell lines. (B) FISH analyses showed either EGFR high polysomy or gene amplification in the indicated cell lines. Green: chromosome 7 centromere; Red: EGFR.
Cell growth of the primary Chinese lung cancer cell lines in two dimension and soft agar
Five adherent NSCLC cell lines were used for further MTT assay and soft agar assay for testing their cell growth ability. Three cell lines with EGFR mutations SH-224, SH-437, and SH-450 grow faster than SH-289 and SH416 (Figure 5A).
Figure 5.
Cell growth of primary Chinese lung cancer cell lines in two dimension and soft agar. (A) The cell growth kinetics of the five adherent NSCLC cell lines are indicated. (B) The anchorage-independent cell growth of the five NSCLC cell lines. CRL-5803 serves as the control.
Consistently, all the three cell lines (SH-224, SH-437, and SH-450) growing faster in two-dimension culture formed recognizable colonies in soft agar. In contrast, no colonies were seen in the two slow-growing cell lines SH-416 and SH-289 (Figure 5B).
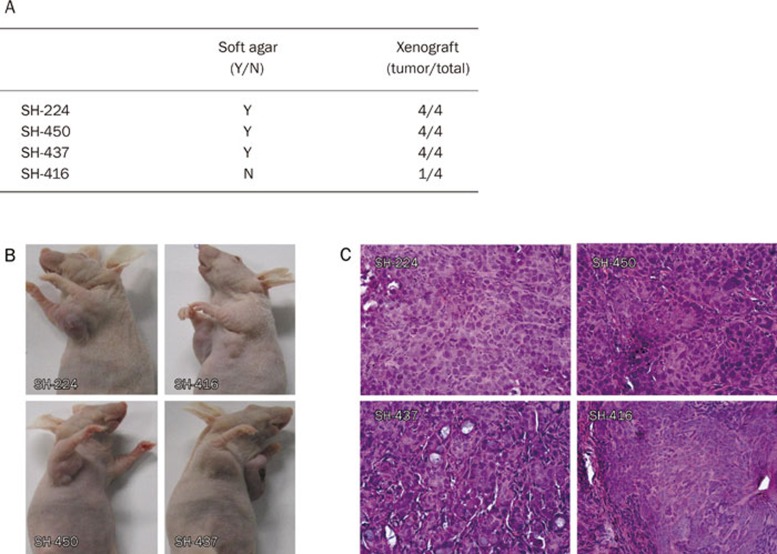
Tumorigenicity of primary Chinese NSCLC cell lines
We further checked the tumor formation ability of these NSCLC cell lines in nude mice. Cell lines SH-224, SH-437, and SH-450 readily formed subcutaneous tumors in nude mice 2 weeks following transplantation. Surprisingly, SH-416, the cell line that failed to form colonies in soft agar, could also form a visible graft in nude mice at a low rate (1/4) (Figure 6A, 6B). This may somehow reflect the difference between in vitro and in vivo systems. Nonetheless, these data also suggest that SH-289 and SH-416 may have a low potential for tumor formation. The tumors grew locally as compact masses. In each instance the histological appearance of the tumors closely resembled the original tumors from which they were derived (Figure 1, 6C).
Figure 6.
Tumor formation ability of primary Chinese NSCLC cell lines in nude mice. (A) The summary of tumorigenicity in four Chinese NSCLC cell lines in nude mice. (B) Representative photos are shown for the xenografts; (C) H&E staining of the tumors from xenografts.
Discussion
The in vitro cell line model system has contributed significantly to cancer research and resulted in several seminal biomedical discoveries. However, the majority of these cancer cell lines are from Western populations, thus dramatically constraining the studies focusing on ethnic differences in cancer biology, drug response or toxicity between Chinese and Caucasian populations. In this study, we have established eight lung cancer cell lines from a Chinese genetic background. These well-characterized cell lines will provide an ideal platform for stimulating studies in the future.
In general, it is considered much more difficult to establish lung cancer cell lines in comparison with other types of epithelia-originated cell lines28. Chemically defined media such as ACL3, ACL4, and HITES were first used for primary lung cancer cell culture with success. We tried here to add 5% FBS to the chemically defined medium ACL4, which stimulates the growth of not only the cancer cells but also the fibroblast. With great effort to eliminate fibroblast growth using scraping methods, we have successfully established eight Chinese lung cancer cell lines. It is possible that serum played an important role in making the floating cells attach to a culture dish. However, the success rate in our study still remains low (4.5%, Table 2). Further work is warranted to optimize the chemically defined medium.
Table 2. Summary of the success rate of all primary cell lines from different lung cancer subtypes.
| Pathology | Success | Total | Success rate |
|---|---|---|---|
| Ad | 3 | 83 | 3.6% |
| SCC | 1 | 43 | 2.3% |
| AS | 1 | 6 | 16.7% |
| LC | 1 | 4 | 25% |
| PDE | 1 | 11 | 9.1% |
| SCLC | 1 | 4 | 25% |
| Others | 0 | 28 | 0 |
| Summary | 8 | 179 | 4.5% |
To our surprise, five of the seven NSCLC cell lines we established harbor EGFR mutations as well as EGFR polysomy or allele amplification. EGFR mutations in all established Chinese NSCLC cell lines reach 71%. Through several decades of efforts, 21 lung cancer cell lines with EGFR kinase domain mutations have been developed worldwide: 12 with exon 19 deletions and 7 with the exon 21 mutation L858R and/or drug-resistant exon 20 mutation T790M (Table 3). Two uncommon mutations L861Q and G719S are also seen. Among these EGFR mutant cell lines, nine are from Caucasian populations, eleven from Japan and one from Hong Kong. Here, we have established a comparable number of NSCLC cell lines with EGFR mutations in a Chinese genetic background. It is noteworthy that the majority of the previously established EGFR mutant cell lines are from lung adenocarcinomas, while two are from squamous cell carcinomas, which are not commonly used. In contrast, our collection of cell lines is composed of various pathologies including adenocarcinoma (3), squamous cell carcinoma (1), adeno-squamous cell carcinoma (1) and large cell carcinoma (1) (Table 1). In addition, SH-437 harbors an exon 21 point mutation P848L, which has not been previously reported in any cell line. Taken together, our effort to establish Chinese lung cancer cell lines was a very successful small step toward our long-term goal of establishing more than 30 Chinese lung cancer cell lines. These cell lines will provide important tools for studying the ethnic differences in lung cancer biology as well as in drug treatment between Chinese and Caucasian populations in the future.
Table 3. Previously established lung cancer cell lines with EGFR kinase domain mutations.
| Cell line | Pathology | EGFR mutation status | References |
|---|---|---|---|
| H1650 | Ad | Exon 19 deletion (746–750) | 29 |
| HCC827 | Ad | Exon 19 deletion (746–750) | 30 |
| HKULC 3 | Ad | Exon 19 deletion (756–760) | 31 |
| PC3 | Ad | Exon 19 deletion (747–749), A750P | 32 |
| PC9 | Ad | Exon 19 deletion (746–750) | 33 |
| PC-14 | Ad | Exon 19 deletion (746–750) | 32 |
| HCC2935 | Ad | Exon 19 deletion (746–751), S752I | 34 |
| HCC4006 | Ad | Exon 19 deletion (747–749), A750P | 35 |
| H820 | Ad | Exon 19 deletion (746–751), T790M | 36 |
| HCC2279 | Ad | Exon 19 deletion (746–750) | 37 |
| RERF-LC-Ad2 | Ad | Exon 19 deletion (747–749), A750P | 32 |
| H1975 | Ad | L858R and T790M | 29, 38 |
| H3255 | Ad | L858R | 39 |
| HCC4011 | Ad | L858R | 34 |
| 11-18 | Ad | L858R | 32 |
| LCSC#1* | Ad | L858R | 32 |
| LC2/Ad | Ad | L858R | 32 |
| KTA-7 | Ad | L861Q | 32 |
| PC-7 | Ad | G719S | 32 |
| RERF-LC-A1 | SCC | L858R | 32 |
| KTSq-1 | SCC | Exon 19 deletion (746–750) | 32 |
Author contribution
Hong-bin JI and Hai-quan CHEN designed the research; Chao ZHENG, Yi-hua SUN, and Xiao-lei YE performed the research; Hong-bin JI and Chao ZHENG wrote the paper.
Acknowledgments
This work was supported by the National Basic Research Program of China (2010CB912102), the National Natural Science Foundation of China (30871284 and 30971461), the Chinese Academy of Sciences (KSCX1-YW-22), and the Science and Technology Commission of Shanghai Municipality (08PJ14105, 09JC1416300). Hong-bin JI is a scholar of the Hundred Talents Program of the Chinese Academy of Sciences.
References
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Gazdar AF, Girard L, Lockwood WW, Lam WL, Minna JD. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst. 2010;102:1310–21. doi: 10.1093/jnci/djq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nat Rev Cancer. 2002;2:315–9. doi: 10.1038/nrc775. [DOI] [PubMed] [Google Scholar]
- Lieber M, Smith B, Szakal A, Nelson-Rees W, Todaro G. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer. 1976;17:62–70. doi: 10.1002/ijc.2910170110. [DOI] [PubMed] [Google Scholar]
- Masuda N, Fukuoka M, Takada M, Kudoh S, Kusunoki Y. Establishment and characterization of 20 human non-small cell lung cancer cell lines in a serum-free defined medium (ACL-4) Chest. 1991;100:429–38. doi: 10.1378/chest.100.2.429. [DOI] [PubMed] [Google Scholar]
- Loh PM, Clamon GH, Robinson RA, White ML, Hukku B, Rossi NP, et al. Establishment and characterization of four new human non-small cell lung cancer cell lines. Cancer Res. 1984;44:3561–9. [PubMed] [Google Scholar]
- Anger B, Bockman R, Andreeff M, Erlandson R, Jhanwar S, Kameya T, et al. Characterization of two newly established human cell lines from patients with large-cell anaplastic lung carcinoma. Cancer. 1982;50:1518–29. doi: 10.1002/1097-0142(19821015)50:8<1518::aid-cncr2820500812>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Baillie-Johnson H, Twentyman PR, Fox NE, Walls GA, Workman P, Watson JV, et al. Establishment and characterisation of cell lines from patients with lung cancer (predominantly small cell carcinoma) Br J Cancer. 1985;52:495–504. doi: 10.1038/bjc.1985.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney DN, Gazdar AF, Bepler G, Guccion JG, Marangos PJ, Moody TW, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45:2913–23. [PubMed] [Google Scholar]
- Simms E, Gazdar AF, Abrams PG, Minna JD. Growth of human small cell (oat cell) carcinoma of the lung in serum-free growth factor-supplemented medium. Cancer Res. 1980;40:4356–63. [PubMed] [Google Scholar]
- Oie HK, Russell EK, Carney DN, Gazdar AF. Cell culture methods for the establishment of the NCI series of lung cancer cell lines. J Cell Biochem Suppl. 1996;24:24–31. doi: 10.1002/jcb.240630504. [DOI] [PubMed] [Google Scholar]
- Gazdar AF, Gao B, Minna JD. Lung cancer cell lines: Useless artifacts or invaluable tools for medical science. Lung Cancer. 2010;68:309–18. doi: 10.1016/j.lungcan.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang TW, Oak CH, Chang HK, Suo SJ, Jung MH. EGFR and KRAS mutations in patients with adenocarcinoma of the lung. Korean J Intern Med. 2009;24:48–54. doi: 10.3904/kjim.2009.24.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller VA. Epidermal growth factor receptor mutations, small-molecule kinase inhibitors, and non-small-cell lung cancer: current knowledge and future directions. J Clin Oncol. 2005;23:2556–68. doi: 10.1200/JCO.2005.07.799. [DOI] [PubMed] [Google Scholar]
- Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc. 2009;6:201–5. doi: 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- Sun YH, Fang R, Gao B, Han XK, Zhang JH, Pao W, et al. Comparable rate of EGFR kinase domain mutation in lung adenocarcinomas from Chinese male and female never-smokers. Acta Pharmacol Sin. 2010;31:647–8. doi: 10.1038/aps.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Sun Y, Zhang J, Ren Y, Fang R, Han X, et al. Spectrum of LKB1, EGFR, and KRAS mutations in chinese lung adenocarcinomas. J Thorac Oncol. 2010;5:1130–5. doi: 10.1097/JTO.0b013e3181e05016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan VH, Moore MM, McLachlan AJ, Piquette-Miller M, Xu H, Clarke SJ. Ethnic differences in drug metabolism and toxicity from chemotherapy. Expert Opin Drug Metab Toxicol. 2009;5:243–57. doi: 10.1517/17425250902800153. [DOI] [PubMed] [Google Scholar]
- Sekine I, Yamamoto N, Nishio K, Saijo N. Emerging ethnic differences in lung cancer therapy. Br J Cancer. 2008;99:1757–62. doi: 10.1038/sj.bjc.6604721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Ren Y, Fang Z, Li C, Fang R, Gao B, et al. Lung adenocarcinoma from east asian never smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol. 2010;28:4616–20. doi: 10.1200/JCO.2010.29.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber DA, Bell DW, Sordella R, Kwak EL, Godin-Heymann N, Sharma SV, et al. Molecular targeted therapy of lung cancer: EGFR mutations and response to EGFR inhibitors. Cold Spring Harb Symp Quant Biol. 2005;70:419–26. doi: 10.1101/sqb.2005.70.043. [DOI] [PubMed] [Google Scholar]
- Yang CH. EGFR tyrosine kinase inhibitors for the treatment of NSCLC in East Asia: present and future. Lung Cancer. 2008;60:S23–30. doi: 10.1016/S0169-5002(08)70102-8. [DOI] [PubMed] [Google Scholar]
- Gazdar AF, Oie HK. Cell culture methods for human lung cancer. Cancer Genet Cytogenet. 1986;19:5–10. doi: 10.1016/0165-4608(86)90365-1. [DOI] [PubMed] [Google Scholar]
- Brower M, Carney DN, Oie HK, Gazdar AF, Minna JD. Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 1986;46:798–806. [PubMed] [Google Scholar]
- Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–55. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- Ali S, Banerjee S, Schaffert JM, El-Rayes BF, Philip PA, Sarkar FH. Concurrent inhibition of NF-kappaB, cyclooxygenase-2, and epidermal growth factor receptor leads to greater anti-tumor activity in pancreatic cancer. J Cell Biochem. 2010;110:171–81. doi: 10.1002/jcb.22523. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sugaya M, Takenoyama M, Osaki T, Yasuda M, Nagashima A, Sugio K, et al. Establishment of 15 cancer cell lines from patients with lung cancer and the potential tools for immunotherapy. Chest. 2002;122:282–8. doi: 10.1378/chest.122.1.282. [DOI] [PubMed] [Google Scholar]
- NCI-Navy Medical Oncology Branch cell line supplement. J Cell Biochem Suppl. 1996;24:1–291. [PubMed] [Google Scholar]
- Girard L, Zochbauer-Muller S, Virmani AK, Gazdar AF, Minna JD. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894–906. [PubMed] [Google Scholar]
- Lam DC, Girard L, Suen WS, Chung LP, Tin VP, Lam WK, et al. Establishment and expression profiling of new lung cancer cell lines from Chinese smokers and lifetime never-smokers. J Thorac Oncol. 2006;1:932–42. [PubMed] [Google Scholar]
- Nagai Y, Miyazawa H, Huqun, Tanaka T, Udagawa K, Kato M, et al. Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res. 2005;65:7276–82. doi: 10.1158/0008-5472.CAN-05-0331. [DOI] [PubMed] [Google Scholar]
- Noro R, Gemma A, Kosaihira S, Kokubo Y, Chen M, Seike M, et al. Gefitinib (IRESSA) sensitive lung cancer cell lines show phosphorylation of Akt without ligand stimulation. BMC Cancer. 2006;6:277. doi: 10.1186/1471-2407-6-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Yamamoto H, Lockwood WW, Valencia I, Soh J, Peyton M, et al. MET gene amplification or EGFR mutation activate MET in lung cancers untreated with EGFR tyrosine kinase inhibitors. Int J Cancer. 2009;124:1778–84. doi: 10.1002/ijc.24150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu H, Takahashi T, Nomura M, Majmudar K, Suzuki M, Lee H, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–6. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- Shimamura T, Li D, Ji H, Haringsma HJ, Liniker E, Borgman CL, et al. Hsp90 inhibition suppresses mutant EGFR-T790M signaling and overcomes kinase inhibitor resistance. Cancer Res. 2008;68:5827–38. doi: 10.1158/0008-5472.CAN-07-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Liang K, Li X, Fan Z. Responses of cancer cells with wild-type or tyrosine kinase domain-mutated epidermal growth factor receptor (EGFR) to EGFR-targeted therapy are linked to downregulation of hypoxia-inducible factor-1 alpha. Mol Cancer. 2007;6:63. doi: 10.1186/1476-4598-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–7. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- Tracy S, Mukohara T, Hansen M, Meyerson M, Johnson BE, Jänne PA. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. 2004;64:7241–4. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]