Hepatocellular carcinoma (HCC) is the fifth most prevalent malignancy worldwide, and it accounts for 85–90% of all primary liver cancers. Owing to the limited therapeutic options available, the 5-year survival rate remains relatively low for patients with HCC. The development of HCC is a multistage process1 that involves changes in the expression of multiple oncogenes, tumor suppressor genes and microRNAs that are involved in the cell cycle, as well as in cell growth, apoptosis, migration and tumor metastasis.2, 3 Finding an accurate biomarker that can predict HCC progression is important for clinicians to be able to determine the optimal therapeutic strategy.
Over the past two decades, alpha-fetoprotein (AFP) has not only been widely used as a biomarker for the regeneration of hepatocytes, but also been recognized clinically to be associated with the development of HCC. Tatarinov and Abelev4 first used AFP as a diagnostic marker for HCC in 1964, and the mounting clinical evidence since this report indicates that AFP elevation is linked to a more aggressive tumor phenotype characterized by vascular invasion, metastasis and poor differentiation.5 However, the diagnostic accuracy of AFP is often questionable because of its low sensitivity; it remains to be determined whether AFP levels in HCC represent anything more than a coincidental epiphenomenon.
miRNA122 is a liver tissue-specific microRNA that is expressed in the developing liver and constitutes 70% of all miRNAs in the adult liver.6 The best-known function of miRNA122 in the mammalian liver is to regulate lipid and cholesterol metabolism, and it is also involved in the replication of the hepatitis C virus.7 Recently, miRNA122 has been reported to be downregulated in the tumor tissues of HCC patients. Downregulation of this microRNA is associated with the pathological features of HCC, including metastasis and recurrence, as well as with poor prognosis. The mechanisms underlying these phenotypic features after miRNA122 downregulation include chromosomal instability, increased anti-apoptotic activity and a high expression of ADAM10/17.8 However, the molecular mechanisms that underlie the miRNA122 downregulation in HCC remain unknown.
It remains unclear whether AFP or miRNA122 is more efficient at predicting HCC disease progression. Kojima et al. 9 report the novel finding that the decreased levels of miRNA122 in HCC are responsible for both the elevation in AFP levels and the more biologically aggressive HCC phenotype, indicating that it is a more accurate predictor of HCC progression.
In this article, we review the literature regarding the important issue of whether AFP expression is uniquely upregulated in miRNA122-silenced cells. If this is the case, what is the nature of the signaling pathway that links these two molecules? It appears unlikely that miRNA122 directly modifies AFP expression, because the 3′-UTR of AFP mRNA does not contain predicted miRNA122 target sequences (http://www.targetscan.org). Mutations in the genes that encode p53, TGF-β10 and ZBTB2011 have been reported to promote the expression of AFP, and Kojima et al. also report that decreased expression of ZBTB20 promoted AFP expression in miRNA122-silenced cells. However, as ZBTB20 also lacks any predicted miRNA122 target sequences, based on computational searches of the 3′-UTR sequence, it is also unlikely that miRNA122 directly regulates ZBTB20 expression.
Through computational searches and luciferase reporter experiments, the authors revealed that miRNA124 directly targeted the ZBTB20 3′-UTR and that the overexpression of miRNA124 in miRNA122-silenced cells decreased ZBTB20 protein expression levels. Finally, using MATCH, a transcription factor binding site search engine, the authors demonstrate multiple CUX1 binding sites in the putative promoter regions of the gene encoding miRNA124. By silencing or ectopically expressing the CUX1 gene, the authors confirmed that CUX1 can directly regulate the expression levels of miRNA124 through binding to its promoter region.
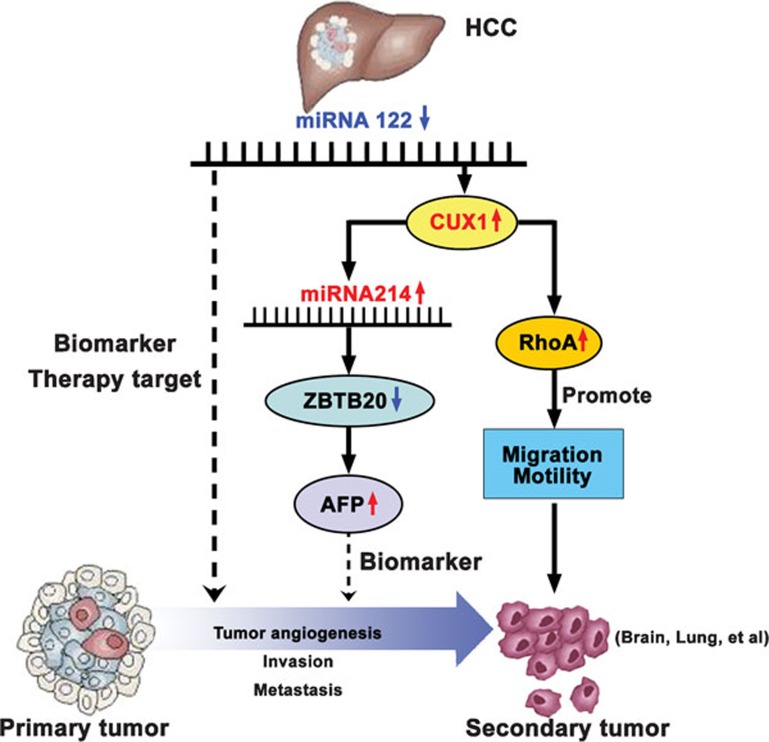
In addition, the authors have confirmed that decreased levels of miRNA122 directly promote the expression of CUX1. Thus, the silencing of miRNA122 leads to an increase in the level of CUX1 protein expression. This causes the repression of ZBTB20 expression through increased miRNA124 expression, which ultimately results in increased AFP expression. The upregulation of CUX1 also enhances RhoA activity, which increases the malignant properties of cancer cells (Figure 1). Finally, the authors proved that this miRNA122/AFP signaling pathway (miRNA122/CUX1/miRNA124/ZBTB20/AFP) also exists in HCC cell lines, transgenic mice and human HCC samples.
Figure 1.
The newly discovered miRNA122-mediated signaling pathway determines HCC progression. In HCC patients, the decreased levels of miRNA122 lead to increased expression of the CUX1 protein, which may subsequently enhance RhoA activity and promote the migration and motility of tumor cells, ultimately leading to the development of secondary or metastatic tumors. In addition, the upregulation of CUX1 inhibits ZBTB20 expression, through increasing miRNA214 expression. Suppression of ZBTB20, in turn, further upregulates AFP expression. AFP, generally, is used as a biomarker in the clinic to predict HCC progression, although its diagnostic accuracy is often questionable owing to its low sensitivity. By contrast, the decreased miRNA122 levels in HCC patients are responsible for both the elevation in AFP levels and the more biologically aggressive HCC phenotype. Thus, miRNA122 likely presents not only a more accurate predictor of HCC progression, but also a novel target for developing an optimal therapeutic strategy for HCC patients. AFP, alpha-fetoprotein; CUX1, cut-like homebox 1; HCC, hepatocellular carcinoma; RhoA, RAS homolog gene family, member A; ZBTB20, zinc finger and BTB domain containing 20.
miRNA122 and its related signaling proteins are markers that reflect a disease that is more biologically aggressive than the AFP marker and represent potential therapeutic targets for HCC patients. These groundbreaking findings unravel the miRNA122-mediated molecular mechanisms that underlie elevated AFP levels in patients with HCC. The two main limitations of the study may suggest additional information. Firstly, the demographic backgrounds of the patients in the cohort were not clear. It is well known that HCC is a complex disease, and many clinical characteristics, such as gender, etiology (for example, HBV, HCV and ethanol), Child-Pugh classification, tumor size, tumor number, tumor location and extrahepatic metastasis, will directly affect the prognosis. Although this study confirmed the associations between the miRNA122, CUX1, miRNA124, ZBTB20 and AFP molecules in this novel signaling pathway and the disease grade in certain HCC patients, the lack of details on patient background might mask the true relationship between miRNA122, AFP and disease progression. Secondly, it remains unclear whether and how miRNA122 levels reflect disease progression in HCC. Although this study proved (by using miRNA-silenced cells and a xenograft model) that the loss of miRNA122 functionality leads to a more malignant phenotype with regard to cell migration and invasion, there remains a lack of direct evidence from human HCC samples. The enrollment of various patient cohorts with a range of severities of progressive disease into clinical studies may allow the further elucidation of whether the loss of miRNA122 expression in patients with HCC is associated with disease progression. More importantly, certain issues remain unaddressed in this study: (i) the identity of the signaling pathway that is associated with the loss of miRNA122 expression in patients with HCC; (ii) the differing expression profile of miRNA122 in patients with HCC with different pathological causes; and (iii) the detailed functions of AFP during the clinical course of HCC. Therefore, further comprehensive studies are required to clarify the role of miRNA122 expression in HCC patients.
Computational analyses have indicated that a single miRNA may influence hundreds of genes in terms of mRNA and protein expression. In this way, if one miRNA signal—such as miRNA122, which targets cyclin G1, ADAM10, ADAM17, SRF, Igf1R, Bcl-w12 and CUX1, amongst others—is identified as a key player in the progression of HCC, then recovery of the function of this miRNA may have a significant therapeutic effect. Given recent technological advances, miRNAs or antagomirs are feasible and safe therapeutic options.8 These studies provide novel targets for the development of new treatments for HCC.8, 9, 12
References
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci (Lond) 2011;121:141–158. doi: 10.1042/CS20110005. [DOI] [PubMed] [Google Scholar]
- Takata A, Otsuka M, Kojima K, Yoshikawa T, Kishikawa T, Yoshida H, et al. MicroRNA-22 and microRNA-140 suppress NF-κB activity by regulating the expression of NF-κB coactivators. Biochem Biophys Res Commun. 2011;411:826–831. doi: 10.1016/j.bbrc.2011.07.048. [DOI] [PubMed] [Google Scholar]
- Abelev GI. Production of embryonal serum alpha-globulin by hepatomas: review of experimental and clinical data. Cancer Res. 1968;28:1344–1350. [PubMed] [Google Scholar]
- Yamamoto K, Imamura H, Matsuyama Y, Kume Y, Ikeda H, Norman GL, et al. AFP, AFP-L3, DCP, and GP73 as markers for monitoring treatment response and recurrence and as surrogate markers of clinicopathological variables of HCC. J Gastroenterol. 2010;45:1272–1282. doi: 10.1007/s00535-010-0278-5. [DOI] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Huang S, He X. The role of microRNAs in liver cancer progression. Br J Cancer. 2011;104:235–240. doi: 10.1038/sj.bjc.6606010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Takata A, Vadnais C, Otsuka M, Yoshikawa T, Akanuma M, et al. MicroRNA122 is a key regulator of α-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nat Commun. 2011;2:338. doi: 10.1038/ncomms1345. [DOI] [PubMed] [Google Scholar]
- Peng SY, Chen WJ, Lai PL, Jeng YM, Sheu JC, Hsu HC. High alpha-fetoprotein level correlates with high stage, early recurrence and poor prognosis of hepatocellular carcinoma: significance of hepatitis virus infection, age, p53 and beta-catenin mutations. Int J Cancer. 2004;112:44–50. doi: 10.1002/ijc.20279. [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang H, Tsai W, Zhang Y, Du Y, Zhong J, et al. Zinc finger protein ZBTB20 is a key repressor of alpha-fetoprotein gene transcription in liver. Proc Natl Acad Sci USA. 2008;105:10859–10864. doi: 10.1073/pnas.0800647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]