Abstract
Aim:
To examine the influence of ginsenoside Rh2 (Rh2), a triterpene saponin extracted from the traditional medicinal plant ginseng, on the expression of miRNAs in human glioma cells.
Methods:
The expression profile of miRNA (miR) was analyzed in human U251, T98MG and A172 glioma cells using a miRNA array and quantitative real-time PCR. Cell viability was assessed using a colorimetric assay (cell counting kit-8). Transfection of miR-128 was performed using Lipofectamine 2000. Caspase 3 activity was determined using a caspase colorimetric assay kit. Apoptosis was assessed using annexin V and propidium iodide staining. Protein expression was determined with Western blot analysis. miRNA-128 targeting activity was measured using a luciferase reporter assay.
Results:
In U251 cells treated with Rh2 (12 μg/mL), 14 of 452 human miRNAs were up-regulated and 12 were down-regulated as detected with the miRNA array assay. The up-regulation of miR-128 by Rh2 was further verified in human U251, T98MG and A172 cells using quantitative real-time PCR. In U251 cells, transfection of a miR-128 inhibitor (50 nmol/L) prevented the overexpression of miR-128 by Rh2, and significantly blunted Rh2-induced cytotoxicity, apoptosis, caspase 3 activation, transcriptional activation of E2F3a, a miR-128 target gene, as well as E2F3a protein expression.
Conclusion:
The anti-proliferative effect of Rh2 in human glioma cells was mediated in part through up-regulation of miRNA-128 expression.
Keywords: ginsenoside Rh2, triterpene saponin, glioma cell, microRNA, miR-128, apoptosis, apoptosis cell proliferation, caspase 3, E2F3a
Introduction
Gliomas are the most common brain tumors in humans. Even when treated with surgery, radiotherapy, chemotherapy, and other intensive regimens, malignant gliomas are incurable1. The tumors are characterized by rapid cell growth and diffuse cellular infiltration into adjacent normal tissues2. Although a number of genetic and molecular lesions have been correlated with glioma progression, a complete understanding of the molecular basis and therapy of these tumors remains elusive.
One novel approach to characterizing the molecular and therapeutic targets of gliomas is based on the expression profiling of miRNAs3. These small RNA molecules are a class of endogenously expressed small noncoding RNA, 18-25 nucleotides in length4. To date, more than 700 miRNAs have been identified in humans5. miRNAs are known to be important in the regulation of many fundamental cellular processes, such as cell proliferation, differentiation and apoptosis6, 7. Following binding to the 3′-untranslated regions (UTRs) of specific mRNAs, miRNAs regulate target gene expression by inducing translational repression or mRNA degradation8. It is estimated that up to 30% of human genes may be regulated by miRNAs8. Moreover, approximately 50% of the known miRNAs were reported to be located in cancer–associated genomic regions9, 10 and miRNA dysregulation has been detected in various cancer cells11. Therefore, aberrations in miRNA expression patterns are thought to be involved in the progression of human cancers12. Since the first report of abnormal miRNA expression in glioblastomas in 2005, there has been an increasing number of reports each year describing miRNA dysregulation and function in various brain tumors13, 14, 15, 16, 17. These findings not only provide new insights into the molecular pathogenesis of gliomas, but also are useful in identifying miRNAs as potential targets in therapeutic intervention.
Ginsenoside Rh2 is a biologically active phytochemical extracted from Ginseng, a commonly used alternative drug taken orally in traditional herbal medicines in China, Korea, Japan and some Western countries18. It is a triterpene saponin, consisting of a steroid nucleus and a sugar moiety18. Rh2 has been reported to have a variety of biological effects, such as reducing blood glucose19 and ameliorating ischemic brain injury20; in addition, it has antiallergic activity21 and antiproliferative effects22. The ability of Rh2 to suppress cell growth has also been observed in glioma cells22, 23. Because Rh2 promotes neoplastic cells to return to a normal cell phenotype, it is expected to be a new type of anticancer agent23. It displays low toxicity, is associated with only a few side effects and is generally regarded as an anticancer nutrient23. Although extensive investigations have shown that Rh2 exerts its antiproliferative effects through induction of an apoptotic pathway24, the role of miRNAs in this process has not yet been explored.
Using an miRNA array to examine miRNA expression in Rh2-treated human glioma cells, we found that Rh2 altered the miRNA expression in human glioma U251 cells. We verified the observed up-regulation of the brain-enriched miR-128 by quantitative real-time PCR in human U251, T98MG and A172 glioma cells. To further investigate the role of miR-128 in Rh2-mediated antiproliferation, we transfected miR-128 inhibitor into glioma cells and observed an abrogation of Rh2-induced miR-128 overexpression, causing significant inhibition of Rh2-induced cytotoxicity, apoptosis, caspase-3 activation, transcriptional activation of E2F3a, a miR-128 target gene, and the expression of E2F3a protein.
Materials and methods
Reagents
Ginsenoside Rh2 (20R-form, >99% purity, HPLC pure) was purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China). The chemical structure of ginsenoside Rh2 is shown in Figure 1.
Figure 1.
Chemical structure of ginsenoside Rh2 (20R-form).
Cell lines and culture conditions
Human U251, T98MG and A172 glioma cells were purchased from the Cell Bank of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). The cells were maintained in RPMI-1640 medium (Gibco Life Technologies, Grand Island, NY, USA), supplemented with 10% fetal bovine serum (Sijiqing, Hangzhou, China), 100 U/mL penicillin and 100 mg/mL streptomycin. All cells were routinely passaged and maintained in a humidified incubator of 5% CO2 at 37 °C.
Cell proliferation assay
Cell viability was assessed colorimetrically using the cell counting kit-8 (CCK-8, Dojindo Laboratories, Tokyo, Japan)25. Human glioma U251 cells (1×105) were seeded in each well of a 96-well plate and incubated for 24 h prior to treatment with different dosages of Rh2 or vehicle. After treatment, 10 μL of the CCK-8 solution was added into each well, and the cells were incubated for an additional 2 h. The absorbance value (A) at 470 nm was read using a microplate reader (Bio-Rad, CA, USA) with a reference wavelength of 630 nm.
miRNA microarray analysis
The U251 cells were cultured for 24 h and then incubated with 12 μg/mL Rh2 for 24 h. Total cellular RNA was isolated from Rh2-treated and –untreated cells using TRIzol reagent (Invitrogen™, Carlsbad, CA, USA) according to the manufacturer's protocol. Five micrograms of total RNA was labeled at the 3′-end with Hy3™ using miRCURY Array Labeling kit (Exiqon, Vedback, Denmark) for 1 h at 0 °C.
The labeled RNA was purified using a Miniprep Kit (QIAgen, Valencia, CA, USA). The resulting sample was diluted with hybridization buffer, and 20 μL of the diluted sample was loaded onto the spotted area of the array and covered with a Bioarray LifterSlip coverslip. The chamber was rotated in a 60 °C water bath for 14 h. The miRCURY LNA miRNA Array 8.1 contained probes for 452 mature human miRNAs printed in quadruplicate. Next, the slides were washed in wash buffer A (2×SSC/0.2% SDS) at 60 °C until the Bioarray LifterSlip coverslip fell off. Then the slides were washed with wash buffer B (1×SSC) twice and wash buffer C (0.2×SSC) once. After drying by centrifugation for 5 min at 1000 round per minute, the slides were scanned with a Genepix 4000B laser scanner (Molecular Devices) and digitized by Genepix Pro 6.0. The resulting ratios of miRNA spots to background were calculated. The data were analyzed by subtracting out the background and normalizing the signals using global Lowess (locally weighted scatter plot smoothing) regression algorithm. All miRNAs having a ratio of 2.0 or greater were included.
Quantitative real-time PCR for miRNA expression
To confirm the miRNA levels obtained from the microarray results, miRNA expression was assessed using quantitative real-time PCR (qRT-PCR). Total cellular RNA was extracted from each of the experimental groups. Reverse transcription was performed using gene specific primers. The sequences of the primer pairs were as follows: miR-128: forward 5′-CGCGCTCACAGTGAACCG-3′ and reverse 5′-GTGCAGGGTCCGAGGT-3′ miR-15b: forward 5′-TAGCAGCACATCATGGTTTACA-3′ and reverse 5′-TCGTGCCAGCGGCTCG-3′ miR-21: forward 5′-CGGGATCCTGGGGTTCGATCTTAACAGGC-3′ and reverse 5′-CGGAATCCCCACAATGCAGCTTAGTTTTCC-3′ miR-25: forward 5′-GTGTTGAGAGGGCGGAGACTT-3′ and reverse 5′-TCAGACCGAGACAAGTGCAA-3′14, 15, 16, 17. U6 was taken as an internal control (forward 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse 5′-CGCTTCACGAATTTGCGTGTCAT-3′). qRT-PCR was performed using a Rotor-Gene 3000 real-time SYBR-green PCR system. The reactions were carried out in a 96-well optical plate at 95 °C for 10 min and then amplified for 10 s at 90 °C, followed by 1 min at 60 °C for 40 cycles. The relative value of each miRNA to U6 RNA was calculated using the 2-△△Ctmethod, where Ct is the number of cycles at which the application reaches a threshold, as determined by SDS software v1.2 (Applied Biosystems Inc). Thermal denaturation was administered at the conclusion of the PCR to determine the number of the products that were present in the reaction. Each reverse transcription and PCR assay were performed in triplicate.
miR-128 inhibitor transfection
The miR-128 inhibitor 5′-AAAGAGACCGGUUCACUGUGA-3′ and miR-128 inhibitor control 5′-CAGUACUUUUGUGUAGUACAAA-3′ were purchased from RiboBio Co Ltd, China. U251 cells in a volume of 100 μL DMEM medium were plated in each well of a 96-well plate. After 24 h of culture, the cells were transfected with 50 nmol/L miR-128 inhibitor or inhibitor control for 48 h using Lipofectamine 2000 (Invitrogen). After transfection, the wells were treated with Rh2 for different lengths of time in order to study cell proliferation, apoptosis, as well as caspase 3 and E2F3a expression.
Luciferase assay
A E2F3a 3′-untranslated region (UTR) reporter plasmid (pmiR-RB-REPORT™ 3′-UTR) was purchased from RiboBio Co Ltd, China. This vector was constructed by ligating a 1127-bp fragment to the type E2F3a 3′-UTR, which encompassed the target sequence for miR-128. The E2F3a 3′-UTR (accession number: NM0019490) was amplified from the genomic DNA using the following primers: forward 5′-AAACAATGCCAGGGTGTCTC-3′, reverse 5′-GCTCACACACGAAATGGCTA-3′. The empty plasmid containing only the E2F3a 3′-UTR was used as a control. The U251 cells were transfected with 50 nmol/L E2F3a 3′-UTR plasmid or empty E2F3a 3′-UTR plasmid for 48 h using Lipofectamine 2000 (Invitrogen). After transfection, the cells were treated with Rh2 for 24 h, and the luciferase assay was performed using TECAN Genios multifunctional microplate reader.
Western blotting
Cells were harvested and lysed in a buffer containing 50 mmol/L Tris-HCI, pH 8.0, 150 mmol/L NaCl, 1% NP40 and protease inhibitors (2 μg/mL leupeptin, 2 μg/mL pepstatin, 2 μg/mL aprotinin and 2 μg/mL PMSF, Sigma) for 30 min on ice. Lysates were centrifuged at 12 000 round per minute at 4 °C for 20 min, and the supernatants were collected. The protein concentrations were determined using the Bradford method. The proteins were separated using SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were incubated with rabbit anti-human E2F3a (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or mouse anti-human β-actin antibody (Sigma, USA).
Apoptosis assay
Apoptosis was determined by annexin V and propidium iodide (PI) staining using the apoptosis detection kit (Beyotime, China ) according to the manufacturer's instructions. Briefly, U251 cells were transfected with 50 nmol/L miR-128 inhibitor or miR-128 inhibitor control for 48 h, and then with 12 μg/mL Rh2 for 48 h. After treatment, the cells were washed with 50 mmol/L cold phosphate buffer (pH 7.5), centrifuged at 1200×g for 5 min, and suspended in binding buffer. The treated cells were incubated with annexin V and propidium iodide for 15 min at room temperature, after which the samples were analyzed for annexin V binding affinity within 1 h by flow cytometry26.
Caspase 3 activity assay
Caspase 3 activity was determined using a caspase colorimetric assay kit (BioVision Research Products, USA) according to manufacturer's protocol. The assay is based on spectrophotometric detection of the chromophore ρ-nitroanilide (ρNA) after cleavage from the labeled substrate DEVD-ρNA. Briefly, 1×106 cells were first treated with 50 nmol/L miR-128 inhibitor or miR-128 inhibitor control for 48 h and then with a different dosage of Rh2 for various time. The cells were collected, washed with ice-cold PBS, and lysed in a lysis buffer. Cell lysates were measured for protease activity using a caspase-specific peptide conjugated with the color molecule ρNA. The caspase-cleaved chromophore ρNA was quantitated with a spectrophotometer at a wavelength of 405 nm27.
Statistical analysis
The results were analyzed by one-way ANOVA or Student's t test. Both analyses were performed using SPSS 13.0 statistical software. A value of P<0.05 was considered statistically significant. All values were expressed in terms of mean±SEM or mean±SD.
Results
Effects of ginsenoside Rh2 on miRNA expression in human glioma U251 cells
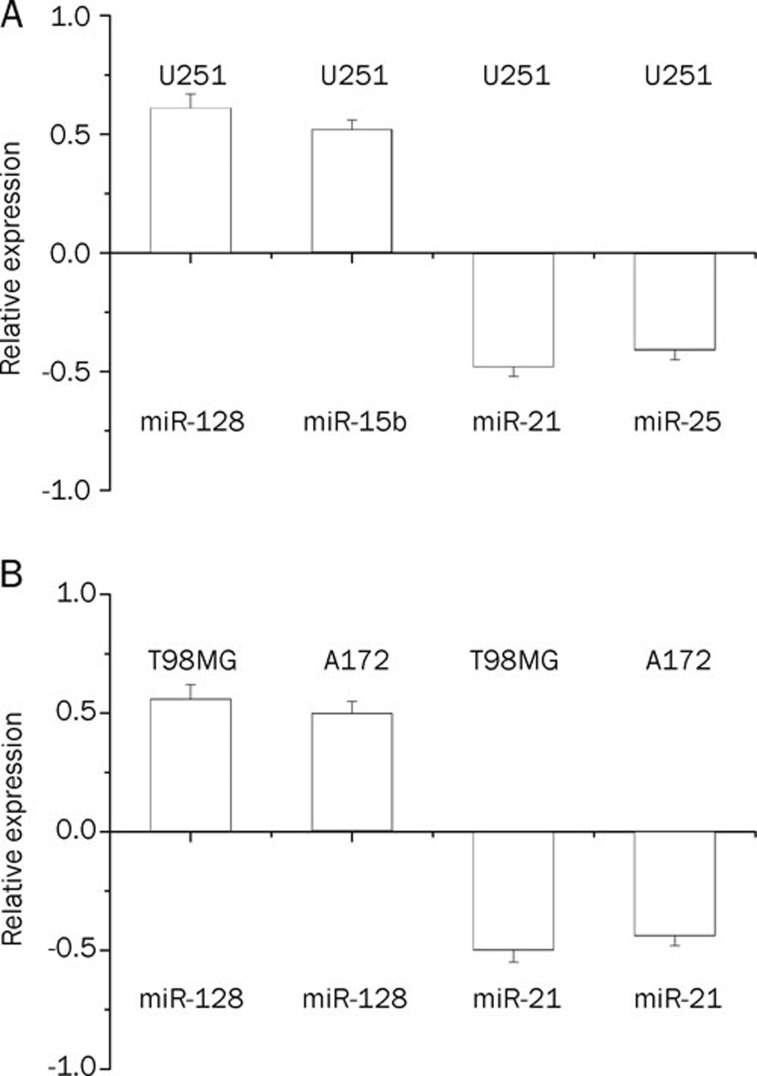
To identify the miRNAs involved in Rh2-induced cytotoxicity, total RNA was extracted from U251 cells treated with 12 μg/mL Rh2 for 24 h for miRNA microarray analysis. The Exiqon microarray (Exiqon, Vedback, Denmark) containing 452 mature human miRNA probes was used to identify the cellular miRNA expression profile. miRNAs up-regulated more than 2-fold or down-regulated by 2-fold by Rh2 treatment were classified as being up-regulated or down-regulated miRNAs, respectively. Table 1 summarizes the results of Rh2-regulated miRNAs and their chromosomal locations are summarized. Of the 452 human miRNAs tested the microarray, 14 miRNAs were up-regulated and 12 miRNAs were down-regulated in the Rh2-treated U251 cells. To further confirm the miRNA array results, we randomly selected the following four miRNAs: the up-regulated miR-128 and miR-15b as well as the down-regulated miR-21 and miR-25, and measured their expression levels by quantitative RT-PCR. As shown in Figure 2A, the results of quantitative RT-PCR for the four selected miRNAs were consistent with the miRNA array results, indicating that the results of miRNA array are reliable. To determine whether Rh2-induced miRNA expression occurred in other glioma cells, we measured the expression of miR-128 and miR-21 in human glioma cell lines, T98MG and A172. We observed consistent results in these two glioma cells as compared to U251 cells (Figure 2B).
Table 1. Microarray analysis of miRNA expression in human gliomas U251 cells treated with ginsenoside Rh2.
| miRNA | Fold (Mean±SEM) | P value | Localization | Up-down regulation |
|---|---|---|---|---|
| miR-15b | 3.15±0.18 | 0.015 | 17q23.1 | up |
| let-7c | 2.03±0.15 | 0.010 | 21q11.2 | up |
| let-7d | 2.97±0.25 | 0.024 | 7q22.1 | up |
| miR-29b | 2.32±0.21 | 0.017 | 6q23.31 | up |
| miR-106b | 2.26±0.17 | 0.005 | 7q22.1 | up |
| miR-125b | 2.88±0.30 | 0.020 | 11q24.1 | up |
| miR-128 | 3.04±0.19 | 0.013 | 2q21.3 | up |
| miR-129 | 2.54±0.24 | 0.014 | 7q32.1 | up |
| miR-137 | 2.13±0.14 | 0.008 | 1q21.3 | up |
| miR-138 | 2.29±0.11 | 0.009 | 3q21.33 | up |
| miR-181a | 2.41±0.22 | 0.011 | 1q32.1 | up |
| miR-181b | 2.62±0.26 | 0.021 | 1q32.1 | up |
| miR-181c | 2.35±0.23 | 0.017 | 19p13.3 | up |
| miR-323 | 2.57±0.18 | 0.013 | 14q32.31 | up |
| miR-16 | 0.41±0.07 | 0.001 | 21q11.2 | down |
| miR-18 | 0.37±0.10 | 0.007 | 9q22.2 | down |
| miR-21 | 0.17±0.04 | 0.000 | 17q23.1 | down |
| miR-25 | 0.23±0.09 | 0.003 | 7q22.1 | down |
| miR-32 | 0.43±0.18 | 0.009 | 11q24.1 | down |
| miR-92 | 0.39±0.11 | 0.005 | 21q11.2 | down |
| miR-107 | 0.37±0.13 | 0.006 | 7q32.1 | down |
| miR-155 | 0.28±0.09 | 0.002 | 3p21.33 | down |
| miR-210 | 0.45±0.11 | 0.005 | 11p15.5 | down |
| miR-218 | 0.37±0.21 | 0.012 | 4p15.31 | down |
| miR-328 | 0.42±0.12 | 0.007 | 16q22.1 | down |
| miR-370 | 0.38±0.07 | 0.003 | 14q32.2 | down |
1. The results presented fold change of signal ratio of 12 μg/mL Rh2-treated cells to untreated control cells.
2. The raw data were normalized and analyzed with software of MatLab version7.4, which produced an average value of the four spot replications of each miRNAs.
3. Chromosome localization of microRNAs as refered in miRBase sequences (http://microrna.sanger.ac.uk).
Figure 2.
Validation of miRNA expression by quantitative real-time PCR. (A) The expressions of up-regulated miR-128 and miR-15b, as well as the down-regulated miR-21 and miR-25 in U251 cells were verified. The results of quantitative RT-PCR for the four selected miRNAs were consistent with the miRNA array results. (B) The expressions of miR-128 and miR-21 in human T98MG and A172 glioma cells were determined. The consistent results in these two giloma cells were observed as compared to U251 cells. The cells were treated with 12 μg/mL Rh2 for 24 h. Data were expressed as means±SEM of 3 independent experiments. Fold change of miRNA expression was presented in log 2 scale.
miRNA-128 mediated Rh2-induced growth inhibition in glioma cells
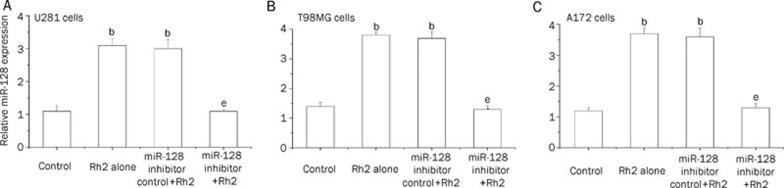
Given that miR-128 has been reported to inhibit glioma cell proliferation17, we sought to further investigated its role in Rh2-induced cytotoxicity. We first investigated the effect of miR-128 inhibitor on miR-128 overexpression induced by Rh2 in human U251, T98MG and A172 glioma cells with qRT-PCR. As shown in Figure 3, inhibition of miR-128 significantly suppressed Rh2-induced miR-128 overexpression, as compared with miR-128 inhibitor control. These results demonstrate that anti-miR-128 treatment was effective for inhibition of miR-128 overexpression.
Figure 3.
Transfection of miR-128 inhibitor significantly suppressed Rh2-induced miR-128 over-expression in U281 cells (A), T98MG cells (B), and A172 cells (C). The effects of miR-128 inhibitor transfection on Rh2-induced miR-128 overexpression in human U251, T98MG and A172 glioma cells were determined using qRT-PCR. The cells were transfected with 50 nmol/L miR-128 inhibitor or inhibitor control for 48 h, and then with Rh2 (12 μg/mL) for 24 h. The experiment was performed three times, each in triplicate. Data were expressed as means±SEM. bP<0.05 compared with control. eP<0.05 compared with Rh2 alone or miR-128 inhibitor control+Rh2.
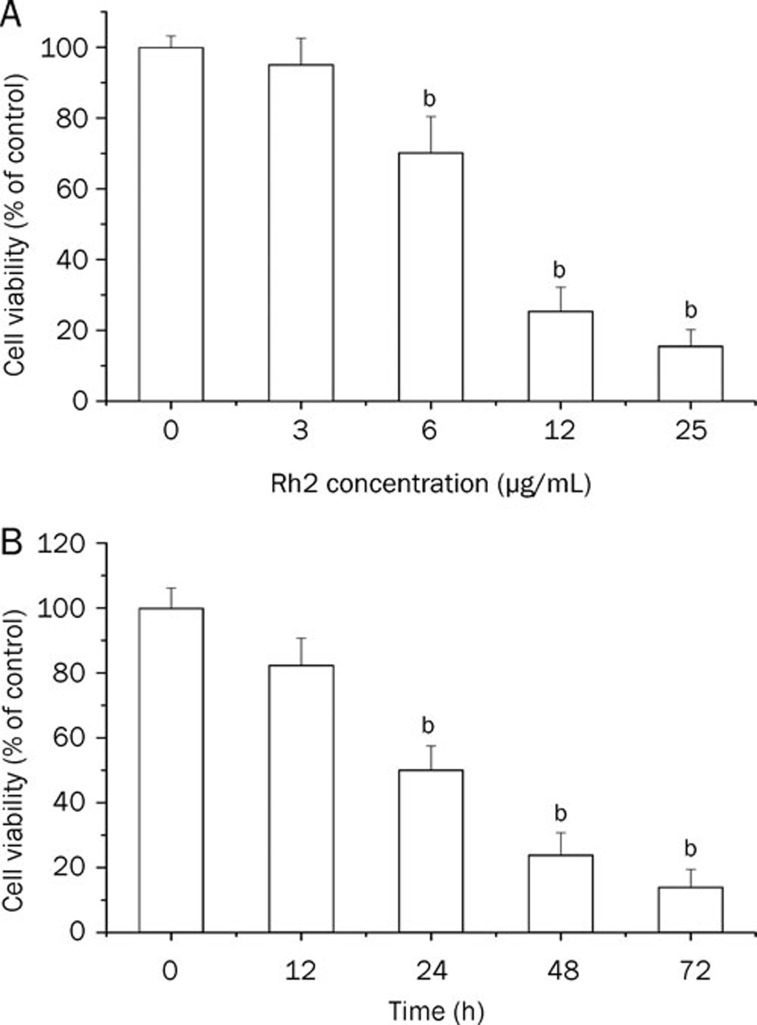
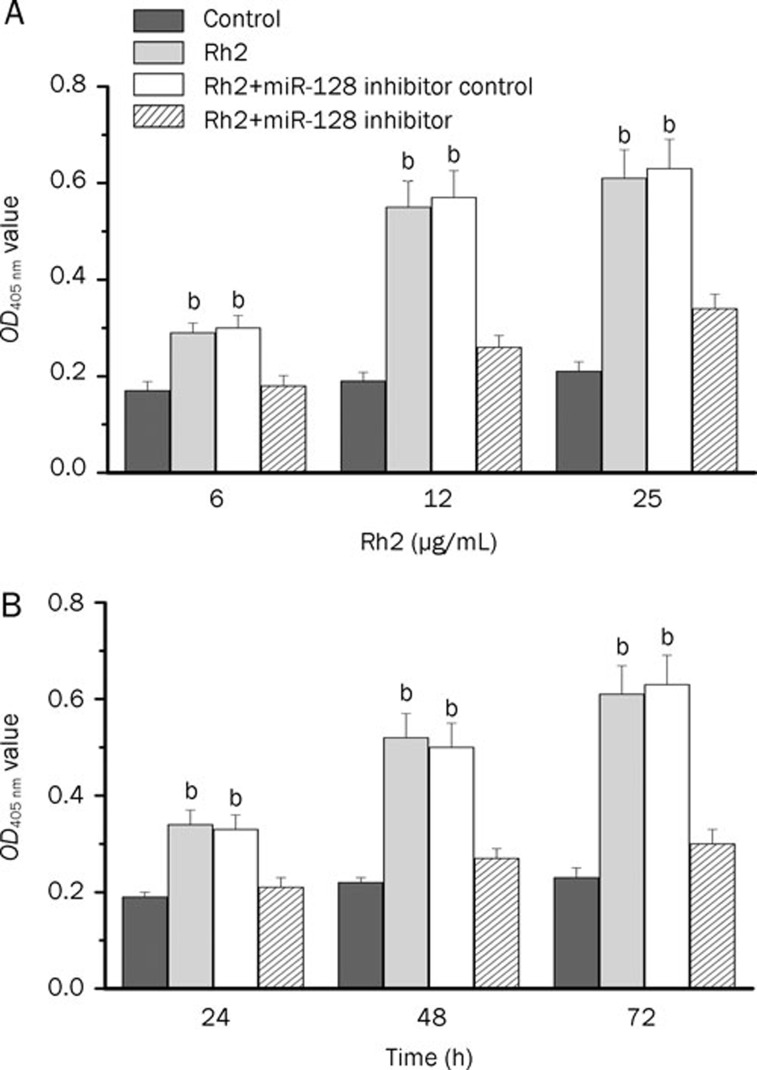
We next examined the antiproliferative effect of Rh2 in human glioma U251 cells. As shown in Figure 4, Rh2 treatment (6, 12, and 25 μg/mL) of U251 cells resulted in a dose-dependent inhibition of cell growth. In a time course study, 12 μg/mL Rh2 resulted in increasing levels of inhibition of cell growth up to 72 h, as measured by the cell counting kit-8 assay.
Figure 4.
Rh2 inhibited viability of human glioma U251 cells determined using a CCK-8 assay. (A) Dose-dependent response. U251 cells were treated with Rh2 (3, 6, 12 and 25 μg/mL) for 48 h. (B) Time-dependent response. U251 cells were treated with Rh2 (12 μg/mL) for 12, 24, 48 and 72 h. The results represented three independent experiments. Data were expressed as means±SEM. bP<0.05 compared with control.
To investigate whether miR-128 could be involved in Rh2-induced growth inhibition, we transfected U251 cells with 50 nmol/L miR-128 inhibitor or inhibitor control for 48 h and then examined the alteration of Rh2-induced cell growth inhibition. Knockdown of miR-128 following transfection of miR-128 inhibitor significantly increased the proliferation of U251 cells treated with Rh2, as compared with the cells transfected with a miR-128 inhibitor control (Figure 5).
Figure 5.
Transfection of miR-128 inhibitor prevented Rh2-induced cytotoxicity in glioma U251 cells. U251 cells were transfected with 50 nmol/L miR-128 inhibitor to knockdown miR-128. A miR-128 inhibitor control was used as negative control. At 48 h after transfection, the cells were treated with 12 μg/mL Rh2 for 24 or 48 h. Cell proliferation was determined using CCK-8 assay. bP<0.05 compared with Rh2+miR-128 inhibitor. The experiment was performed three times, each in triplicate.
miRNA-128 mediated Rh2 induction of apoptosis in glioma U251 cells
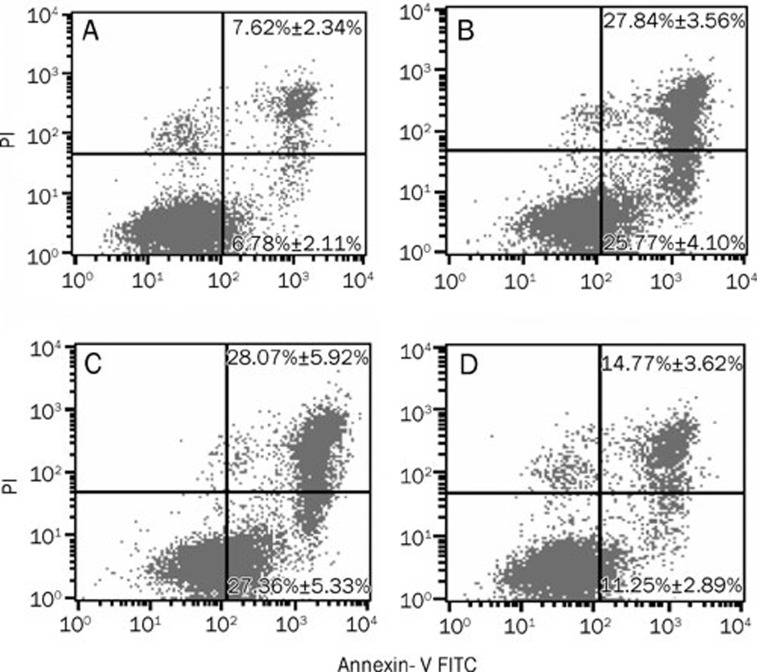
Rh2 is known to exert its tumor growth inhibitory effect through the induction of apoptosis28, 29. We further examined whether Rh2-induced overexpression of miR-128 was able to inhibit apoptosis induced by this compound. The rate of apoptosis was measured by flow cytometry with annexin V and PI staining following Rh2 treatment. U251 cells were treated with 50 nmol/L miR-128 inhibitor or miRNA-128 inhibitor control for 48 h prior to treatment with 12 μg/mL Rh2 for 48 h. Transfection of the miR-128 inhibitor significantly prevented Rh2-induced apoptosis in U251 cells. The percentages of early apoptosis cells were 6.78%±2.11%, 25.77%±4.10%, 27.36%±5.33% and 11.25%±2.89% in the control, Rh2 alone, Rh2+miR-128 inhibitor control, and Rh2+miR-128 inhibitor group, respectively (Figure 6 A–D, the lower right quadrant). The percentages of late apoptotic cells were 7.62%±2.34%, 27.84%±3.56%, 28.07%±5.92% and 14.77%±3.62% in the control, Rh2 alone, Rh2+miR-128 inhibitor control, and Rh2+miR-128 inhibitor group, respectively (Figure 6 A–D, the upper right quadrant).
Figure 6.
Transfection of miR-128 inhibitor inhibited Rh2-induced apoptosis in U251 cells. The cells were transfected with 50 nmol/L miR-128 inhibitor or inhibitor control for 48 h and then with Rh2 (12 μg/mL) for 48 h. Apoptosis was evaluated by annexin V and propidium iodide staining. The LR and UR quadrants indicated the percentage of early and late apoptotic cells, respectively, in control (A), Rh2 alone (B), Rh2+miR-128 inhibitor control (C) and Rh2+miR-128 inhibitor group (D). The experiment was performed three times, each in triplicate. Each point represented the means±SEM. There were significant differences between Rh2 alone or Rh2+miR-128 inhibitor control and control group, as well as between Rh2 alone and Rh2+miR-128 inhibitor group (P<0.05).
miRNA-128 mediated Rh2 induction of caspase 3 activity in glioma U251 cells
The previous studies have shown that the process of Rh2-induced apoptosis requires the activation of caspase 3 protease28. To assess whether miR-128 overexpression is involved in Rh2-induced caspase 3 activation in U251 cells, we measured caspase 3 activities after the cells were treated with Rh2 alone or Rh2 plus miR-128 inhibitor. Results from the colorimetric assay demonstrated that caspase 3 activity increased significantly in a dose- and time-dependent manner in Rh2-treated U251 cells (Figure 7). We also found that the knockdown of miR-128 by transfection of a miR-128 inhibitor significantly inhibited Rh2-induced caspase 3 activation, as compared to cells treated with miR-128 inhibitor control (Figure 7).
Figure 7.
Transfection of miR-128 inhibitor inhibited Rh2-induced caspase 3 activation in U251 cells. (A) Transfection of miR-128 inhibitor inhibited the caspase 3 activation induced by treatment with 6, 12, and 25 μg/mL Rh2 for 48 h. The cells were transfected with 50 nmol/L miR-128 inhibitor or inhibitor control for 48 h, and then with Rh2 for 48 h. (B) Transfection of miR-128 inhibitor inhibited the caspase 3 activation induced by treatment with 12 μg/mL Rh2. The cells were transfected with 50 nmol/L miR-128 inhibitor or inhibitor control for 48 h, and then with Rh2 for 24, 48, and 72 h. Caspase 3 avtivity was determined using colorimetric assay. The experiment was performed three times each in triplicate. Mean±SEM. bP<0.05 compared with control or miR-128 inhibitor+Rh2.
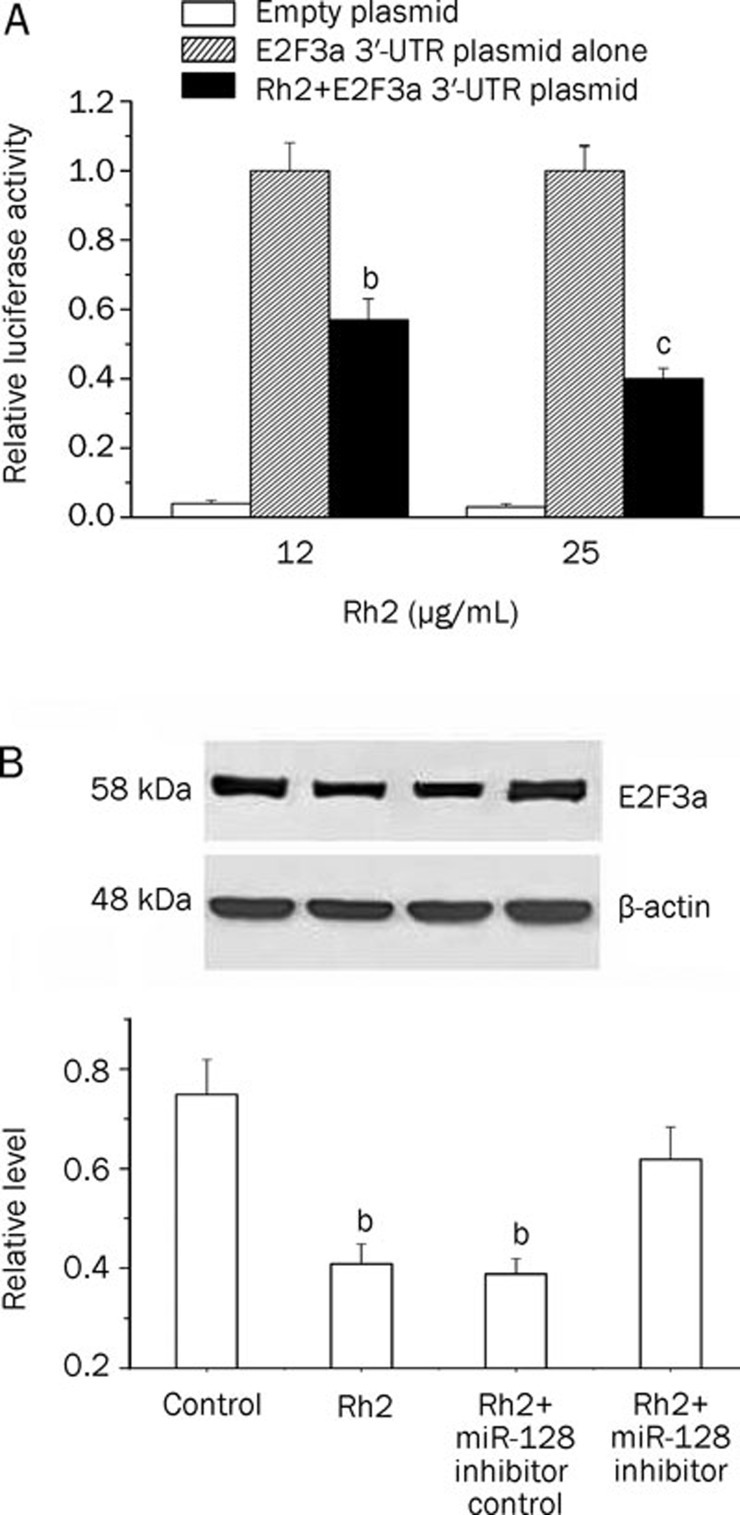
Rh2 suppressed the activity of luciferase reporter containing the E2F3a-3′-UTR and the level of E2F3a protein in glioma U251 cells
E2F3a is a validated target of miR-128, and has been reported to mediate miR-128-induced glioma cell growth inhibition17. To investigate whether overexpression of miR-128 induced by Rh2 affects E2F3a expression, the activity of a luciferase reporter containing the E2F3a-3′-UTR sequence was determined in U251 cells. The protein level of E2F3a was also assessed in U251 cells. We found that cells treated with 12 and 25 μg/mL Rh2 had suppressed luciferase activity (Figure 8A), indicative of transcriptional inhibition, as well as decreased the E2F3 protein expression in U251 cells (Figure 8B). To further explore whether miR-128 inhibitor prevents Rh2-induced inhibition of E2F3a expression, cells were transfected with 50 nmol/L miR-128 inhibitor for 48 h prior to 12 μg/mL Rh2 for 24 h. We also found that transfection of miR-128 inhibitor prevented Rh2-induced inhibition of E2F3a expression, as compared with cells transfected with miR-128 inhibitor control (Figure 8B).
Figure 8.
Rh2 suppressed the activity of luciferase reporter containing the E2F3a-3′-UTR and the level of E2F3a protein in U251 cells. (A) Rh2 inhibited the luciferase activity of E2F3a-3′-UTR. The activity of luciferase reporter containing the E2F3a-3′-UTR sequence was measured by TECAN Genios multifunctional microplate reader. The cells were transfected with 50 nmol/L E2F3a 3′-UTR plasmid or empty E2F3a 3′-UTR plasmid for 48 h using Lipofectamine 2000 (Invitrogen). After transfection, the cells were treated with Rh2 for 24 h, and the luciferase assay was performed. The experiment was performed three times, each in triplicate. Mean±SEM. bP<0.05, cP<0.01 compared to E2F3a 3′-UTR plasmid alone. (B) Transfection of miR-128 inhibitor prevented Rh2-induced inhibition of E2F3a protein expression in U251 cells, as compared with Rh2+miR-128 inhibitor control. The cells were transfected with 50 nmol/L miR-128 inhibitor for 48 h and then with 12 μg/mL Rh2 for 24 h. β-actin was used as a loading control in Western blot. bP<0.05 as compared to control or Rh2+miR-128 inhibitor.
Discussion
Many miRNAs have been reported to have an oncogenic or a tumor suppressor function and to be involved in cell proliferation, growth and apoptosis11, 30. However, little is known about the ralationship between miRNA expression and Rh2-induced anti-proliferation. The present study demonstrated that Rh2, a natural glycoside from the Ginseng, altered miRNA expression in human glioma U251 cells, including 14 up-regulated and 12 down-regulated miRNAs. We verified that miR-128 was up-regulated in human U251, T98MG and A172 glioma cells by quantitative real-time PCR. Among the miRNAs regulated by Rh2, some have been reported to be tumor suppressors. For instance, miR-128 and miR-181 function as tumor suppressors and trigger growth inhibition in human glioma cells. In addition, knockdown or reduction of miR-21 led to glioma cell apoptosis and cell growth inhibition, reduced invasiveness and suppressed tumorigenicity31. Our findings indicate that miRNAs play important roles in Rh2-induced antiproliferation in glioma cells.
miR-128 is a brain-enriched miRNA whose overexpression inhibits glioma cell proliferation17. In our study, miR-128 was up-regulated by Rh2. To further investigate miR-128's role in Rh2-induced cytotoxicity, a miR-128 inhibitor was used to diminish miR-128 overexpression induced by Rh2 and led to an increase in U251 cell proliferation. Our study demonstrated that Rh2 could inhibit the proliferation of glioma U251 cells in a dose- and time-dependent manner. Moreover, we found that miR-128 inhibitor significantly inhibited miR-128 up-regulation and restored cell viability in Rh2-treated cells, implying that miR-128 overexpression is one of the principal mechanisms by which Rh2 mediates anti-proliferation in U251 cells.
Activation of apoptotic pathways is a key mechanism by which chemotherapeutic drugs kill cancer cells. Rh2 has been reported to induce apoptosis in glioma cells28. Here, we demonstrated that Rh2 could induce apoptosis of glioma U251 cells in a dose- and time-dependent manner, illustrated by annexin V and propidium iodide staining. Moreover, we found that treatment with a miR-128 inhibitor markedly inhibited Rh2-induced apoptosis in these cells. Previous studies have shown that Rh2 could induce apoptosis through activation of the caspase pathway in human neuroblastoma28. Consistent with the previous finding, our results showed that Rh2 significantly increased caspase 3 activity in U251 cells. We also found that knockdown of miR-128 expression prevented Rh2-induced caspase 3 activation. These results suggest that miR-128-mediated apoptosis is a key mechanism in Rh2-induced cytotoxicity in U251 cells.
In addition, our results showed that knockdown of Rh2-induced miR-128 overexpression by transfection of miR-128 inhibitor only partially prevented Rh2-induced anti-proliferation in glioma U251 cells. However, in our initial experiments, we transfected a miR-128 mimic into U251 cells and observed a reduced proliferation of the cells. In cells treated with a miR-128 mimic in addition to Rh2, the compound did have an anti-proliferation effect, although there was no significant difference compared with miR-128 mimic control (data not shown). These results suggest that there may be additional miRNAs that also mediate Rh2-induced cytotoxicity in glioma cells. For instance, miR-21 has been found to function as an oncogene and to be abundant and up-regulated in glioma cells32, 33. Taken together with our findings, these results suggest that the fold change values of miR-21 was larger than those of miR-128. In fact, the expression levels of miR-128 were lower than those of miR-21 in our microarray results. Therefore, miR-21 appears to be a better candidate for study. In the present study, we focused on miR-128 because it is a brain-enriched miRNA and has strong anti-proliferative roles in glioma development. However, our results do not exclude the involvement of miR-21 in Rh2-induced cytotoxicity. Further studies are needed to investigate the molecular mechanism of miR-21 in Rh2-induced anti-proliferative effects.
Computational algorithms showed several hundreds of putative targets of miR-128 (http://www.microrna.org). A recent study also showed that the transcription factor E2F3a is a direct target of miR-12817. Considering that overexpression of miR-128 can inhibit proliferation of glioma cells by targeting E2F3a, we wondered whether Rh2 could affect E2F3a via induction of miR-128 overexpression. We found that Rh2 inhibited E2F3a protein expression using Western blotting, an effect that was reversed following treatment with an miR-128 inhibitor. These results suggest that Rh2-induced inhibition of glioma cell proliferation may be mediated by negatively regulating the expression of E2F3a, a miR-128 target gene. E2F3a has been found to stimulate cell proliferation, apoptosis and carcinogenesis in a transgenic mouse model34. E2F3a protein expression negatively correlated with the expression level of miR-128 in glioma cells17. The sequence of miR-128 and its target site in the 3′-UTR of E2F3a is shown in Figure 9, as predicted by Miranda software (http://www.microrna.org). The 5′-end of miR-128 is complementary to the 3′-UTR of E2F3a, which mediates post-transcriptional negative regulation of E2F3a via RNA duplex formation.
Figure 9.
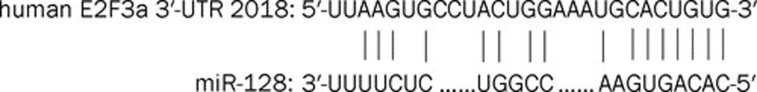
Sequence of miR-128 and its target site in the 3′-UTR of E2F3a.
Except for E2F3a, miR-128 has been reported to exert its anti-proliferative and stem cell self-renewal effects in glioma cells through targeting Bmi-1, a stem cell renewal factor33. In our study, we could not investigate the effect of Rh2 on Bmi-1 and stem cell self-renewal effects in glioma cells. Therefore, it would be interesting to examine the effects of Rh2 has on stem cell self-renewal through miR-128-mediated Bmi-1 pathway in human glioma neurosphere cultures that possess features of glioma “stem-like” cells.
In conclusion, the present study shows that ginsenoside Rh2 exerts its anti-proliferative effect in U251 glioma cells in part by up-regulation of miRNA-128 expression. This finding provides new insight into the understanding of the molecular mechanism of Rh2-mediated cytotoxicity.
Author contribution
Hua FENG and Nan WU designed research; Nan WU and Guo-cai WU performed research; Rong HU and Mei LI contributed new analytical tools and reagents; Rong HU and Mei LI analyzed data; and Nan WU wrote the paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No 30872660).
References
- Franceschi E, Tosoni A, Bartolini S, Mazzocchi V, Fioravanti A, Brandes AA. Treatment options for recurrent glioblastoma: pitfalls and future trends. Expert Rev Anticancer Ther. 2009;9:613–9. doi: 10.1586/era.09.23. [DOI] [PubMed] [Google Scholar]
- Claes A, Idema AJ, Wesseling P. Diffuse glioma growth: a guerilla war. Acta Neuropathol. 2007;114:443–58. doi: 10.1007/s00401-007-0293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang JC, Kwok WK, Chen Z, Ng HK. Oncogenic role of microRNAs in brain tumors. Acta Neuropathol. 2009;117:599–611. doi: 10.1007/s00401-009-0525-0. [DOI] [PubMed] [Google Scholar]
- Mendes ND, Freitas AT, Sagot MF. Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res. 2009;37:2419–33. doi: 10.1093/nar/gkp145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova J, Slaby O, Vyzula R, Michalek J. MicroRNA involvement in glioblastoma pathogenesis. Biochem Biophys Res Commun. 2009;386:1–5. doi: 10.1016/j.bbrc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Hagen JW, Lai EC. microRNA control of cell-cell signaling during development and disease. Cell Cycle. 2008;7:2327–32. doi: 10.4161/cc.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–41. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlhut Espinosa CE, Slack FJ. The role of microRNAs in cancer. Yale J Biol Med. 2006;79:131–40. [PMC free article] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu W, Chao T, Zhang Y, Yan X, Gong Y, et al. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272:197–205. doi: 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, et al. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100:1234–42. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- Xia H, Qi Y, Ng SS, Chen X, Chen S, Fang M, et al. MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem Biophys Res Commun. 2009;380:205–10. doi: 10.1016/j.bbrc.2008.12.169. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan X, et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med. 2009;87:43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]
- Shibata S. Chemistry and cancer preventing activities of ginseng saponins and some related triterpenoid compounds. J Korean Med Sci. 2001. pp. S28–37. [DOI] [PMC free article] [PubMed]
- Lai DM, Tu YK, Liu IM, Chen PF, Cheng JT. Mediation of beta-endorphin by ginsenoside Rh2 to lower plasma glucose in streptozotocin-induced diabetic rats. Planta Med. 2006;72:9–13. doi: 10.1055/s-2005-916177. [DOI] [PubMed] [Google Scholar]
- Park EK, Choo MK, Oh JK, Ryu JH, Kim DH. Ginsenoside Rh2 reduces ischemic brain injury in rats. Biol Pharm Bull. 2004;27:433–6. doi: 10.1248/bpb.27.433. [DOI] [PubMed] [Google Scholar]
- Park EK, Choo MK, Kim EJ, Han MJ, Kim DH. Antiallergic activity of ginsenoside Rh2. Biol Pharm Bull. 2003;26:1581–4. doi: 10.1248/bpb.26.1581. [DOI] [PubMed] [Google Scholar]
- Kim HE, Oh JH, Lee SK, Oh YJ. Ginsenoside RH-2 induces apoptotic cell death in rat C6 glioma via a reactive oxygen- and caspase-dependent but Bcl-X(L)-independent pathway. Life Sci. 1999;65:PL33–40. doi: 10.1016/s0024-3205(99)00252-0. [DOI] [PubMed] [Google Scholar]
- Zeng XL, Tu ZG. In vitro induction of differentiation by ginsenoside Rh2 in SMMC-7721 hepatocarcinoma cell line. Pharmacol Toxicol. 2003;93:275–83. doi: 10.1111/j.1600-0773.2003.pto930605.x. [DOI] [PubMed] [Google Scholar]
- Cheng CC, Yang SM, Huang CY, Chen JC, Chang WM, Hsu SL. Molecular mechanisms of ginsenoside Rh2-mediated G1 growth arrest and apoptosis in human lung adenocarcinoma A549 cells. Cancer Chemother Pharmacol. 2005;55:531–40. doi: 10.1007/s00280-004-0919-6. [DOI] [PubMed] [Google Scholar]
- Wen XY, Wu SY, Li ZQ, Liu ZQ, Zhang JJ, Wang GF, et al. Ellagitannin (BJA3121), an anti-proliferative natural polyphenol compound, can regulate the expression of MiRNAs in HepG2 cancer cells. Phytother Res. 2009;23:778–84. doi: 10.1002/ptr.2616. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li J, Gu J, Huang B, Zhao Y, Zheng D, et al. Potentiation of (–)-epigallocatechin-3-gallate-induced apoptosis by bortezomib in multiple myeloma cells. Acta Biochim Biophys Sin (Shanghai) 2009;41:1018–26. doi: 10.1093/abbs/gmp094. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Liu WD, Yang HZ, Zhang Y, Fang ZG, Liu PQ, et al. Inactivation of PI3k/Akt signaling pathway and activation of caspase-3 are involved in tanshinone I-induced apoptosis in myeloid leukemia cells in vitro. Ann Hematol. 2010;89:1089–97. doi: 10.1007/s00277-010-0996-z. [DOI] [PubMed] [Google Scholar]
- Kim YS, Jin SH. Ginsenoside Rh2 induces apoptosis via activation of caspase-1 and -3 and up-regulation of Bax in human neuroblastoma. Arch Pharm Res. 2004;27:834–9. doi: 10.1007/BF02980175. [DOI] [PubMed] [Google Scholar]
- Kim YS, Jin SH, Lee YH, Kim SI, Park JD. Ginsenoside Rh2 induces apoptosis independently of Bcl-2, Bcl-xL, or Bax in C6Bu-1 cells. Arch Pharm Res. 1999;22:448–53. doi: 10.1007/BF02979151. [DOI] [PubMed] [Google Scholar]
- Hagen JW, Lai EC. microRNA control of cell-cell signaling during development and disease. Cell Cycle. 2008;7:2327–32. doi: 10.4161/cc.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhang J, Jia Q, Ren Y, Wang Y, Shi L, et al. Reduction of miR-21 induces glioma cell apoptosis via activating caspase 9 and 3. Oncol Rep. 2010;24:195–201. doi: 10.3892/or_00000846. [DOI] [PubMed] [Google Scholar]
- Shi L, Chen J, Yang J, Pan T, Zhang S, Wang Z. MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 2010;1352:255–64. doi: 10.1016/j.brainres.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–30. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- Paulson QX, McArthur MJ, Johnson DG. E2F3a stimulates proliferation, p53-independent apoptosis and carcinogenesis in a transgenic mouse model. Cell Cycle. 2006;5:184–90. doi: 10.4161/cc.5.2.2307. [DOI] [PubMed] [Google Scholar]