Abstract
Aim:
To investigate the relationship of fibroblast growth factor receptor 4 (FGFR4) gene polymorphisms with the response of Chinese patients with non-small cell lung cancer (NSCLC) to chemotherapy.
Methods:
A total of 629 patients with Stage III (A+B) or IV NSCLC, as well as 729 age- and gender-matched healthy controls were recruited. All the patients received platinum-based chemotherapy, and the therapeutic effects were evaluated. Three polymorphisms in the FGFR4 gene (rs351855G/A, rs145302848C/G, and rs147603016G/A) were genotyped, and the association between the 3 polymorphisms and the chemotherapy effect was analyzed using SPSS software, version 16.0.
Results:
The genotype frequencies of rs145302848C/G and rs147603016G/A were not significantly different between NSCLC patients and healthy controls on one hand, and between the responders and non-responders to the chemotherapy on the other hand. The distribution of AA genotype and A-allele of rs351855G/A was significantly lower in NSCLC patients than in healthy controls. Using patients with the GG genotype as a reference, the AA carrier had a significantly reduced risk for the development of NSCLC after normalizing to age, sex and smoking habits. In NSCLC patients, this genotype occurred more frequently in the responders to the chemotherapy than in non-responders. The chance of being a responder was significantly increased with the AA genotype as compared to G genotype. The AA genotype of rs351855G/A had a better prognosis compared with GA and GG genotype carriers: the overall survival of patients with the AA genotype of rs351855G/A was significantly longer than those with the GG+GA genotype (21.1 vs 16.5 months).
Conclusion:
The rs351855G/A polymorphisms of FGFR4 gene can be used to predict the occurrence, chemotherapy response and prognosis of NSCLC.
Keywords: non-small cell lung cancer, platinum-based chemotherapy, fibroblast growth factor receptor 4, gene polymorphism, response to chemotherapy, prognosis, Chinese population
Introduction
Lung cancer remains a leading cause of cancer death worldwide. Despite recent improvements in treatment, the prognosis remains very poor. Platinum-based chemotherapy is the standard first-line chemotherapy treatment for advanced non-small cell lung cancer (NSCLC). However, drug resistance is a major problem during chemotherapy1,2,3. In clinical practice, the chemotherapy response is quite different among patients4. Recent studies have showed that the genetic factor is a major determinant for the chemotherapy response in NSCLC patients5,6,7,8. Some genes have been reported to be associated with the response to chemotherapy in NSCLC patients. However, the ideal candidate gene predicting the chemotherapy response has not been identified.
Fibroblast growth factor receptor 4 (FGFR4) belongs to the tyrosine kinase receptor family, and its activation mediates cell proliferation, survival, migration, and resistance to apoptosis9,10. FGFR4 is differentially expressed in different tissues and cell types and has distinct differences in receptor functions11,12. Several genetic polymorphisms were identified in the FGFR4 gene, such as rs351855G/A (Gly388Arg), which is a nonsynonymous SNP located in the transmembrane domain of the fgfr4 protein. The rs145302848C/G and rs147603016G/A polymorphisms are both located in the 5′-upstream region of FGFR4, which indicates that they may potentially regulate expression13,14,15,16. However, it is currently unknown whether the polymorphisms of FGFR4 affect the response to chemotherapy in NSCLC patients.
Materials and methods
Patients
A total of 629 patients with Stage III (A+B) or IV NSCLC that was confirmed cytologically or histologically were enrolled into this study. To avoid the potential influence of poor clinical conditions on chemotherapy response, other eligibility criteria were included as follows: normal blood chemistry (hemoglobin >9 g/dL, neutrophil count >1500/mm3 and platelet count >100 000/mm3), hepatic function (bilirubin <1.5 times the normal upper limit, aspartate aminotransferase and alanine aminotransferase <2 times the normal upper limit) and renal function (creatine clearance rate >50 mL/s), and a normal electrocardiogram at the beginning of treatment. The exclusion criteria included symptomatic brain metastases, spinal cord compression, uncontrolled massive pleural effusion, previously received chemotherapy and other chronic disease. A total of 729 age- and gender-matched healthy controls were recruited from the physical examination center of Sir Run Run Shaw Hospital (Hangzhou, China). The controls were free of any cancer, respiratory system disease or other chronic disease. The study was approved under the ethics committees of Sir Run Run Shaw Hospital, and written informed consent was obtained from each participant. One hundred and nine patients were followed up for the overall survival period. The patients involved in the follow-up population were selected randomly, although patients who accepted a second-line target therapy were excluded.
Chemotherapy regimens and therapeutic effect evaluation
All of the patients received the previously described platinum-based chemotherapy. All of the chemotherapeutic drugs were administered intravenously, and the treatment cycles were repeated every 3–4 weeks. Patient responses to the treatment were determined after four cycles by the WHO criteria, which classified the response into four categories: complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). CR was defined as a complete disappearance of all measurable lesions. PR required at least a 50% reduction in measurable lesions. Patients with SD had less than a 50% decrease or no more than a 25% increase in the size of measurable lesions. PD was assigned to patients when the measurable lesions increased by more than 25% or when new lesions appeared. For data analysis, CR and PR were combined as responders and SD and PD were grouped as non-responders.
Genotyping analyses
Genomic DNA was extracted from the peripheral blood lymphocytes. The primers were as follows: rs351855-F: 5′-GACCGCAGCAGCGCCCGAGGCCAG-3′, R: 5′-AGAGGGAAGAGGGAGAGCTTCTG-3′ rs145302848-F: 5′-CAGAGGAGGACCCCACATG-3′, R: 5′-TGGAGTCAGGCTGTCACATG-3′ rs147603016-F: 5′-CCGCAGCAGCGCCCGAGGCC-3′, R: 5′-GAAGCGGGAGAGCTTCTGC-3′. PCR products containing the three polymorphic sites were then sequenced using a 3130 sequence platform (Applied Biosystems, Foster City, CA, USA) and the conditions recommended in the manufacturer's instructions. The sequence data were read with the DNSstar software.
Statistical analyses
The main endpoint of the study was overall survival, which was calculated from the start of the treatment to the date of the last follow-up or death. The tumor response was evaluated every 2 cycles of chemotherapy according to the Response Evaluation Criteria in Solid Tumor Group (RECIST). Demographic and clinical variables were compared across the genotypes, using Fisher's exact test or Pearson's χ2 test. A Bonferroni test was used to correct the result of multiple tests. Survival curves were plotted using the Kaplan-Meier method and were compared with the log-rank test. The association between each genotype and the survival rate was estimated by computing the hazard ratios and their 95% confidence intervals by multivariate Cox's regression models, where the most frequent allele was assumed to be the reference. Statistical significance was set at P=0.05. All of the tests were two-sided, and the analyses were conducted with SPSS (Statistical Package for the Social Sciences) software, version 16.0.
Results
Patient characteristics are shown in Table 1. The mean age and gender were similar between NSCLC and controls. The NSCLC group had a higher prevalence of smokers than the control group. In total, 311 patients were identified to have adenocarcinomas and 318 to have squamous cell carcinomas. For cancer staging, 331 patients had stage III disease and 298 had stage IV diseases. Of these patients, 219 had well differentiated tumor while 360 had moderately differentiated tumors and 269 had poorly differentiated tumors.
Table 1. Characteristic of patients.
| Characteristics | NSCLC | Control | Pa | ||
|---|---|---|---|---|---|
| Age (years) | 67.2±5.3 | 67.4±5.7 | NSb | ||
| Gender | |||||
| Male | 378 | 60.10% | 437 | 59.95% | NSb |
| Female | 251 | 39.90% | 292 | 40.05% | |
| Smoke status | |||||
| Non-smokers | 311 | 49.44% | 276 | 37.86% | <0.001 |
| Smoker | 318 | 50.56% | 453 | 62.14% | |
| Histology | |||||
| Squamous cell carcinoma | 318 | 46.73% | |||
| Adenocarcinoma | 311 | 53.27% | |||
| Stage | |||||
| III | 331 | 52.62% | |||
| IV | 298 | 47.38% | |||
| Differentiation | |||||
| Well | 219 | 46.73% | |||
| Moderate | 360 | 37.38% | |||
| Poor | 269 | 15.89% | |||
| Chemotherapy regimens | |||||
| DDP/CBP+TAX/TXT/DOC | 268 | 46.73% | |||
| DDP/CBP+GEM | 253 | 37.38% | |||
| DDP/CBP+NVB | 108 | 15.89% | |||
aBy fisher exact T test.
bNS: not significant.
The genotype frequencies of the FGFR4 gene in NSCLC and controls are presented in Table 2. The genotype frequencies of all SNPs in the controls were in Hardy-Weinberg equilibrium (P>0.05). The genotype frequencies of rs145302848C/G and rs147603016G/A were not significantly different between NSCLC patients and controls. For rs351855G/A polymorphisms, NSCLC patients had a lower prevalence of AA than control subjects (P<0.001). For allele comparison, NSCLC subjects had lower A-allele frequency than controls as well (P<0.001). With GG as a reference, the AA carriers had a significantly reduced risk for the development of NSCLC after normalizing to age, gender, and smoking habit (OR=0.507, P<0.001). Using the G allele as a reference, the OR for the A-allele carriers was 0.730 (P<0.001) (Table 1).
Table 2. The genotype frequencies of FGFR gene in NSCLC and controls.
| NSCLC | Control | Adjusted | 95% CI | Adjusted | |||||
|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | OR | P valuea,b | ||||
| rs351855G/A | GG | 193 | 30.68 | 163 | 22.36 | Ref | |||
| GA | 331 | 52.62 | 391 | 53.64 | 0.715 | 0.554 | 0.922 | 0.030 | |
| AA | 105 | 16.69 | 175 | 24.01 | 0.507 | 0.368 | 0.697 | 0.000 | |
| G | 717 | 57.00 | 717 | 49.18 | Ref | ||||
| A | 541 | 43.00 | 741 | 50.82 | 0.730 | 0.627 | 0.850 | 0.000 | |
| rs145302848C/G | CC | 163 | 25.91 | 179 | 24.55 | Ref | |||
| CG | 320 | 50.87 | 365 | 50.07 | 0.963 | 0.742 | 1.249 | 0.775 | |
| GG | 146 | 23.21 | 185 | 25.38 | 0.867 | 0.640 | 1.174 | 0.435 | |
| C | 646 | 51.35 | 723 | 49.59 | Ref | ||||
| G | 612 | 48.65 | 735 | 50.41 | 0.932 | 0.801 | 1.084 | 0.360 | |
| rs147603016G/A | GG | 167 | 26.55 | 189 | 25.93 | Ref | |||
| GA | 304 | 48.33 | 366 | 50.21 | 0.940 | 0.726 | 1.216 | 0.638 | |
| AA | 158 | 25.12 | 174 | 23.87 | 1.028 | 0.762 | 1.387 | 0.508 | |
| G | 638 | 50.72 | 744 | 51.03 | Ref | ||||
| A | 620 | 49.28 | 714 | 48.97 | 1.013 | 0.871 | 1.178 | 0.871 | |
aBinary logistic regression was applied. Data was adjusted with gender, age, tumor histology, disease stage, and chemotherapy regimen.
bCorrected with Bonferoni correction.
All of the patients who received platinum-based chemotherapy were subdivided into responder and non-responder groups. Of these patients, 291 (46.3%) responded to chemotherapy (CR+PR) and 338 (53.7%) showed no response to chemotherapy (SD+PD). The polymorphic genotypes and the allele frequency of rs145302848C/G and rs147603016G/A were not significantly different between the patients who responded and those who did not respond to the platinum-based treatment (both P>0.05). However, for rs351855G/A, the genotypes and allele frequency were substantially different between the treated responders and non-responders. The AA genotype distribution was significantly higher in responders than that in non-responders (P=0.003). The A-allele frequency was also significantly higher in responders than that in non-responders (P=0.001). A logistic regression analysis showed that patients of the AA genotype had a significantly increased chance of responding to the treatment compared with the patients of the GG genotype [OR=2.86 (1.72–4.76), 95% CI, P<0.001] after normalizing for gender, age, tumor histology, disease stage, and chemotherapy regimens (Table 3). The OR for the A allele carriers was 1.40 compared with the G allele carriers (P=0.003).
Table 3. The chemotherapy response according to the rs351855G/A genotypes.
| NSCLC | Control | Adjusted | 95% CI | Adjusted | |||||
|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | OR | P valuea | ||||
| rs351855G/A | GG | 90 | 30.93 | 103 | 30.47 | 1.00 | |||
| GA | 126 | 43.30 | 205 | 60.65 | 0.70 | 0.49 | 1.01 | 0.055 | |
| AA | 75 | 25.77 | 30 | 8.88 | 2.86 | 1.72 | 4.76 | <0.001 | |
| G | 306 | 52.58 | 411 | 60.80 | 1.00 | ||||
| A | 276 | 47.42 | 265 | 39.20 | 1.40 | 1.12 | 1.75 | 0.003 | |
aBinary logistic regression was applied; data was adjusted with gender, age, tumor histology, disease stage, and chemotherapy regimen.
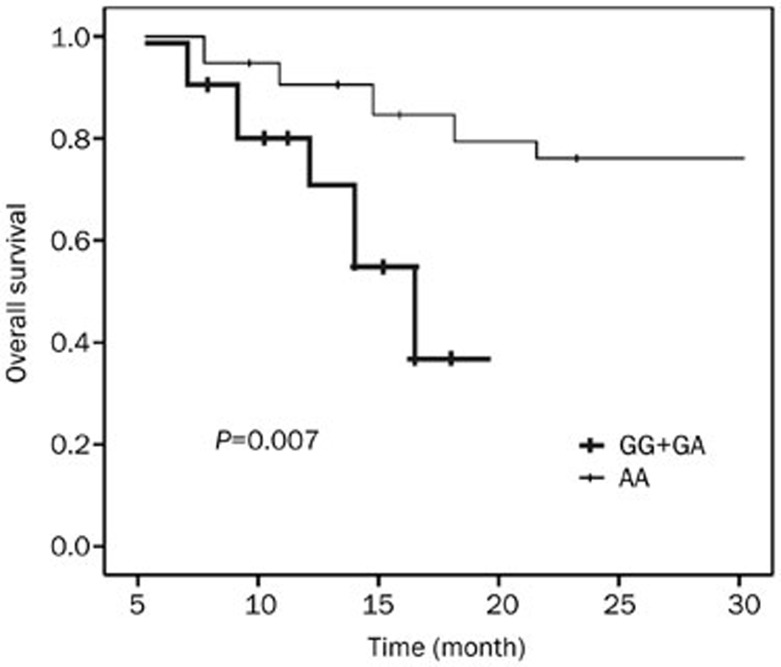
The distribution of three polymorphisms in a select population and the ratio of responders/non-responders were not different significantly from the whole NSCLC population. For the overall survival, the median follow-up duration for all the 109 cohort patients was 18.6 months (range 6-30 months). Log-rank analyses showed that the overall survival in patients with the AA genotype of rs351855G/A was significantly longer than that in those with the GG+GA genotype (21.1 vs 16.5 months, P=0.007, Figure 1). The overall survival time in carriers of the rs145302848C/G and rs147603016G/A polymorphisms was similar (both P>0.05). Multivariate Cox proportional hazards regression models were performed to estimate the hazard ratios (HR) for overall survival and 95% CIs, adjusting for age, gender, smoking habit, histology, stage, and chemotherapy status. The HR for the AA genotype of rs351855G/A was 0.572 (95% CI: 0.33–0.98) compared with GG+GA carriers (P=0.007). Besides rs351855G/A polymorphisms, the other risk factors influencing prognosis included smoking (HR=1.562, 95% CI: 1.114–2.784, P=0.002), chemotherapy response (HR=1.562, 95% CI: 1.114–2.784, P=0.002) and cancer stage (HR=1.896, 95%CI: 1.312–2.765, P=0.003).
Figure 1.
Survival curve of NSCLC patients, divided with genotype of rs351855. Compared with GG+GA group, survival of AA group was significantly longer, with P=0.007.
Discussion
This study investigated the role of FGFR4 genetic polymorphisms in the susceptibility and chemotherapy response in advanced-stage NSCLC patients who underwent platinum-based chemotherapy treatment. Among the three tested FGFR4 genetic polymorphisms, we found that the rs351855G/A was significantly associated with the risk of developing NSCLC. In addition, this rs351855G/A also determined the chemotherapy response and prognosis. To the best of our knowledge, no study has reported the association of FGFR4 genetic polymorphisms with chemotherapy response in NSCLC patients. The results of this study suggest that the rs351855G/A polymorphisms can predict the occurrence, chemotherapy response and prognosis of NSCLC.
The polymorphism rs351855 in the FGFR4 gene encodes FGFR4 containing either glycine (Gly, GGG) or arginine (Arg, AGG) at codon 38817. Compared with the Gly388 variant, the Arg388 variation in the transmembrane domain (exon 9) of the FGFR4 protein results in FGFR4 stabilization and increases phosphorylation after ligand binding18.
Rs351855 has been shown to increase the susceptibility of developing several types of autoimmune, inflammatory, or vascular disease including ischemic stroke19, coronary artery disease20, and irritable bowel syndrome21. Hepatocellular carcinoma has been linked to rs351855G22. However, a European population-based meta-analysis indicated that the A-allele was a risk factor for cancer23. Moreover, in a basic in vitro study, both alleles of rs351855 were required for function: the G-allele affects tumor growth, and the A-allele affects migration24. In the current study, our data support the potential risks associated with the G-allele polymorphism, which is consistent with the previous Chinese population-based hepatocellular carcinoma study. It remains unclear whether the ethnic difference or the double mutation caused the inconsistent result. Additional basic molecular evidence is required, or a much larger population-based study should be conducted.
The FGFR4 gene polymorphisms have been associated with disease progression and poor prognosis in several tumor types, including colon, breast, prostate, head and neck, soft tissue sarcomas, melanoma and lung cancer25,26,27,28. A polymorphism at rs351855 in the FGFR4 gene has also been associated with chemotherapy in some types of cancers. In a study of patients receiving neoadjuvant chemotherapy for primary breast cancer, FGFR4 Arg388 predicted a pathologic complete response. In the same study, the FGFR4 Arg388 mutation was associated with a higher risk of axillary lymph node involvement, which is consistent with a well-known paradox of poor prognostic factors predicting a good response to neoadjuvant chemotherapy29. The FGFR4 388Arg allele also predicts prolonged survival and platinum sensitivity in patients with FIGO Stage II–IV epithelial ovarian cancer17.
In the present study, we determined that the effects of FGFR4 genetic polymorphisms could also be extended to NSCLC patients, although the detailed mechanism still needs to be explored. The most significant goal of our study was to provide a potential molecular marker to predict the response of NSCLC patients to chemotherapy. However, in a previous study that involved 465 patients and a 5-year follow-up, Matakidou et al found that there was little evidence to suggest that the Gly388Arg polymorphism of FGFR4 represented a robust marker of lung cancer pathological parameters and prognosis24. This discrepancy might be due to the different subjects that were enrolled. We enrolled only NSCLC patients; however, Matakidou et al also enrolled SCLC patients. Additionally, the ethnic differences between Japanese and Chinese populations may also produce inconsistent results.
Several limitations in this study need to be addressed. This study was a single-center cohort investigation on a relatively small scale, and thus, replication studies with large independent cohorts are warranted. Second, we did not detect the FGFR4 protein in either serum or cancer tissue of NSCLC subjects. The detection of FGFR4 expression would help to clarify the effect of the genetic variations of FGFR4 on the chemotherapy response. Third, the size of the tested population was not big enough to ensure a detailed analysis of the different treatments. We believe that this type of clinical data mining would be helpful to determine the mechanism of FGFR4.
Author contribution
Hong-mei FANG and Ying-zhi FANG designed the study and wrote the manuscript; Li-juan ZHOU and Han-ying ZHOU conducted the sample recruitment and genotype experiment; and Gang TIAN conducted the data analysis.
References
- Watanabe H, Yamamoto S, Kunitoh H, Sekine I, Yamamoto N, Ohe Y, et al. Tumor response to chemotherapy: the validity and reproducibility of RECIST guidelines in NSCLC patients. Cancer Sci. 2003;94:1015–20. doi: 10.1111/j.1349-7006.2003.tb01394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima E, Katou H. Adjuvant chemotherapy for resectable non-small cell lung cancer (NSCLC) Kyobu Geka. 2008;61:4–8. [PubMed] [Google Scholar]
- Perng RP, Yang CH, Chen YM, Chang GC, Lin MC, Hsieh RK, et al. High efficacy of erlotinib in Taiwanese NSCLC patients in an expanded access program study previously treated with chemotherapy. Lung Cancer. 2008;62:78–84. doi: 10.1016/j.lungcan.2008.02.023. [DOI] [PubMed] [Google Scholar]
- Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer. 2011;71:3–10. doi: 10.1016/j.lungcan.2010.08.022. [DOI] [PubMed] [Google Scholar]
- Shiraishi K, Kohno T, Tanai C, Goto Y, Kuchiba A, Yamamoto S, et al. Association of DNA repair gene polymorphisms with response to platinum-based doublet chemotherapy in patients with non-small-cell lung cancer. J Clin Oncol. 2010;28:4945–52. doi: 10.1200/JCO.2010.30.5334. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Ligorio C, Toschi L, Rossi E, Trisolini R, Paioli D, et al. EGFR and HER2 gene copy number and response to first-line chemotherapy in patients with advanced non-small cell lung cancer (NSCLC) J Thorac Oncol. 2007;2:423–9. doi: 10.1097/01.JTO.0000268676.79872.9b. [DOI] [PubMed] [Google Scholar]
- Wu J, Liu J, Zhou Y, Ying J, Zou H, Guo S, et al. Predictive value of XRCC1 gene polymorphisms on platinum-based chemotherapy in advanced non-small cell lung cancer patients: A systematic review and meta-analysis. Clin Cancer Res. 2012;18:3972–81. doi: 10.1158/1078-0432.CCR-11-1531. [DOI] [PubMed] [Google Scholar]
- Rosell R, Felip E, Taron M, Majo J, Mendez P, Sanchez-Ronco M, et al. Gene expression as a predictive marker of outcome in stage IIB-IIIA–IIIB non-small cell lung cancer after induction gemcitabine-based chemotherapy followed by resectional surgery. Clin Cancer Res. 2004;10:4215s–4219s. doi: 10.1158/1078-0432.CCR-040006. [DOI] [PubMed] [Google Scholar]
- Galzie Z, Kinsella AR, Smith JA. Fibroblast growth factors and their receptors. Biochem Cell Biol. 1997;75:669–85. [PubMed] [Google Scholar]
- Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer. 2000;7:165–97. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J. 2011;437:199–213. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- Kornmann M, Beger HG, Korc M. Role of fibroblast growth factors and their receptors in pancreatic cancer and chronic pancreatitis. Pancreas. 1998;17:169–75. doi: 10.1097/00006676-199808000-00010. [DOI] [PubMed] [Google Scholar]
- Spinola M, Leoni VP, Tanuma J, Pettinicchio A, Frattini M, Signoroni S, et al. FGFR4 Gly388Arg polymorphism and prognosis of breast and colorectal cancer. Oncol Rep. 2005;14:415–9. [PubMed] [Google Scholar]
- Falvella FS, Frullanti E, Galvan A, Spinola M, Noci S, De Cecco L, et al. FGFR4 Gly388Arg polymorphism may affect the clinical stage of patients with lung cancer by modulating the transcriptional profile of normal lung. Int J Cancer. 2009;124:2880–5. doi: 10.1002/ijc.24302. [DOI] [PubMed] [Google Scholar]
- Ho CK, Anwar S, Nanda J, Habib FK. FGFR4 Gly388Arg polymorphism and prostate cancer risk in Scottish men. Prostate Cancer Prostatic Dis. 2010;13:94–6. doi: 10.1038/pcan.2009.49. [DOI] [PubMed] [Google Scholar]
- Xu B, Tong N, Chen SQ, Hua LX, Wang ZJ, Zhang ZD, et al. FGFR4 Gly388Arg polymorphism contributes to prostate cancer development and progression: a meta-analysis of 2618 cases and 2305 controls. BMC Cancer. 2011;11:84. doi: 10.1186/1471-2407-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marme F, Hielscher T, Hug S, Bondong S, Zeillinger R, Castillo-Tong DC, et al. Fibroblast growth factor receptor 4 gene (FGFR4) 388Arg allele predicts prolonged survival and platinum sensitivity in advanced ovarian cancer. Int J Cancer. 2012;131:E586–91. doi: 10.1002/ijc.27329. [DOI] [PubMed] [Google Scholar]
- Wang J, Yu W, Cai Y, Ren C, Ittmann MM. Altered fibroblast growth factor receptor 4 stability promotes prostate cancer progression. Neoplasia. 2008;10:847–56. doi: 10.1593/neo.08450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HF, Zhao KJ, Yang PF, Fang YB, Zhang YH, Liu JM, et al. Association between fibroblast growth factor receptor 4 Gly388Arg polymorphism and ischaemic stroke. J Int Med Res. 2012;40:1708–14. doi: 10.1177/030006051204000509. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Liu T. Fibroblast growth factor receptor 4 polymorphisms and coronary artery disease: a case control study. Mol Biol Rep. 2012;39:8679–85. doi: 10.1007/s11033-012-1723-8. [DOI] [PubMed] [Google Scholar]
- Wong BS, Camilleri M, Carlson PJ, Odunsi-Shiyanbade S, McKinzie S, Busciglio I, et al. Pharmacogenetics of the effects of colesevelam on colonic transit in irritable bowel syndrome with diarrhea. Dig Dis Sci. 2012;57:1222–6. doi: 10.1007/s10620-012-2035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhou Y, Lu M, An Y, Li R, Chen Y, et al. Association between fibroblast growth factor receptor 4 polymorphisms and risk of hepatocellular carcinoma. Mol Carcinog. 2012;51:515–21. doi: 10.1002/mc.20805. [DOI] [PubMed] [Google Scholar]
- Frullanti E, Berking C, Harbeck N, Jezequel P, Haugen A, Mawrin C, et al. Meta and pooled analyses of FGFR4 Gly388Arg polymorphism as a cancer prognostic factor. Eur J Cancer Prev. 2011;20:340–7. doi: 10.1097/CEJ.0b013e3283457274. [DOI] [PubMed] [Google Scholar]
- Heinzle C, Gsur A, Hunjadi M, Erdem Z, Gauglhofer C, Stattner S, et al. Differential effects of polymorphic alleles of FGF receptor 4 on colon cancer growth and metastasis. Cancer Res. 2012;72:5767–77. doi: 10.1158/0008-5472.CAN-11-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Tsuchiya N, Yuasa T, Inoue T, Kumazawa T, Narita S, et al. Polymorphisms of fibroblast growth factor receptor 4 have association with the development of prostate cancer and benign prostatic hyperplasia and the progression of prostate cancer in a Japanese population. Int J Cancer. 2008;123:2574–9. doi: 10.1002/ijc.23578. [DOI] [PubMed] [Google Scholar]
- Liwei L, Chunyu L, Jie L, Ruifa H. Association between fibroblast growth factor receptor-4 gene polymorphism and risk of prostate cancer: a meta-analysis. Urol Int. 2011;87:159–64. doi: 10.1159/000329069. [DOI] [PubMed] [Google Scholar]
- Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, Schmitt M, et al. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 2002;62:840–7. [PubMed] [Google Scholar]
- Spinola M, Leoni V, Pignatiello C, Conti B, Ravagnani F, Pastorino U, et al. Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J Clin Oncol. 2005;23:7307–11. doi: 10.1200/JCO.2005.17.350. [DOI] [PubMed] [Google Scholar]
- Marme F, Werft W, Benner A, Burwinkel B, Sinn P, Sohn C, et al. FGFR4 Arg388 genotype is associated with pathological complete response to neoadjuvant chemotherapy for primary breast cancer. Ann Oncol. 2010;21:1636–42. doi: 10.1093/annonc/mdq017. [DOI] [PubMed] [Google Scholar]