Abstract
Interleukin-6 (IL-6) is a pleiotropic, pro-inflammatory cytokine produced by various types of cells, including macrophages. Within the IL-6 gene promoter region, the signature binding motif of CBF1/Su(H)/Lag-1 (CSL), a key DNA-binding protein in the Notch signaling pathway, was identified and found to overlap with a consensus nuclear factor (NF)-κB-binding site. Notch signaling is highly conserved and is involved in the regulation of biological functions in immune cells. In this study, we investigated the role of Notch signaling in the regulation of the IL-6 transcript in murine macrophages. The upregulation of Notch1 protein levels and the appearance of cleaved Notch1 (Val1744) correlated well with the increased IL-6 mRNA expression levels in murine primary bone marrow-derived macrophages (BMMφ) after activation by lipopolysaccharide (LPS) together with interferon-gamma (IFN-γ). Treatment of BMMφ with the γ-secretase inhibitor IL-CHO to suppress the transduction of Notch signaling resulted in a partial decrease in the level of IL-6 mRNA and the amount of IL-6 protein produced. In contrast, the overexpression of a constitutively activated intracellular Notch1 protein (NIC) in the RAW264.7 macrophage-like cell line resulted in significantly higher IL-6 transcript expression levels than in cells transfected with the empty vector control. The NF-κB inhibitor completely abrogated IL-6 mRNA expression induced by the overexpression of NIC. Chromatin immunoprecipitation (ChIP) using an anti-Notch1 antibody demonstrated that Notch1 is associated with the IL-6 promoter in RAW264.7 cells activated by LPS/IFN-γ but not in unstimulated cells. Taken together, these results strongly suggest that Notch1 positively regulates IL-6 expression via NF-κB in activated macrophages.
Keywords: CSL, interleukin-6, macrophages, NF-κB, Notch signaling
Introduction
Interleukin-6 (IL-6) is a pleiotropic cytokine that plays a major role in the regulation of the immune system, hematopoiesis, inflammation and oncogenesis.1 It is mainly produced by cells of the immune system, including T cells, B cells and macrophages. Hyperproduction of IL-6 has been associated with various chronic diseases, such as rheumatoid arthritis and Castleman's disease.2 Therefore, IL-6 and its receptors are one of the most investigated systems for the potential therapeutic treatment of such chronic diseases.
During an acute infection, innate immune cells produce large quantities of IL-6 upon encountering microbial components that are ligands for pattern recognition receptors, such as the Toll-like receptors (TLRs), as part of the acute inflammatory response. At the transcriptional level, IL-6 mRNA expression is regulated by various transcription factors, including AP1, C/EBP and nuclear factor (NF)-κB.3, 4 The IL-6 promoter region, an approximately 1.2-kb region lying upstream (5′) of the IL-6-coding sequence, contains the elements necessary for regulating IL-6 transcription upon activation by various stimuli.5 Cooperative regulation between these transcription factors has been reported for the transcriptional regulation of IL-6.3, 4 The NF-κB family of proteins is believed to be a key regulator of IL-6 transcription, and NF-κB conserved binding motifs were identified between −73 and −64 bp upstream of the transcriptional start site in humans.6, 7 At the site of NF-κB-binding motifs, a CBF1/Su(H)/Lag-1 (CSL) protein-binding motif has been identified, and CSL has been shown to bind to this region.5, 8 CSL is a DNA-binding protein that plays a central role in the evolutionarily-conserved Notch signaling pathway. When Notch signaling is not active, CSL functions as a transcriptional repressor by recruiting transcriptional co-repressors, such as NcoR and histone deacetylase, to the promoters of target genes. Upon the activation of Notch, CSL is converted to a transcriptional activator when it forms a complex with the intracellular Notch receptor and recruits the scaffold protein (MAML) and transcriptional coactivators, such as CBP/p300, to the promoters, thereby driving the transcription of target genes.9
Previously, we and others have reported the involvement of Notch signaling in regulating inflammatory responses in macrophages upon stimulation with lipopolysaccharide (LPS) or costimulation with LPS plus interferon-gamma (IFN-γ).10, 11, 12 Upon stimulation with TLR ligands, human and murine macrophages activate Notch signaling, and this signaling in turn regulates optimal inflammatory responses, partially through the NF-κB and p38 pathways.10, 11 However, the involvement of Notch signaling and IL-6 is still not entirely understood. Originally, it was suggested that CSL acted as a negative regulator of IL-6 transcription because overexpression of a constitutively active Notch1 did not induce activation of the IL-6 promoter in a reporter assay in human non-immune cell lines.5, 8 In contrast, CSL-deficient murine macrophages produced less IL-6 upon LPS stimulation.10 Therefore, in this study, we investigated the direct involvement of Notch signaling in regulating the transcription of IL-6 in murine bone marrow-derived macrophages (BMMφ) and an established murine macrophage-like cell line. We report here that Notch1 is associated with the IL-6 promoter and that activated Notch1 is sufficient to induce IL-6 transcription via the NF-κB pathway.
Materials and methods
Primary murine macrophages and cell line
The murine macrophage-like RAW264.7 cell line (ATCC No. TIB-71) and primary BMMφ from female C57BL/6J mice (National Laboratory Animal Center, Mahidol University, Salaya, Thailand) were used in this study. All procedures involving laboratory animals were conducted according to the guidelines issued by Chulalongkorn University. The animal protocols used in this study were approved by the Animal Care and Use Committee of Faculty of Science, Chulalongkorn University (Protocol Rev. No. 1023001). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (HyClone, Logan, UT, USA) supplemented with 10% (v/v) fetal bovine serum (FBS) (HyClone), 100 U/ml penicillin (General Drugs House Co. Ltd, Bangkok, Thailand), 0.4 mg/ml streptomycin (M&H Manufacturing Co. Ltd, Samut Prakan, Thailand), 1% (w/v) sodium pyruvate (HyClone) and 1% (w/v) N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid (HyClone) at 37 °C and incubated in a humidified 5% (v/v) CO2 incubator.
Reagents
For the inhibition of Notch signaling, the γ-secretase IL-CHO (a kind gift from Professor Todd Golde, University of Florida, Gainesville, FL, USA) was used as previously described.11 Cells were pretreated with 25 µM IL-CHO (dissolved in dimethylsulfoxide (DMSO)) or the vehicle control (DMSO) for 1 h before stimulation as indicated in the figures. LPS from Salmonella (Sigma Aldrich, St Louis, MO, USA) and (IFNg) (R&D Systems, Minneapolis, MN, USA) were used for the stimulation of macrophages as indicated in the figures. BAY-11 (Merck Whitehouse Station, NJ, USA) dissolved in DMSO was used to inhibit the NF-κB signaling pathway.
Cell transfection
Overexpression of the truncated intracellular form of Notch1 (NIC) in RAW264.7 cells was achieved by using the plasmid pcDNA3 containing the intracellular Notch1 (NIC) encoding the sequence corresponding to amino acid residues 1759 to 2556 (pcDNA3NIC), with transfection of the empty pcDNA3 plasmid used as a vector control (both were kind gifts from Professor Barbara Osborne from the University of Massachusetts in Amherst, MA, USA). To eliminate endotoxin contamination, all plasmids were prepared using the Endo-free Plasmid Maxi kit (Qiagen, Hilden, Germany). Transient transfection was performed with the FuGeneHD transfection reagent (Roche, Mannheim, Germany) according to the manufacturer's instructions. The overexpression of NIC was confirmed by western blotting.
Western blotting
Cells were subjected to cell lysis, and the total protein was then extracted as previously described.13 Protein concentrations were measured using the bicinchoninic acid protein assay (Pierce, Rockford, IL, USA) according to the manufacturer's instructions. The primary antibodies used in this study were as follows: rabbit anti-Notch1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit anti-cleaved Notch1 (Val1744) antibody (Cell Signaling Technology, Beverly, MA, USA) and mouse anti-β-actin (Chemicon International, Temecula, CA, USA). The secondary antibodies used were donkey anti-rabbit IgG and sheep anti-mouse IgG, both conjugated to horseradish peroxidase (Amersham Biosciences, Little Chalfont, UK). The signals were detected by chemiluminescence.
Reverse transcription PCR (RT-PCR) and quantitative RT-PCR
Total RNA from cells, treated as indicated in the figures, was extracted using TriZol reagent (Invitrogen, Paisley, UK). The obtained total RNA (500 ng to 1 µg) was used to generate cDNA using random hexamer primers (Qiagen) and reverse transcriptase, as recommended by the supplier's instructions (Fermentas, Glen Burnie, MD, USA). PCR amplification of the 474-bp murine IL-6 gene was performed with MIL-6F/R primers (Table 1), with amplification of a 380-bp fragment of the β-actin gene (using βACTF/R primers) as a control (Table 1). The quantitative PCR (qPCR) amplification was performed using an MJ Mini personal Thermal cycler (BioRad, Richmond, VA, USA), with Maxima SYBR Green/ROX qPCR Master Mix (Fermentas, Burlington, Ont., Canada) using 2 µl cDNA according to the manufacturer's protocol (Fermentas). The primers used for the qPCR amplification are shown in Table 1. The relative expression levels were calculated and analyzed by 2−ΔΔCT.14
Table 1. Primers used in this study.
| Primer name | Target | Sequence (5′–3′) | Sizea | Useb |
|---|---|---|---|---|
| MIL-6F | mIL-6 gene | CATGTTCTCTGGGAAATCGTGG | 474 bp | RT-PCR of the IL-6 gene |
| MIL-6R | AACGCACTAGGTTTGCCGAGTA | |||
| QMIL-6F QMIL-6R | mIL-6 gene fragment | CTCTGGGA AATCGTGGAAATG AAGTGCATCATCGTTGTTCATACA | 75 bp | qRT-PCR (IL-6 transcript levels) |
| QBACTF QBACTR | β-actin gene fragment | ACCAACTGGGACGACATGGAGAA GTGGTGGTGAAGCTGTAGCC | 380 bp | qRT-PCR (cDNA reference control) |
| CSLF CSLR | CSL-containing region of the mil-6 gene | TCGATGCTAAACGACGTCAC TCAATTCCAGAAACCGCTATG | 230 bp | qPCR detection of CSL-bound DNA (ChIP assay) |
Expected amplicon size in bp using these primer pairs.
Application of these primer pairs in this report.
Abbreviations: ChIP, chromatin immunoprecipitation; CSL, CBF1/Su(H)/Lag-1; qPCR, quantitative PCR; qRT-PCR, quantitative PCR with reverse transcription.
ELISA for measuring IL-6
Secreted IL-6 levels produced by macrophages activated by LPS (100 ng/ml) plus recombinant mouse IFN-g (rmIFN) (10 ng/ml) were measured with the Ready-Set-Go! Mouse IL-6 kit according to the manufacturer's instructions (eBioscience, San Diego, CA, USA).
Chromatin immunoprecipitation (ChIP)
The murine macrophage-like RAW264.7 cell line (1×107 cells) was activated by the addition of LPS (100 ng/ml) plus rmIFN-γ (10 ng/ml). The EZ Magna ChIP Chromatin Immunoprecipitation Kit (Millipore, Billerica, MA, USA) was used according to the manufacturer's protocol for the ChIP assay. The antibody used in the ChIP assay was rabbit anti-Notch1 antibody (Santa Cruz Biotechnology). The presence of DNA fragments containing the target IL-6 promoter region was detected by qPCR with CSLF/R primers (Table 1), which are specific for the IL-6 promoter region that contains the putative CSL-binding sites, yielding an expected amplicon of 230 bp. Control amplification was performed using DNA input as a reference.
Statistical analysis
A one-way ANOVA (F test) and multiple comparisons (Fisher's least significant difference) were used to analyze the data. Values of P<0.05 were considered to be statistically significant.
Results
Expression of the Notch1 protein and IL-6 transcript levels in stimulated BMMφ
In order to examine the regulatory role of Notch signaling in IL-6 mRNA expression, we first investigated the expression profile of both Notch1, the main receptor expressed during the activation of macrophages, and IL-6 mRNA at various time points. Notch1 was upregulated in LPS/IFN-γ-stimulated BMMφ as early as 3 h after the initial activation, and this upregulation lasted until 24 h after the initial activation (Figure 1a). The upregulation of Notch1 appeared to be slower than macrophages correspondingly costimulated with LPS plus rmIFN-γ (data not shown). Cleaved Notch1 (Val1744), an indicator of the activation of the Notch signaling pathway, also appeared in the BMMφ activated with LPS plus rmIFN-γ after 1 h and then decreased thereafter to almost basal levels by 24 h (Figure 1a). By contrast, IL-6 mRNA expression was detected in BMMφ as early as 1 h after LPS/IFN-γ costimulation and peaked at 6 h after stimulation before decreasing; however, the transcript levels were still significantly elevated after 24 h (Figure 1b). The upregulation of IL-6 by LPS stimulation alone was statistically lower than its upregulation in cells receiving a combination of LPS and rmIFN-g. Therefore, we decided to use LPS and rmIFN-g as the stimuli for macrophages in this study unless otherwise indicated. The correlation between Notch1 protein expression and IL-6 mRNA levels prompted us to ask whether Notch signaling regulates IL-6 transcript expression levels in this setting.
Figure 1.
Expression of Notch1 and IL-6 in stimulated BMMφ. BMMφ cells were stimulated with LPS (100 ng/ml) plus rmIFN-g (10 ng/ml) for the indicated durations before cell lysates and total RNA extracts were prepared. (a) The expression levels of Notch1 and cleaved Notch1 (Val1744) in the cell lysates were determined by western blotting, with β-actin as the loading control. The blots shown are representative of two independent experiments. (b) The IL-6 mRNA expression level, determined by qRT-PCR of the total RNA, is shown after normalization to β-actin. The results represent two independent experiments each performed in triplicate and are shown as mean±s.d. BMMφ, bone marrow-derived macrophages; IFN, interferon; IL, interleukin; LPS, lipopolysaccharide; qRT-PCR, quantitative reverse transcription PCR.
Effect of the γ-secretase inhibitor (IL-CHO) and NF-κB inhibitor on IL-6 transcript and IL-6 protein expression levels
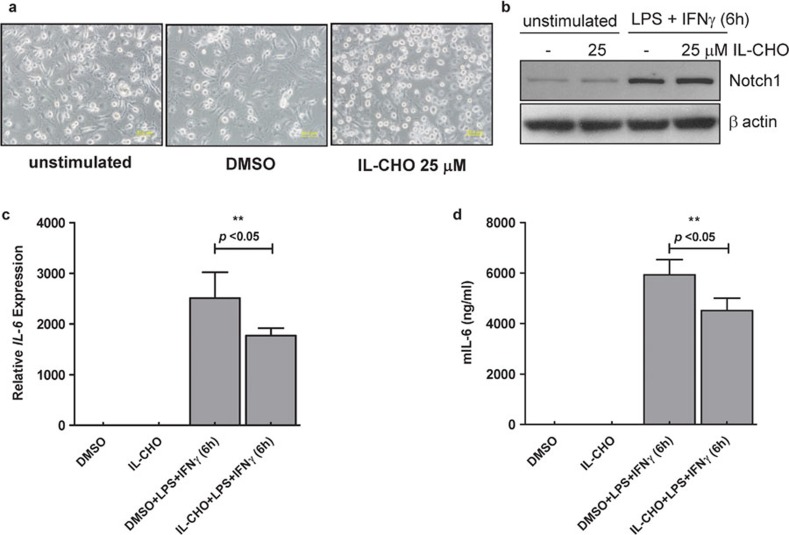
To address the involvement of Notch signaling in IL-6 mRNA expression, we took a pharmacological approach using the γ-secretase inhibitor IL-CHO. Treatment of BMMφ cells with IL-CHO (25 µM) led to a slightly different cell morphology from that seen in the mock-treated cells, where IL-CHO-treated cells were often rounder than control cells (Figure 2a). However, IL-CHO treatment did not have any effect on the expression levels of the uncleaved Notch1, which, at 6 h of treatment, was still elevated as highly in the IL-CHO-treated cells as that seen in the mock-treated control cells (Figure 2b).
Figure 2.
Effect of IL-CHO treatment on IL-6 expression in BMMφ. BMMφ cells were pretreated with IL-CHO (25 µM in 1% (v/v) DMSO) or mock vehicle control (1% (v/v) DMSO) for 1 h before stimulation with LPS (100 ng/ml) plus rmIFN-g (10 ng/ml) for 6 h. (a) Cell morphologies were observed by phase contrast microscopy after 1 h of treatment, and cell lysates and total RNA extracts were prepared after 6 h of treatment. (b) The expression level of Notch1 protein in the cell lysates was determined by western blotting, with β-actin as the loading control. The blots shown are representative of two independent experiments. (c) The IL-6 mRNA expression level, determined by qRT-PCR, is shown after normalization to β-actin. (d) IL-6 levels in the culture supernatant were measured by ELISA 6 h after stimulation. The results for both (c) and (d) are shown as mean±s.d. and are derived from two independent experiments, each performed in triplicate. Means with asterisks above them are significantly different (P<0.05). BMMφ, bone marrow-derived macrophages; DMSO, dimethylsulfoxide; IFN, interferon; IL, interleukin; LPS, lipopolysaccharide; qRT-PCR, quantitative reverse transcription PCR.
We next asked whether IL-CHO treatment had any effect on IL-6 mRNA expression levels and secreted IL-6 protein production. As shown in Figure 2c and d, IL-CHO treatment at the same dose (25 µM), which suppressed the cleavage of Notch1, significantly reduced both the level of IL-6 mRNA and the level of secreted IL-6 in the culture supernatant (both P≤0.05). However, this was not a complete decrease, as it did not reduce either mRNA or protein levels to a basal level. Significant levels of IL-6 transcripts and secreted IL-6 protein were still present, suggesting that Notch signaling may not be the sole regulator of IL-6 transcription in activated murine primary BMMφ.
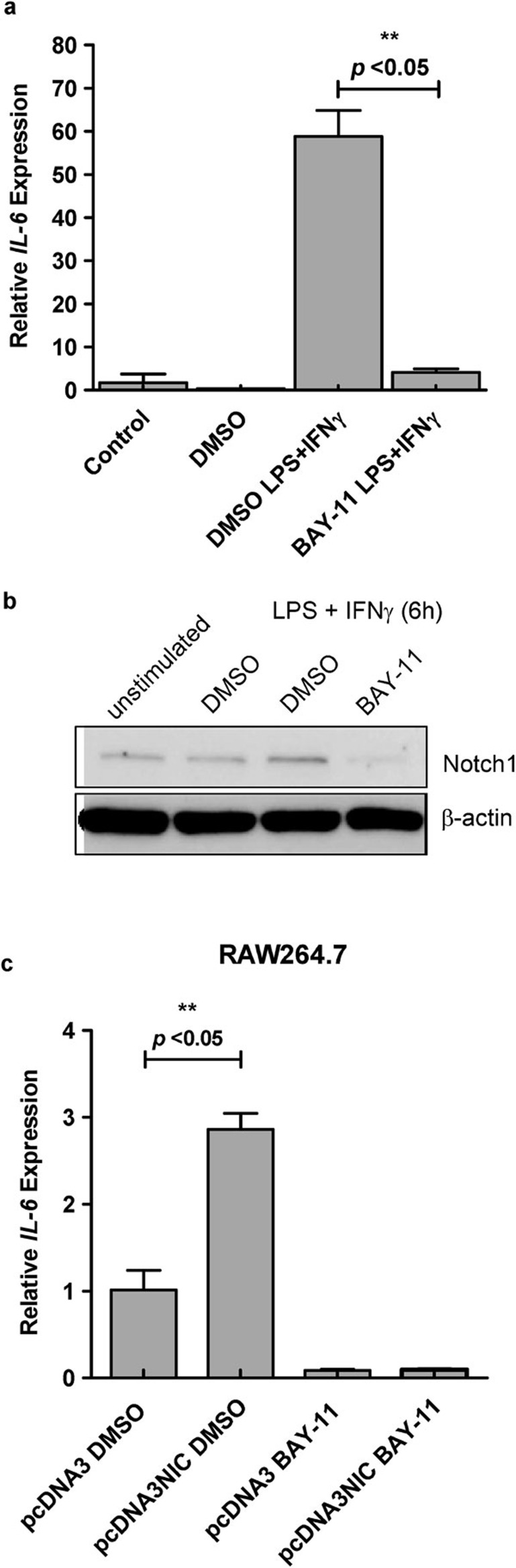
Because the NF-κB pathway is a major regulator of IL-6 transcription and consensus NF-κB sites can be found in the IL-6 promoter, we treated cells with the NF-κB inhibitor BAY-11 and found an almost complete downregulation of IL-6 transcripts upon activation with LPS/IFN-γ (Figure 3a). Interestingly, when the level of Notch1 was examined after BAY-11 treatment, it was found to be almost completely downregulated to the basal level (Figure 3b). These results indicated that NF-κB signaling is the major regulator of transcription of IL-6 and that this regulation may be mediated in part through signaling via the Notch receptor.
Figure 3.
Effect of NF-κB inhibitor on IL-6 mRNA expression and Notch1 expression in BMMφ. (a) BMMφ were pretreated with BAY-11 (10 µM) or DMSO (as a vehicle control) for 1 h before stimulation with LPS (100 ng/ml) and IFN-γ (10 ng/ml) for 6 h. Total RNA was isolated and analyzed for IL-6 mRNA by qRT-PCR. The results are shown as mean±s.d. and are derived from two independent experiments, each performed in triplicate. Means with asterisks above them are significantly different (P<0.05). (b) Cell lysates from cells treated as described above were analyzed for Notch1 expression by western blotting, with β-actin as the control. (c) RAW264.7 cells were transiently transfected with the empty vector (pcDNA3) or the NIC-encoding vector (pcDNA3NIC) for 24 hr and then treated with BAY-11 (10 µM) or DMSO for 3 h. Total RNA was isolated and analyzed for IL-6 mRNA by qRT-PCR. The results are shown as mean±s.d. Means with asterisks above them are significantly different (P<0.05). BMMφ, bone marrow-derived macrophages; DMSO, dimethylsulfoxide; IFN, interferon; IL, interleukin; LPS, lipopolysaccharide; NF, nuclear factor; qRT-PCR, quantitative reverse transcription PCR.
Effect of overexpression of constitutively active Notch1 (NIC) on IL-6 transcription
Because γ-secretase has multiple substrates, the effects of its inhibition by IL-CHO may unexpectedly interfere with other signaling pathways. Therefore, we used an overexpression approach to confirm the likely involvement of Notch signaling in the regulation of IL-6 transcript expression. Plasmids encoding NIC or the empty vector control were used to transiently transfect RAW264.7 cells, and the expression of the NIC protein product was confirmed by western blotting (Figure 4a). Without any stimulation, the IL-6 transcript expression levels were low in all three different cell types at 24 h after transfection, but they were significantly higher in the NIC-transfected cells than in either the control (pcDNA3)-transfected or the untransfected control cells (P<0.05) (Figure 4b).
Figure 4.
Expression of IL-6 in the NIC-overexpressing RAW264.7 macrophage cell line. (a) RAW264.7 cells were transiently transfected with empty vector (pcDNA3) or NIC-encoding vector (pcDNA3NIC). Cells were left unstimulated or stimulated with LPS (100 ng/ml) for the indicated duration before cell lysates were prepared and analyzed for the expression of Notch1 by western blotting, using β-actin as the loading control. The blots shown are representative of three independent experiments. (b) Transiently transfected RAW264.7 cells were left unstimulated for 24 h before total RNA was extracted. The il-6 transcript levels were then measured by qRT-PCR and normalized to β-actin. (c) RAW264.7 cells were transiently transfected with empty vector (pcDNA3) or NIC-encoding vector (pcDNA3NIC) for 12 h and stimulated with LPS (100 ng/ml) for 6 h. Total RNA from the treated cells was analyzed for IL-6 mRNA expression by qRT-PCR. The results are shown as mean±s.d. and are derived from two independent experiments, each performed in triplicate. Means with asterisks above them are significantly different (P<0.05). IL, interleukin; LPS, lipopolysaccharide; qRT-PCR, quantitative reverse transcription PCR.
Because LPS can activate the NF-κB pathway through TLR4 stimulation, we asked whether NF-κB activation by LPS treatment and Notch signaling can cooperatively induce IL-6 expression. Upon LPS stimulation, a slight increase in Notch1 expression was observed in RAW264.7 cells transfected with the control vector (Figure 4a). After activation with LPS for 6 h, the IL-6 transcript levels were markedly higher under all three conditions than in the unstimulated cells, as was expected. More importantly, NIC overexpressing cells showed a significantly higher IL-6 mRNA level than empty vector-transfected cells in the presence of LPS stimulation (Figure 4c). Although the relative levels of IL-6 mRNA in resting NIC overexpressing macrophages were much lower than those in LPS-stimulated cells, the increase in IL-6 mRNA after overexpression of NIC alone strongly suggests that Notch signaling may be sufficient to enhance or initiate the transcription of the IL-6 gene, and that LPS together with NIC overexpression synergistically increased IL-6 mRNA expression.
To evaluate the involvement of the NF-κB pathway in NIC-mediated IL-6 transcription, we treated cells overexpressing NIC or mock-transfected RAW264.7 cells with the NF-κB inhibitor BAY-11 and measured the level of IL-6 transcripts. As shown in Figure 3c, treatment with BAY-11 completely abrogated the expression of IL-6 mRNA to a basal level. This result strongly suggests that Notch signaling mainly induces IL-6 transcription via the NF-κB pathway.
Association of Notch1 with the IL-6 promoter in stimulated macrophages
Within the minimal 5′ promoter region that is sufficient for the induction of IL-6 gene transcription, a completely matched CSL-binding site (5′-CATGGGAA-3′) was previously identified between −67 and −60 bp from the transcription initiation site, and this site was found to overlap with an NF-κB-binding site located between −73 and −64 bp (Figure 5a).7 When the nucleotide sequences from the murine and human IL-6 promoter are aligned, it is apparent that the potential CSL-binding site is completely conserved in both species. Previously, the CSL protein was shown to directly bind to this CSL-binding sequence in the IL-6 promoter,5, 7, 8 but the potential association of Notch with CSL on this promoter has not been addressed. Because the results presented here have so far indicated that Notch signaling could directly, at least in part, switch on IL-6 transcription, we therefore carried out a ChIP assay using the RAW264.7 macrophage-like cell line to test whether Notch associates with the IL-6 promoter. Upon activation by LPS/IFN-γ, RAW264.7 cells highly upregulated Notch1, which peaked at 6 h after stimulation but remained at high levels up to 24 h after stimulation (Figure 5b). In addition, the appearance of cleaved Notch1 was detected starting at 6 h and peaked at 12 h after stimulation (Figure 5b). When the expression level of IL-6 mRNA was examined, the peak was observed at 6 h after LPS/IFN-γ stimulation but declined by 24 h later, whereas LPS stimulation alone induced only a weak increase in the IL-6 transcript level at 6 and 12 h after stimulation (Figure 5c). Thus, we used unstimulated cells and LPS/IFN-γ stimulation for 12 h as the assay conditions for the ChIP assay. When anti-Notch1 antibodies and normal rabbit IgG were used to immunoprecipitate sheared chromatin, qPCR amplification revealed that the IL-6 promoter containing the CSL-binding region was detected in the ChIP at a significantly higher level when using anti-Notch1 antibodies than the control sera (Figure 5d). In addition, the association of Notch1 with the IL-6 promoter was not detected in unstimulated cells (Figure 5d). This result strongly suggested that the receptor Notch1 is associated with the IL-6 promoter only in LPS/IFN-γ-stimulated RAW267.4 macrophage-like cells.
Figure 5.
ChIP assay for binding of Notch1 to the IL-6 promoter in macrophages. (a) Schematic representation of the human and murine IL-6 promoters. Binding regions for NF-κB and the CSL protein are shown. (b–d) RAW264.7 cells were stimulated with LPS (100 ng/ml) plus IFN-γ (10 ng/ml) for the indicated durations. Cell lysates and total RNA extracts were then made, and ChIP assays were performed. (b) The Notch1 and cleaved Notch1 (Val1744) expression levels in cell lysates are shown, as determined by western blotting using β-actin detection as the loading control. The blots shown are representative of two independent experiments. (c) The IL-6 mRNA levels are shown, as detected by RT-PCR of the total RNA. (d) A ChIP assay was performed using normal rabbit IgG or rabbit anti-Notch1 antibodies in unstimulated or LPS/IFN-γ-stimulated RAW264.7 cells. The association of Notch1 with the IL-6 gene promoter region was measured in the immunoprecipitate by qPCR relative to that for input DNA. The results are shown as mean±s.d. and are derived from two independent experiments, each performed in triplicate. Means with asterisks above them are significantly different (P<0.05). ChIP, chromatin immunoprecipitation; CSL, CBF1/Su(H)/Lag-1; IFN, interferon; IL, interleukin; LPS, lipopolysaccharide; NF, nuclear factor; qPCR, quantitative PCR; RT-PCR, reverse transcription PCR.
Discussion
The Notch signaling pathway has been established to be involved in the inflammatory responses of macrophages, and the suppression of Notch signaling by pharmacological inhibitors has been shown to negatively affect various pro-inflammatory mediators, such as tumor-necrosis factor-α, inducible nitric oxide synthase and IL-6. The effects of the γ-secretase inhibitor IL-CHO has been shown to correlate well with the results obtained from CSL-deficient mice.10, 13 These results support the critical roles of Notch signaling in regulating in flammatory processes inactivated macrophages. Although Notch signaling has been shown to regulate these processes via the NF-κB, p38 mitogen-activated protein kinase (MAPK) and signal transducer and activator of transcription 1 (STAT1) pathway,11, 12, 15 whether it directly regulates any of these responses has not been thoroughly investigated.
In this study, we examined the regulatory mechanisms of Notch signaling-mediated IL-6 gene expression in LPS/IFN-γ-activated murine BMMφ and an established murine macrophage-like cell line (RAW264.7) in detail. As previously reported, treatment with the γ-secretase inhibitor IL-CHO decreased both IL-6 mRNA and IL-6 protein levels in LPS/IFN-γ-stimulated macrophages. More importantly, overexpression of constitutively active Notch1 alone is sufficient to induce IL-6 mRNA transcript levels. This result strongly suggests that Notch signaling alone is sufficient to switch on IL-6 mRNA transcription. Upon stimulation with only LPS, a significantly increased IL-6 mRNA level was maintained in activated Notch1-overexpressing cells. Therefore, Notch signaling may function cooperatively with other signaling pathways for optimal IL-6 expression. Interestingly, overexpression of constitutively active Notch1 in the RAW264.7 cell line induced a much lower (1320-fold) mRNA transcript level than when cells were stimulated with LPS, implying that Notch signaling alone may only minimally induce IL-6 expression.
Previously, a CSL-binding site in the IL-6 promoter region was identified, and CSL was shown to directly bind to this site, which overlaps with the NF-κB-binding site. However, using an in vitro binding assay, it was reported that CSL binds to this site with a lower affinity than NF-κB.5 In addition, overexpression of constitutively active Notch1 in a human non-immune cell line did not induce an IL-6 reporter construct.8 Thus, CSL was proposed to negatively regulate IL-6 transcription by blocking the access of the NF-κB complexes to this promoter. In our study, we used primary murine BMMφ and a murine macrophage-like cell line, which are cell types capable of producing IL-6 upon stimulation, unlike human non-immune cells. The discrepancies between our results and previous reports may thus lie in the different types of cells used, the species from which the cells were derived (human vs. murine) or both. Notch signaling is well known to function in a context-dependent manner, and the control of expression of the pleiotropic IL-6 gene may be differently regulated in different cell types, especially between primary BMMφ and non-immune cells.
Interestingly, when we inhibited the NF-κB pathway using a pharmacological approach, we found a dramatic decrease in the Notch1 expression level. This result correlated well with the complete downregulation of the IL-6 mRNA level upon LPS/IFN-γ stimulation. This is the first report indicating that the NF-κB pathway may engage in crosstalk with Notch signaling by regulating Notch1 expression in macrophages. A recent study by Foldi et al. demonstrated that stimulation with TLR agonists led to upregulation of the Notch ligand Jagged1, which is mediated by NF-κB.16 The authors suggested that this event initiates the Notch signaling cascade during a pro-inflammatory response.
Multiple Notch molecules are expressed in macrophages, and their expression levels are differentially regulated upon activation. In murine primary macrophages, Notch1 and Notch2 are the dominantly expressed receptors. Silencing Notch1 in RAW264.7 did not lead to a dramatic decrease in the IL-6 mRNA levels upon stimulation (data not shown), which implies that other Notch molecules (such as Notch2) may compensate for the decrease in Notch1 expression.
Hu et al.10 have reported that the transcription factor products of the Hey1 and Hes1 genes, which are target genes of the Notch signaling pathway, negatively regulate pro-inflammatory responses, including the production of IL-6. These authors directly deleted or overexpressed Hey1 and Hes1 in macrophages, but the molecular mechanism by which these proteins control the expression of their target genes was not evaluated. When we overexpressed constitutively active Notch1 (NIC), which spontaneously induces the upregulation of Hes1,17 we did not observe a negative effect on IL-6 transcription. The reason for these discrepancies is unknown; however, many genes are under the control of the Notch signaling pathway, and these genes may cross-regulate each other. Therefore, the results from manipulating Hes1/Hey1 and the Notch receptors may not necessarily be similar to each other.
When the RAW264.7 cell line was transiently transfected to overexpress NIC, treatment with an NF-κB inhibitor resulted in the complete downregulation of IL-6, with or without LPS stimulation. These results suggest a strong link between Notch signaling and the NF-κB pathway in regulating IL-6 expression. Notch signaling is reported to activate the NF-κB pathway, and in turn, the NF-κB pathway sustains the activation of Notch signaling via the induction of Notch ligands.16, 18, 19 In addition, Notch and NF-κB may cooperatively bind to the IL-6 promoter to promote the optimal expression of IL-6. Therefore, it is likely that Notch signaling and NF-κB interact to regulate IL-6 expression in macrophages.
In conclusion, we demonstrated in this report that Notch signaling directly regulates IL-6 mRNA expression by associating with the promoter region of IL-6. Although this mechanism cannot optimally induce IL-6 expression, compared to an inflammatory stimulus, it illustrates a direct regulatory mechanism in which inflammatory gene expression is controlled by the Notch signaling pathway.
Acknowledgments
This work was partly supported by the Thailand Research Fund (TRF) Grant No. RSA5280014 and by the Thai Government Stimulus Package 2 (TKK2555), under the Project for Establishment of Comprehensive Center for Innovative Food, Health Products and Agriculture. Financial supports from the TRF through the Royal Golden Jubilee Ph.D. Program (PHD/0337/2551) and Research Foundation Enhancement (Ratchadaphiseksomphot Endowment Fund to authors are also acknowledged.
References
- Kishimoto T. Interleukin-6: from basic science to medicine—40 years in immunology. Annu Rev Immunol. 2005;23:1–21. doi: 10.1146/annurev.immunol.23.021704.115806. [DOI] [PubMed] [Google Scholar]
- Kishimoto T. IL-6: from laboratory to bedside. Clin Rev Allergy Immunol. 2005;28:177–186. doi: 10.1385/CRIAI:28:3:177. [DOI] [PubMed] [Google Scholar]
- Xiao W, Hodge DR, Wang L, Yang X, Zhang X, Farrar WL. NF-kappaB activates IL-6 expression through cooperation with c-Jun and IL6-AP1 site, but is independent of its IL6-NFkappaB regulatory site in autocrine human multiple myeloma cells. Cancer Biol Ther. 2004;3:1007–1017. doi: 10.4161/cbt.3.10.1141. [DOI] [PubMed] [Google Scholar]
- Xiao W, Hodge DR, Wang L, Yang X, Zhang X, Farrar WL. Co-operative functions between nuclear factors NFkappaB and CCAT/enhancer-binding protein-beta (C/EBP-beta) regulate the IL-6 promoter in autocrine human prostate cancer cells. Prostate. 2004;61:354–370. doi: 10.1002/pros.20113. [DOI] [PubMed] [Google Scholar]
- Plaisance S, Vanden Berghe W, Boone E, Fiers W, Haegeman G. Recombination signal sequence binding protein Jkappa is constitutively bound to the NF-kappaB site of the interleukin-6 promoter and acts as a negative regulatory factor. Mol Cell Biol. 1997;17:3733–3743. doi: 10.1128/mcb.17.7.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libermann TA, Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol Cell Biol. 1990;10:2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa K, Mori A, Yamamoto K, Okudaira H. Transcriptional roles of CCAAT/enhancer binding protein-beta, nuclear factor-kappaB, and C-promoter binding factor 1 in interleukin (IL)-1beta-induced IL-6 synthesis by human rheumatoid fibroblast-like synoviocytes. J Biol Chem. 1998;273:7620–7627. doi: 10.1074/jbc.273.13.7620. [DOI] [PubMed] [Google Scholar]
- Palmieri M, Sasso MP, Monese R, Merola M, Faggioli L, Tovey M, et al. Interaction of the nuclear protein CBF1 with the kappaB site of the IL-6 gene promoter. Nucleic Acids Res. 1999;27:2785–2791. doi: 10.1093/nar/27.13.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr Opin Cell Biol. 2007;19:166–175. doi: 10.1016/j.ceb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Hu X, Chung AY, Wu I, Foldi J, Chen J, Ji JD, et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity. 2008;29:691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaga T, Buranaruk C, Rengpipat S, Fauq AH, Golde TE, Kaufmann SH, et al. Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur J Immunol. 2008;38:174–183. doi: 10.1002/eji.200636999. [DOI] [PubMed] [Google Scholar]
- Monsalve E, Ruiz-Garcia A, Baladron V, Ruiz-Hidalgo MJ, Sanchez-Solana B, Rivero S, et al. Notch1 upregulates LPS-induced macrophage activation by increasing NF-kappaB activity. Eur J Immunol. 2009;39:2556–2570. doi: 10.1002/eji.200838722. [DOI] [PubMed] [Google Scholar]
- Palaga T, Miele L, Golde TE, Osborne BA. TCR-mediated Notch signaling regulates proliferation and IFN-gamma production in peripheral T cells. J Immunol. 2003;171:3019–3024. doi: 10.4049/jimmunol.171.6.3019. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Monsalve E, Perez MA, Rubio A, Ruiz-Hidalgo MJ, Baladron V, Garcia-Ramirez JJ, et al. Notch-1 up-regulation and signaling following macrophage activation modulates gene expression patterns known to affect antigen-presenting capacity and cytotoxic activity. J Immunol. 2006;176:5362–5373. doi: 10.4049/jimmunol.176.9.5362. [DOI] [PubMed] [Google Scholar]
- Foldi J, Chung AY, Xu H, Zhu J, Outtz HH, Kitajewski J, et al. Autoamplification of Notch signaling in macrophages by TLR-induced and RBP-J-dependent induction of Jagged1. J Immunol. 185:5023–5031. doi: 10.4049/jimmunol.1001544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipo C, Golde TE, Osborne BA, Miele LA. Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab Invest. 2008;88:11–17. doi: 10.1038/labinvest.3700700. [DOI] [PubMed] [Google Scholar]
- Song LL, Peng Y, Yun J, Rizzo P, Chaturvedi V, Weijzen S, et al. Notch-1 associates with IKKalpha and regulates IKK activity in cervical cancer cells. Oncogene. 2008;27:5833–5844. doi: 10.1038/onc.2008.190. [DOI] [PubMed] [Google Scholar]