Abstract
E3 ubiquitin ligases play a crucial role in regulating immune receptor signaling and modulating immune homeostasis and activation. One emerging family of such E3s is the Pelle-interacting (Peli) proteins, characterized by the presence of a cryptic forkhead-associated domain involved in substrate binding and an atypical RING domain mediating formation of both lysine (K) 63- and K48-linked polyubiquitin chains. A well-recognized function of Peli family members is participation in the signal transduction mediated by Toll-like receptors (TLRs) and IL-1 receptor. Recent gene targeting studies have provided important insights into the in vivo functions of Peli1 in the regulation of TLR signaling and inflammation. These studies have also extended the biological functions of Peli1 to the regulation of T-cell tolerance. Consistent with its immunoregulatory functions, Peli1 responds to different immune stimuli for its gene expression and catalytic activation. In this review, we discuss the recent progress, as well as the historical perspectives in the regulation and biological functions of Peli.
Keywords: Peli, Pellino, ubiquitination, NF-kB, TLR, T-cell tolerance
Introduction
Ubiquitination is a post-translational mechanism of protein modification that has long been recognized as a mechanism mediating protein degradation in the 26S proteasome.1 This hot research field has become even hotter during recent years due to the involvement of ubiquitination in various non-degradative molecular events, including protein–protein interactions, intracellular protein trafficking, DNA repair and activation of signaling molecules.2 Ubiquitination involves the covalent attachment of the small ubiquitin protein to the lysine residues of substrate proteins, as either monomers (monoubiquitination) or polymers (polyubiquitination).1 Formation of polyubiquitin chains involves the linkage of the C-terminus of one ubiquitin to an internal lysine residue of another ubiquitin. Since each ubiquitin has seven lysines, all of which can be used for the chain linkage, there are a large variety of polyubiquitin chains. In addition, ubiquitins can be connected through head-to-tail ligation, yielding linear ubiquitin chains. The different ubiquitin chains may mediate distinct biological functions. In particular, lysine 48 (K48)-linked polyubiquitin chains primarily target proteins for proteasomal degradation, whereas K63-linked polyubiquitin chains mediate non-degradative functions, including assembly of signaling complexes and activation of protein kinases.2
The ubiquitination reaction is catalyzed by the sequential action of three enzymes: the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzymes (E2s) and the ubiquitin ligases (E3s).1 This reaction is reversible, since the ubiquitin chains can be deconjugated by a family of ubiquitin-specific proteases, termed deubiquitinases.3 The specificity of ubiquitination is determined by E3s, a large family of proteins that interact with both specific substrates and E2s and function to transfer ubiquitin from E2s to the substrates.1 Based on their functional domains, E3s are divided into two major classes: the RING (really interesting new gene) finger E3s and HECT (homologous to E6-AP C-terminus) domain E3s, although atypical E3s have also been identified.4 The RING finger E3s are by far the largest and also most complex family of ubiquitin ligases, with about 579 members being annotated in the human genome.5 Consistently, the RING E3s have been linked to a large variety of physiological and pathological processes.6 Among the newly characterized RING E3s are the Pelle-interacting (Peli, also called Pellino) proteins, a family of evolutionally conserved immune regulators thought to regulate signaling from Toll-like receptors (TLRs) and IL-1 receptor (IL-1R).7, 8 Recent gene targeting studies have provided new insights into the in vivo biological functions of Peli.9, 10 Important progress has also been made to elucidate the biochemical mechanism mediating signal-induced Peli activation.11, 12, 13 In this review, we discuss the recent progress on the regulation and biological functions of Peli in the context of immune receptor signaling and inflammation.
E3s in the regulation of TLR signaling and NF-κB activation
An important function of K63 type of ubiquitination is mediating activation of the NF-κB signaling pathway.14, 15 NF-κB represents a family of transcription factors that participates in the regulation of multiple biological processes, including immune response and inflammation, cell growth and survival, and oncogenesis.16, 17, 18 NF-κB is normally sequestered in the cytoplasm via physical association with specific inhibitory proteins, IκBs. Activation of NF-κB typically involves degradation of a prototypical IκB member, IκBα, and rapid nuclear translocation of NF-κB, although a non-canonical pathway is also involved for NF-κB activation in specific biological processes.19 The IκBα degradation is triggered through its phosphorylation by a multisubunit IκB kinase (IKK) composed of two catalytic subunits (IKKα and IKKβ) and a regulatory subunit, NEMO (NF-κB essential modulator).16, 17 IKK mediates NF-κB activation by integrating signals from a large variety of innate immune receptors as well as the T- and B-cell antigen receptors. The activation of IKK by most immune receptors requires transforming growth factor-β (TGF-β)-activated kinase 1 (Tak1), a ubiquitin-dependent kinase that activates both IKK and MAP kinases (MAPKs).20, 21, 22 Like IKK, Tak1 exists as a complex containing two adaptor proteins: Tab1 and Tab2 (or its homologue Tab3). A common feature of Tab2 (or Tab3) and NEMO is the presence of a ubiquitin-binding domain that specifically binds to K63-linked polyubiquitin chains.14, 15, 23 Such ubiquitin-binding activity is crucial for the recruitment of Tak1 and IKK to upstream signaling adaptors that are conjugated with K63-linked polyubiquitin chains. Within the signaling complexes, both Tak1 and the IKK regulatory subunit NEMO are also conjugated with K63-linked ubiquitin chains.14, 24 It is generally believed that the binding of and/or conjugation with ubiquitin chains may trigger conformational changes in the kinases and, thereby, lead to their catalytic activation.
The ubiquitin-dependent NF-κB activation has been extensively studied in the TLR/IL-1R signaling pathways. TLRs form one of the major families of pattern-recognition receptors, which also include the RIG-I-like receptors, the NOD-like receptors and the C-type lectin receptors.25 By recognizing specific pathogen-associated molecular patterns, the pattern-recognition receptors mediate induction of innate immunity to form the first line of host defense against infections. Both human and mice have multiple TLR members (10 for human and 12 for mice), which detect and respond to a large array of pathogen-associated molecular patterns, ranging from surface molecules to nucleic acids of the invading microbes.25 In response to ligand stimulation, TLRs signal through intracellular adaptors, including the common adaptors MyD88 and TRIF. While TRIF mediates signaling from TLR3 and TLR4, MyD88 transduces signals from all TLRs, except TLR3, as well as from the IL-1R. A hallmark of the MyD88-dependent TLR/IL-1R pathway is the involvement of evolutionarily conserved IL-1R-associated kinases (IRAKs), including IRAK1, IRAK2, IRAK3 (also named IRAK-M) and IRAK4.25 In response to ligand stimulation, IRAK family members are rapidly recruited to the IL-1R/TLR, via the adaptor MyD88, and mediate activation of a downstream factor TRAF6. TRAF6 functions as both an adaptor and a K63-specific E3 ubiquitin ligase and amplifies the IL-1/TLR signal by stimulating several downstream signaling pathways, including those leading to activation of NF-κB and three families of MAPKs, extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK) and p38. These signaling pathways act cooperatively in the induction of genes encoding proinflammatory cytokines and chemokines. The TRIF-dependent TLR pathway also targets the activation of NF-κB, and this is dependent on an adaptor, receptor-interacting protein 1 (RIP1).26 TRIF stimulates the conjugation of K63-linked polyubiquitin chains to RIP1, which in turn is important for recruitment and activation of IKK.26 In addition to the NF-κB pathway, TRIF stimulates activation of the IKK-related kinases, TBK1 and IKKε, and their downstream transcription factor IRF-3, leading to induction of the type I interferons (IFNs).25 This antiviral pathway also requires K63 ubiquitination, which is mediated by the E3 ubiquitin ligase TRAF3.27, 28
In addition to TRAF6 and TRAF3, several other E3 ubiquitin ligases have been implicated in the positive or negative regulation of TLR/IL-1R signaling.29, 30, 31, 32 In particular, a recent study suggests a role for the cIAP1 and cIAP2 in the regulation of MyD88-dependent TLR signaling.31 cIAPs are RING E3s with both K63 and K48 types of ubiquitin ligase activity. In response to lipopolysaccharide (LPS)-mediated TLR4 stimulation, cIAPs are recruited to the MyD88 signaling complex, where they mediate K48 ubiquitination and degradation of TRAF3. Although TRAF3 is required for type I IFN signaling,27, 28 it serves as a negative regulator in the MyD88-dependent TLR pathway.33 TRAF3 degradation is essential for the dissociation of the MyD88 signaling complex from the receptor, a step that is required for stimulating the TRAF6-dependent activation of Tak1 and MAPKs.31 The recent demonstration of Peli proteins as RING E3s adds another family of E3s that regulates TLR signaling and inflammation.
Peli proteins as evolutionally conserved immune regulators
Discovery of Peli as a Drosophila immune regulator
Lower animals like insects do not have adaptive immune system, but they have efficient innate immunity to deal with microbial infections.34, 35 Early studies by Hans Boman and colleagues demonstrated that in response to bacterial infection, the silkmoth Hyalophora cecropia rapidly produces a set of antimicrobial peptides that form an important part of its humoral immunity.36 A connection between the insect antibacterial response and mammalian innate immunity was first indicated by the finding that H. cecropia has an NF-κB-like transcription factor, cecropia immune factor (CIF), which responds to bacteria and appears to mediate the induction of the antibacterial genes.37, 38 This idea was later on confirmed by the identification of an NF-κB analogue, Dif (dorsal-related immunity factor) from Drosophila melanogaster.39 Dif, together with Dorsal and Relish, form the NF-κB family in Drosophila that mediates the induction of various antimicrobial genes.34
Activation of Drosophila NF-κB is mediated by pattern recognition receptors, Toll and immune deficiency, which respond to fungi and bacteria, respectively.34, 35 The Toll pathway of Drosophila is analogous to the mammalian IL-1R/TLR pathway, whereas the immune deficiency pathway resembles the mammalian tumor-necrosis factor receptor (TNFR) pathway. In particular, the signal transduction of Toll involves recruitment of several intracellular components, including Drosophila MyD88, Tube and Pelle. Pelle is a homologue of mammalian IRAK1, characterized by a kinase domain and an N-terminal death domain. Although Tube has no obvious counterpart in the mammalian system, a recent study suggests that Tube originates from an ancestral gene related to IRAK4.40 Consistently, Tube functionally resembles IRAK4 to interact with and activates Pelle.41 Thus, the Drosophila Tube/Pelle pair may be considered the functional equivalent of mammalian IRAK4/IRAK1.
Drosophila Peli (Pellino, hereafter named dPeli) was identified as a Pelle-interacting protein in yeast two-hybrid screening aiming to identify downstream signaling molecules connecting Pelle to the activation of NF-κB.42 dPeli is a protein of 424 amino acids that shares about 50% identity and 70% homology with its counterpart in Caenorhabditis elegans, suggesting Peli to be an evolutionarily conserved gene. Interestingly, the dPeli–Pelle interaction requires the kinase activity of Pelle, and Peli may preferentially bind to autophosphorylated Pelle.42 The function of dPeli in Drosophila innate immunity has been studied recently using dPeli mutant flies (Pli).43 Pli flies have impaired induction of the antibacterial peptide Drosomycin and reduced survival in response to infection by the gram-positive bacterium Micrococcus luteus. Consistently, overexpression of dPeli induces the expression of Drosomycin. Overall, these findings suggest that dPeli is a positive regulator of Drosophila innate immunity against certain strains of gram-positive bacteria.
Mammalian Peli members
The mammalian Peli members were initially identified based on database search,44 and subsequent cloning revealed two highly homologous mouse Peli members (Peli1 and Peli2).45 The function of Peli was first reported in a study with the mouse Peli2. Peli2 is abundantly expressed in non-lymphoid organs but only weakly expressed in spleen and thymus. Analogous to dPeli, Peli2 interacts with IRAK-1, which occurs in an IL-1 inducible manner. In a mouse embryonic fibroblast cell line (C3H10T1/2) and a human embryonic kidney cell line (HEK293), reduction of Peli2 expression by antisense RNA inhibits induction of the chemokine gene IL-8 by IL-1 and the TLR4 ligand LPS in a NF-κB-dependent manner.46 The more detailed mechanism of Peli function was revealed in another study using HEK 293 cells stably expressing the IL-1R.47 In response to IL-1 stimulation, Human Peli1 interacts with both IRAK1 and IRAK4 upon IL-1 stimulation. It appears that Peli1 functions as a scaffold protein involved in the formation of an intermediate IL-1R signaling complex composed of IRAK4, IRAK1 and TRAF6.47 It has been proposed that Peli1 may induce the release of the IRAK complex from the receptor,47 a step required for the activation of downstream signaling events by IL-1 and TLRs.48 RNAi-mediated Peli1 knockdown suggests that this Peli member is required for IL-1-stimulated activation of NF-κB and induction of IL-8. It is currently unclear whether Peli1 indeed is required for mediating the release of IRAK complex from IL-1R or TLRs and how Peli1 could possibly regulate this important step of IL-1R/TLR signaling.
Despite the uncertainties regarding the precise mechanism of Peli function, it is clear that interaction with IRAK1/4 and IRAK-associated factors is a common property of the different Peli members. Like the human Peli1 and mouse Peli2, human Peli2 binds to IRAK1 and IRAK4,49 as well as to TRAF6 and Tak1.50 Similarly, the two isoforms of Peli3, Peli3a and Peli3b, interact with IRAK1, TRAF6, Tak1, as well as to the NF-κB-inducing kinase.51, 52 It is important to note though that these binding assays were performed in transfected mammalian cells. Since the different Peli-interacting proteins are often present in the same complexes, it is likely that some of these proteins may associate with Peli indirectly via other proteins, like IRAK1.
In contrast to the consistent findings on the Peli-interacting factors, the functional studies of Peli members have generated some discrepancies. For example, whereas one study suggests that Peli2 activates NF-κB,46 another study suggests that Peli2, but not Peli1, mediates activation of the MAPK JNK and induction of its downstream transcription factors AP-1 and Elk-1.50 Yet, another study suggests that Peli2 is not required for IL-1-induced NF-κB target gene expression and that overexpressed Peli2 does not reproducibly activate MAPK nor induce reporter gene expression through specific transcription factors.49 The biological functions of Peli3 seem to be also complex. Some studies reveal that both isoforms of Peli3 activate MAPKs (ERK, JNK and p38) and the transcription factors AP-1 and CREB, although they do not activate NF-κB.51, 52 On the other hand, another study demonstrates that the shorter isoform of Peli3 (Peli3b) negatively regulates IL-1-stimulated activation of NF-κB and JNK.53 This latter study suggests that Peli3b mediates K63 ubiquitination of IRAK1 at K134. Since this site is also used for conjugating K48-linked ubiquitin chains, the Peli3b-mediated K63 ubiquitination of IRAK1 inhibits the K48 ubiquitination and degradation of IRAK1 upon IL-1 stimulation. The degradation of IRAK1 seems to be required for IL-1-stimulated activation of Tak1 and its downstream signaling events, since a proteasome inhibitor (MG132) inhibits IRAK1 degradation and also blocks activation of IKK.53 This model is intriguing, although more studies are required for further validation using different cell systems, particularly primary cells derived from Peli3 knockout (KO) mice. In terms of the discrepancies between the different studies, it is possible that at least some of the differences were attributed to the use of different experimental systems.
Peli proteins are E3s with a new RING and a cryptic forkhead-associated (FHA) domain
E3 function of Peli
Peli proteins were initially thought to serve as scaffold proteins that regulate the dynamic assembly of intermediate signaling complexes in the IL-1R pathway.47 Although how Peli exerts such functions remains incompletely understood, the discovery of Peli proteins as E3 ubiquitin ligases provides critical insights into the biochemical mechanism of Peli action.54 Initial sequence analyses reveal that the C-terminal portion of Peli members contain two conserved Cys-Gly-His motifs and two conserved Cys-Pro-X-Cys motifs, reminiscent of C3HC4 RING-like zinc finger domains.44, 47 By sequence alignments with the C3HC4 RING consensus sequence,55 Beyaert and colleagues demonstrate that although Peli proteins do not contain a typical C3HC4 RING, they possess a closely related motif termed CHC2CHC2 RING.54
The E3 function of Peli protein was first demonstrated by the finding that all Peli members were able to induce the polyubiquitination of IRAK1 when overexpressed in mammalian cells.54 This function of Peli proteins is dependent on the presence of an intact RING-like structure. Since the RING domain is dispensable for Peli interaction with IRAK1, these findings strongly suggest that Peli members function as RING E3s that mediate IRAK1 ubiquitination.54 The E3 function of Peli proteins was later on confirmed by using a cell-free ubiquitination system.56, 57 These later studies also reveal that Peli can function together with the Ubc13-Uev1a dimeric E2 ubiquitin-conjugating enzyme to catalyze K63-linked polyubiquitin chains.56, 57 However, Peli does not seem to be an exclusively K63-specific E3 ubiquitin ligase, since it also catalyzes the production of K48 and K11 types of polyubiquitin chains when working with other E2s, such as UbcH3, UbcH4, UbcH5a and Ubc5b.57 The ability of Peli to utilize different E2s suggests a high level of versatility in Peli function. As will be discussed in a later section, both the K63- and K48-specific E3 functions of Peli1 have been linked to important physiological functions.9, 10 Which E2s are required for in vivo functions of Peli and how the chain specificity of Peli-mediated ubiquitination is regulated remains an intriguing topic of investigation.
Although Peli proteins clearly have E3 ligase activity, their mode of action is not well understood. It has been shown that the K63-linked polyubiquitin chains catalyzed by Peli1 and Ubc13-Uev1a are not attached to IRAK1 or Peli1 in vitro, but rather stay as free forms.11, 57 This is in sharp contrast to the efficient conjugation of K63 polyubiquitin chains to IRAK1, when it is coexpressed with Peli in cells.54, 56, 57 Since Peli can mediate ubiquitination together with different E2s, it is thought that Peli-mediated IRAK1 ubiquitination may be a two-step process involving two different E2s. Peli1 may first act with a different E2 to mediate IRAK1 monoubiquitination and then pair with Ubc13-Uev1a to extend the ubiquitin chains with K63 linkage.11 Although this idea needs to be tested by additional experiments, the poor in vitro ubiquitin-conjugating activity of the Peli1/Ubc13/Uev1a system has also been observed with another substrate, RIP1, which is efficiently ubiquitinated by Peli1 in vivo.9 Despite these uncertainties, the accumulating studies have more and more firmly confirmed the E3 ubiquitin ligase function of Peli proteins. The functional significance of the RING-like domain of Peli is also emphasized by a recent finding that this structural domain is evolutionally conserved from human to sponge Peli proteins.58
Substrate binding by an FHA domain
In addition to their E2-binding domain (RING or HECT), single-subunit E3s also contain a domain that mediates substrate binding. It is known that the C-terminal RING domain of Peli is not involved in the Peli–IRAK1 interaction.54 Initial sequence analyses did not reveal any notable structural domains within the N-terminal region of Peli that binds IRAK1. However, a structural study based on X-ray crystallography led to the discovery of a cryptic FHA domain located in the N-terminal region (amino acids 15–275) of Peli2.59 A unique feature of FHA domains is their interaction with protein motifs containing a phosphorylated threonine residue.60 Compared with the typical FHA domains, the Peli2 FHA domain contains two insertion regions that form a wing-like appendage. Nevertheless, the cryptic FHA domain of Peli2 has the conserved β-strands and all of the key amino-acid residues known to be involved in phosphothreonine binding.59 Indeed, the FHA domain of Peli2 binds to phosphothreonine-containing peptides, as well as to the IRAK1 protein expressed in HEK293 cells, and these interactions rely on the conserved ligand-binding amino-acid residues.59 Interestingly, an earlier biochemical study identified a motif of Peli3 (amino acids 43–47; KYGEL) that is critical for binding of Peli3 to IRAK1.52 This motif, which is equivalent to amino acids 18–22 of Peli2, happens to form the first β-strand of the FHA core.59 This finding further suggests that FHA is a functional domain in all Peli members.
The discovery that Peli contains a phosphothreonine-binding domain provides molecular basis to the previous findings that Peli–IRAK1 interaction is induced by IL-1R/TLR signals46, 47, 51 and requires the kinase activity and phosphorylation of IRAK1.42, 49, 54 Collectively, the results of this structural study, along with those of the previous biochemical analyses, suggest an intriguing model for the inducible interaction between Peli and IRAK1. In response to IL-1R/TLR signals, IRAK1 becomes phosphorylated by IRAK4, and phosphorylated IRAK1 recruits Peli via FHA–phosphothreonine interaction. The recruited Peli then catalyzes K63 ubiquitination of IRAK1, thereby allowing the recruitment and activation of downstream kinases Tak1 and IKK. Since Peli also interacts with various other proteins, future studies should examine whether the same or different mechanisms mediate Peli interaction with other substrates or regulatory proteins.
Mechanisms of Peli regulation
Activation by phosphorylation
How the function of E3s is regulated is still poorly understood. Nevertheless, evidence is emerging that phosphorylation plays an important role in regulating the E3 activity and ligand-binding function of E3s.61, 62, 63, 64 Several recent studies demonstrate that the E3 ligase activity of Peli proteins is also regulated by phosphorylation.11, 12, 13, 57 It was noted initially that IRAK1 and IRAK4 phosphorylate Peli members in transfected cells.49, 54 Using recombinant proteins and in vitro ubiquitination assays, Cohen and colleagues demonstrate that the phosphorylation of Peli proteins greatly stimulates their E3 ligase activity.11, 57 How phosphorylation activates Peli remains unclear, but it appears that this mechanism regulates the general E3 function, instead of specific E2 usage, since Peli1 phosphorylation stimulates its E3 function regardless of which E2 is used.11
The functional study of Peli1 phosphorylation is complicated by the presence of multiple potential phosphorylation sites.11 At least in vitro, IRAK1 and IRAK4 phosphorylate Peli1 at up to 10 serines or threonines, the simultaneous mutation of all of which is required to prevent Peli1 activation.11 A caveat of such mutagenesis approach is the possible cause of conformational changes that could have affected the IRAK binding or E3 ubiquitin ligase activity of Peli1. Nevertheless, this in vitro study provides intriguing insights that may help predict the mechanism of Peli regulation. Most interestingly, the major phosphorylation sites of Peli1 are clustered within two regions: the first region (amino acides 76–86) correlates with the wing-like appendage of the FHA domain and the second (containing threonine-288 and serine-293) is located just before the RING domain. Since FHA binds to phosphothreonine-containing motifs, it would be interesting to test whether phoshorylation of Peli1 promotes its dimerization or intramolecular interaction, which in turn could serve as a mechanism of Peli activation. It is also possible that phosphorylation of the FHA region of Peli may promote its interaction with IRAK1 or other substrates/regulators.
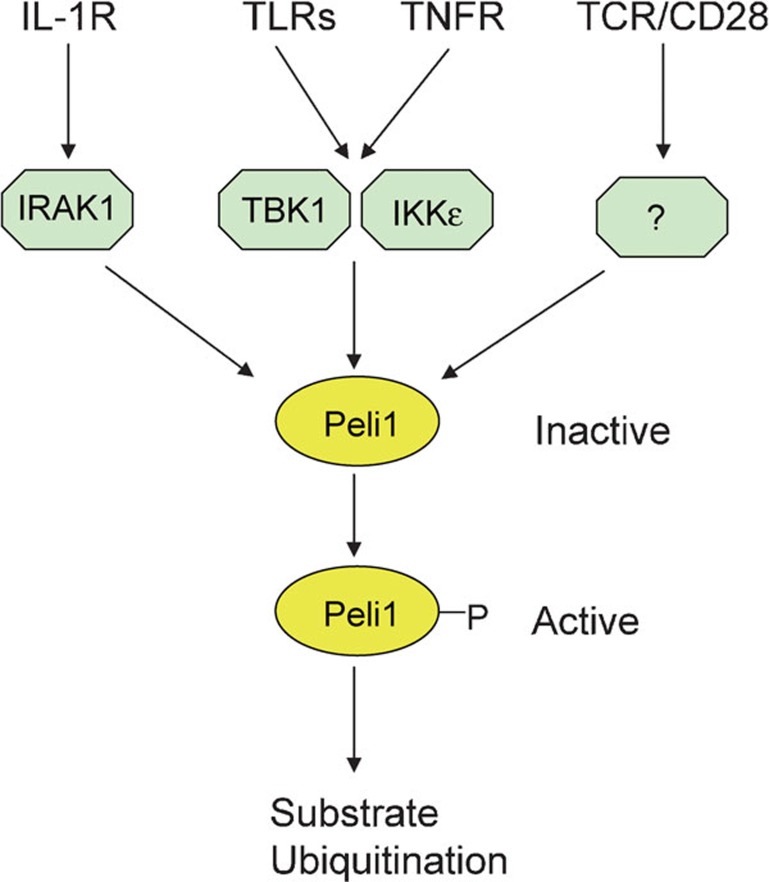
Recent studies reveal that in addition to IL-1, many other immune stimuli, such as ligands of TRIF-dependent and MyD88-dependent TLRs and tumor-necrosis factor-α (TNF-α), stimulate the phosphorylation and activation of Peli112, 13 (Figure 1). Interestingly, despite the implication of IRAK1 as the major kinase of Peli, IRAK1 is required only for Peli1 activation by IL-1, whereas the IKK-related kinases, TBK1 and IKKε, mediate the phosphorylation and activation of Peli1 in the TNFR and TLR pathways. Traditionally thought to be TRIF-dependent kinases, TBK1 and IKKε have now been shown to also respond to signals from the MyD88-dependent TLRs, as well as the TNFR.12, 13 Since both IL-1R- and MyD88-dependent TLRs activate IRAK1, it is surprising that IRAK1 is important only for the IL-1R pathway. Whether this is a cell type-specific function or involves a yet-to-be-defined mechanism remains to be investigated. Since Peli1 is also activated in T cells,10 it raises the question of which kinase(s) mediates Peli1 phosphorylation and activation by the T-cell receptor (TCR) and the T-cell costimulatory molecule CD28 (Figure 1).
Figure 1.
Signal-induced Peli1 activation. Peli1 exists as an inactive form but becomes activated in response to signals from different immune receptors. Peli1 activation involves its phosphorylation by IRAK1 and the IKK-related kinases TBK1 and IKKε. Whereas IRAK1 mediates Peli1 activation by IL-1R, TBK1 and IKKε mediate Peli1 activation by TLRs and TNFR. It remains unclear how Peli1 is activated in T cells by the TCR and CD28 signals. IKK, IκB kinase; IRAK, IL-1R-associated kinase; Peli1, Pelle-interacting protein 1; TCR, T-cell receptor; TLR, Toll-like receptor; TNFR, tumor-necrosis factor receptor.
Ubiquitination and sumoylation
When co-expressed with IRAK1 in mammalian cells, Peli not only induces ubiquitination of IRAK1, but also becomes ubiquitinated itself.52 Since IRAK1 activates the E3 ligase of Peli, it is likely that the Peli ubiquitination represents auto-ubiquitination, although the involvement of other E3 ligases is also possible. At least in vitro, IRAK1-activated Peli1 undergoes auto-ubiquitination in the presence of the E2 UbcH4.11, 57 Since Peli can conjugate K48- and K11-linked polyubiquitin chains, known to mediate protein degradation, the auto-ubiquitination of Peli may contribute to its inducible degradation as shown for Peli3.7, 52 The Peli1 auto-ubiquitination occurs at three major K residues, including K169, K202 and K266. K169 and K202 are located in the wing-like appendage and the FHA core, respectively, whereas K266 is located C-terminal to the FHA domain.11 It would be interesting to examine whether Peli1 auto-ubiquitination interferes with its binding to IRAK1 or other substrates. If this is indeed the case, it will suggest that Peli1 ubiquitination may serve as a mechanism to both terminate its function and target its proteolysis.
A recent study reveals that Peli1 is also conjugated with the small ubiquitin-related modifier (SUMO) both in vitro and in transfected cells.65 This finding is potentially interesting, since accumulating evidence suggests crosstalks between sumoylation and ubiquitination.66 On the one hand, SUMO may interfere with ubiquitination by competing with ubiquitin for attaching to the same lysine residues. On the other hand, sumoylation can prime certain proteins for ubiquitination. Mutagenesis analyses identified five lysine residues (K202, K266, K295, K297 and K303) as potential SUMO acceptor sites of Peli1. Of note, two of these residues (K202 and K266) are also used as ubiquitination acceptor sites, suggesting the possible involvement of direct crosstalks between sumoylation and ubiquitination in the regulation of Peli1 function. However, the functional significance of Peli1 sumoylation has not been investigated. It also remains to be examined whether this modification of Peli1 is induced during TLR/IL-1R signaling or the activation of other cellular pathways.
Transcriptional regulation
Another frequently seen mechanism of E3 regulation is through the control of gene expression. An early study identified Peli1 as one of the TRIF-dependent genes induced by the TLR4 ligand LPS in dendritic cells.67 This finding has been confirmed and extended by a recent work demonstrating that Peli1 is induced at both RNA and protein levels in response to stimulation by both LPS and the TLR3 ligand polyIC.12 These TRIF-dependent TLRs are known to activate the IKK-related kinases, TBK1 and IKKε, which in turn induce the nuclear translocation of the transcription factor IFN-regulatory factor 3 (IRF3) and induction of type I IFNs.68, 69 Peli1 induction requires TBK1/IKKε and the downstream transcription factor IRF3, but it does not involve the type I IFNs.12 It is thus likely that Peli1 is a target gene of IRF3. Since Peli1 is also induced in T cells by the TCR and CD28 costimulatory signals,10 it raises the question of which signaling pathway and transcription factor mediates the T cell-specific Peli1 induction. The induction of Peli1 in T cells appears to serve as an autoregulatory mechanism to prevent aberrant NF-κB activation and T-cell activation. As will be discussed later, Peli1 negatively regulates T-cell activation by targeting the ubiquitination and degradation of a major NF-κB family member, c-Rel.10 However, since Peli1 is a positive regulator in innate immune cells, it is currently unclear how the inducible expression of Peli1 contributes to the regulation of TLR signaling.
The expression level of Peli may also be subject to regulation at post-transcriptional levels, as suggested by the recent identification of Peli1 as a target of a microRNA, miR-21.70 The level of Peli1 mRNA is inversely correlated with that of miR-21 during liver regeneration. Sequence analyses using the TargetScan program revealed a putative target sequence of miR-21 in the 3′-untranslated region of Peli1. Insertion of the 3′-untranslated region of Peli1 downstream of a luciferase reporter gene renders the reporter sensitive to miR-21-mediated inhibition. Overexpression of miR-21 in HEK293 cells also inhibits the induction of the NF-κB reporter gene.70 Although this finding is potentially interesting, it is yet to be examined whether the level of endogenous Peli1 mRNA and protein is indeed regulated by miR-21 and whether miR-21-mediated NF-κB inhibition is mediated through Peli1 down regulation.
In vivo functions of Peli1 revealed by gene targeting studies
Gene targeting in mice is a powerful approach to study the physiological functions of genes. However, one technical issue of this approach in studying the function of gene families is the functional redundancy among family members. The extensive sequence/structural homology between the Peli family members predicts functional redundancy. However, some important non-redundant functions of Peli1 have recently been uncovered through the study of Peli1 KO mice.9, 10
Regulation of TRIF-dependent TLR signaling
The Peli1 KO mice are grossly normal and do not have prominent abnormalities in the development of immune organs and cells.9 However, these mutant animals display reduced sensitivity to endotoxin shock in an acute inflammation model that involves injection of a low-dose of LPS together with 𝒹-galactosamine (a liver-specific translation inhibitor that increases LPS sensitivity).9 In this model, the lethality is primarily mediated through LPS-induced production of TNF-α and also involves other proinflammatory cytokines such as IL-6 and IL-12. The reduced lethality of Peli1 KO mice suggests impaired inflammatory responses to LPS. Indeed, the serum level of proinflammatory cytokines is greatly (TNF-α and IL-6) or moderately (IL-12) reduced in the mutant animals upon LPS/𝒹-galactosamine challenge. The Peli1 deficiency also diminished the in vivo response of mice to polyIC injection in both cytokine production and lethality. In vitro studies further demonstrate a role for Peli1 in mediating LPS- and polyIC-induced proinflammatory gene expression in several cell types, most prominently B cells and MEFs.9 LPS and polyIC stimulate TLR4 and TLR3, respectively, which are known to induce the TRIF-dependent TLR pathway leading to the induction of both proinflammatory cytokines and type I IFNs.25 However, Peli1 is dispensable for IFN induction by both LPS and polyIC, suggesting the selective involvement of Peli1 in the proinflammatory axis of TRIF-dependent TLR signaling.
One major surprise in the Peli1 KO mouse study is the finding that Peli1 is dispensable for IL-1-induced gene expression and signaling, since in vitro studies suggest a critical role for Peli family members in the regulation of IRAKs and IL-1R signaling. Peli1-deficient MEFs do not show any notable defect in IL-1-stimulated expression proinflammatory genes.9 Similarly, the Peli1 KO and control mice produce comparable levels of IL-6 in response to IL-1 injection. One possible explanation to these findings is that Peli1 may be functionally redundant with other Peli1 members in the regulation of IRAKs. IRAK1 and IRAK4 are also involved in MyD88-dependent TLR signaling pathway. However, Peli1 appears to be largely dispensable for gene induction by the MyD88-dependent TLRs. This conclusion is not absolute, though, since the Peli1 deficiency does affect the function of some MyD88-dependent TLRs, such as TLR9 and TLR2/6, depending on the cell types.9 Of note, IRAK1 deficiency also does not cause an overall defect in the MyD88-dependent TLR signaling, although it affects some specific aspects of TLR signaling.71, 72 Thus, another possibility is that Peli1 is required for the activation of IRAK1, but not that of IRAK4.
The TRIF-dependent TLR pathway activates two major downstream signaling axes, which lead to activation of NF-κB and IRF3, respectively. As discussed already, the IRF3 activation is mediated by the IKK-related kinases, TBK1 and IKKε. An important signaling factor that connects TRIF-dependent TLR signals to NF-κB is the adaptor molecule RIP1.73 The signaling function of RIP1 involves its conjugation of K63-linked polyubiquitin chains, which appear to facilitate the recruitment and activation of downstream kinases, Tak1 and IKK.26 Interestingly, the polyIC-induced RIP1 ubiquitination is largely blocked in Peli1-deficient MEFs.9 Consistently, Peli1 is required for polyIC-stimulated activation of IKK and NF-κB MEFs and B cells. On the other hand, the Peli1 deficiency only partially inhibited the activation of IKK and NF-κB by LPS, reflecting the fact that TLR4 also mediates IKK/NF-κB activation via the MyD88-dependent TLR pathway. Furthermore, Peli1 is dispensable for IKK/NF-κB activation by IL-1R and MyD88-dependent TLRs, further emphasizing a role for Peli1 in regulating the TRIF-dependent TLR signaling (Figure 2). It is important to note, however, the phenotype of Peli1 deficiency in the TRIF-dependent TLR signaling varies among cell types, which is prominent in B cells and MEFs and only moderate in bone marrow-derived macrophages and dendritic cells.9 Clearly, more work is warranted to examine the potential functional redundancy of the Peli family members in both the IL-1R and TLR signaling pathways. While generation of triple KO mice, lacking all three Peli members, is the ultimate goal, another approach is to find the cell types that predominantly express Peli1 over the other Peli members. As will be discussed below, this approach has led to the discovery of an unexpected function of Peli1 in the regulation of T-cell activation.10 More comprehensive gene expression analyses may lead to the identification of relevant cell types for the study of TLR/IL-1R signaling. Nevertheless, the current findings suggest that Peli1 mediates RIP1 ubiquitination and plays a critical role in mediating NF-κB activation by TRIF-dependent TLR pathway (Figure 2).
Figure 2.
Peli1 mediates IKK activation by the TRIF-dependent TLR pathway. Based on the signaling adaptors, TLR signaling can be classified into two major pathways: the MyD88- and TRIF-dependent pathways. TLR3 is the only TLR that exclusively signals through TRIF, whereas TLR4 signals through both TRIF and MyD88. The rest of the TLRs, as well as IL-1R, signal through MyD88. An important mechanism of TLR signaling is K63 type ubiquitination of signaling adaptors, including TRAF6, RIP1 and TRAF3, which mediate activation of downstream signaling events. Activation of IKK/NF-κB by the TRIF-dependent TLR pathway, but not that of the MyD88 pathway, depends on Peli1. Peli1 functions by mediating ubiquitination of RIP1 and, thereby, activating IKK and NF-κB. The TRAF3-regulated signaling pathway may crosstalk with the RIP1 pathway through induction of Peli1 gene expression and TBK1-/IKKε-mediated Peli1 activation. IKK, IκB kinase; Peli1, Pelle-interacting protein 1; RIP1, receptor-interacting protein 1; TCR, T-cell receptor; TLR, Toll-like receptor.
Regulation of T-cell activation and autoimmunity
As discussed above, both in vitro and in vivo studies have established Peli as a family of innate immune regulators. However, since Peli possesses both K63 and K48 E3 ligase activities, it suggests the involvement of Peli in different biological functions. Indeed, a recent study has revealed an unexpected role for Peli1 in regulating T-cell activation and homeostasis.10 This finding was triggered by a gene expression analysis to compare the relative expression levels of different Peli family members in innate and adaptive immune cells. Notably, both innate immune cells (macrophages and dendritic cells) and MEFs express at least two of the three Peli members at comparable levels, further indicating potentially functional redundancy of Peli in certain aspects of TLR/IL-1R signaling. Interestingly, however, Peli1 is unique among the family members in that it is highly expressed in B and T lymphocytes. Although B cells also have substantial expression of Peli3, T cells only express a negligible level of Peli2 and Peli3 compared to the strikingly high level of Peli1 expression.10 Moreover, the level of Peli1 is further elevated following T-cell activation. These findings suggest a role for Peli1 in regulating T-cell activation.
Based on the critical role of Peli1 in mediating TLR signaling, it would predict that loss of Peli1 in T cells might impair T-cell activation and cause immune deficiency. Surprisingly, the opposite function of Peli1 was discovered using the Peli1 KO mice.10 The Peli1-deficient T cells are hyper-responsive to in vitro stimulation by agonistic TCR/CD28 antibodies (anti-CD3 and anti-CD28) or T-cell mitogens (phorbyl myristate acetate and ionomycin). The mutant T cells appear to also undergo activation in vivo, since the Peli1 KO mice contain substantially increased frequencies of CD4 and CD8 T cells with memory or effector phenotypes. Under normal conditions, T cells are tightly controlled so that they are responsive to foreign antigens, but are tolerant to antigens derived from self-tissues or commensal microbes.74 Since the animals are housed under specific pathogen-free conditions, the in vivo T-cell activation in Peli1 KO mice is very likely due to the loss of peripheral T-cell tolerance. Indeed, the activated T cells seem to be self-reactive and infiltrate into various nonlymphoid organs and cause tissue inflammation. At old ages, the Peli1 KO mice also have other symptoms of autoimmunity, including elevated antinuclear autoantibodies and immunoglobulin deposition in kidney glomeruli.10
Peripheral T-cell tolerance is mediated by both intrinsic and extrinsic mechanisms; the intrinsic mechanism involves negative regulation of the TCR/CD28 signaling by T-cell intrinsic molecules, whereas the extrinsic mechanism involves the immunosuppressive function of T regulatory (Treg) cells and other suppressive cells.75, 76, 77, 78 Thus, autoimmunity may occur due to either Treg defect or impaired negative regulation of TCR/CD28 signaling. Peli1 is not required for the generation or in vitro suppressive function of Treg cells. Although it is unclear whether Peli1 mediates the in vivo function of Treg cells, mixed bone marrow adoptive transfer study clearly demonstrates a role for this E3 in mediating the T-cell intrinsic mechanism of tolerance. This finding is consistent with the hyper-responsive phenotype of the Peli1-deficient T cells to in vitro stimulation.
Loss of Peli1 does not substantially influence the TCR-proximal signaling events, such as activation of the protein tyrosine kinase Zap70, or the activation of downstream kinases, IKK and ERK. The early phase activation of NF-κB, following T-cell activation by agonistic anti-TCR and anti-CD28 antibodies or mitogens, is also largely normal in the Peli1-deficient T cells. However, the Peli1 deficiency results in aberrant activation of late-phase NF-κB. This is due to a specific role for Peli1 in the negative regulation of c-Rel, an NF-κB family member that is activated with delayed kinetics and is critically involved in T-cell activation.79 Peli1 appears to negatively regulate c-Rel by mediating the ubiquitination and proteolysis of activated c-Rel (Figure 3). In response to TCR/CD28 signals, c-Rel undergoes degradative ubiquitination in wild-type T cells, which was detectable only when the cells were incubated with a proteasome inhibitor. This molecular event is dependent on Peli1. At least in a transfection model, Peli1 induces c-Rel ubiquitination with K48 linkage. It remains to be further examined whether Peli1 functions as the direct E3 of c-Rel or indirectly induces c-Rel ubiquitination through another E3. Nevertheless, Peli1 physically interacts with c-Rel under both endogenous and transfected conditions. Controlling c-Rel activation may be an important mechanism by which Peli1 mediates intrinsic T-cell tolerance, since c-Rel is crucial for the activation and differentiation of T cells and for the prevention of T-cell tolerance.80, 81, 82, 83, 84, 85, 86
Figure 3.
Peli1 negatively regulates T-cell activation by mediating c-Rel degradation. In T cells, c-Rel is an important NF-κB member that regulates IL-2 gene expression and T-cell activation. c-Rel functions as either an homodimer or heterodimers associated with different NF-κB members, such as p50 and RelA. Upon IKK-mediated degradation of IκB, the c-Rel-NF-κB dimers translocate to the nucleus, where they participate in the transactivation of IL-2 and other target genes. The TCR/CD28 signals also stimulate the expression of Peli1 via a yet-to-be-characterized transcription factor(s), and the accumulated Peli1 negatively regulates the late-phase activation of NF-κB by mediating ubiquitination and degradation of c-Rel. It is currently unknown whether Peli1 exerts this function in the cytoplasm or nucleus, but Peli1 is located in both of these cellular compartments (Chang M et al., unpubl. data). IKK, IκB kinase; Peli1, Pelle-interacting protein 1; RIP1, receptor-interacting protein 1; TCR, T-cell receptor; TLR, Toll-like receptor.
Interplay with TGF-β signaling components
In addition to directly manipulating the key molecules in NF-κB signaling cascade, Peli1 can also mediate crosstalk between the NF-κB and TGF-β pathways, which are known to antagonize each other under inflammatory settings.87, 88, 89 TGF-β belongs to a superfamily of cytokines, which also includes bone morphogenic proteins and activin. TGF-β is widely expressed in immune cells and plays unique and pivotal regulatory roles in the regulation of immune responses.90, 91 The TGF-β signaling pathway involves sequential activation of the receptor complex (type I and II receptors) and the signal transducers, Smads, upon receptor-ligand binding.87, 92 There are eight mammalian Smads, which can be subgrouped as receptor-activated Smads, common Smad and inhibitory Smads (I-Smads) according to their function. Smad2 and Smad3 are the two receptor-activated Smads selectively activated by TGF-β signaling, which form a complex with the common Smad, Smad4, and corporate with other transcription factors or cofactors to regulate downstream gene expression. The TGF-β signaling pathway is tightly controlled by the I-Smads, Smad6 and Smad7, which are induced in response to TGF-β stimulation and, thereby, form a feedback regulatory loop.92, 93, 94, 95 Smad6 and Smad7 attenuate TGF-β signaling by interacting with TGF-β receptor I, thus interfering with Smad2/3 phosphorylation and subsequent activation.96, 97 The I-Smads also recruit ubiquitin ligases to TGF-β receptor I, leading to the polyubiquitination and degradation of the receptor.98
The crosstalk between TGF-β signaling and other signal transduction pathways is at least partially mediated by the I-Smads. Multiple inflammatory cytokines, such as IFN-γ, TNF-α and IL-1β, induce Smad7 protein expression and, thereby, counteract on the immunosuppression controlled by TGF-β signaling.99, 100 It has also been observed that TGF-β signaling antagonizes NF-κB activation induced by proinflammatory cytokines. In this regard, recent studies indicate that association between I-Smads and Peli1 is critical for attenuating IL-1R-/TLR-induced NF-κB activation by TGF-β.88, 89 It appears that TGF-β induces the expression of both Smad6 and Smad7, which compete with IRAK1 and IRAK4 for engaging Peli1, thereby interfering with the formation of the MyD88/IRAK4/IRAK1/Peli1/TRAF6 signaling complex required for IKK/NF-κB activation. Indeed, the interaction of Smad6/7 with Peli1 blocks IL-1R-/TLR-stiumulated IκBα degradation, NF-κB nuclear translocation and subsequent induction of inflammatory genes in both HEK293 cells and primary macrophages.88, 89 Both the induction of I-Smads and their physical interactions with Peli1 are required for TGF-β-mediated negative regulation of NF-κB. The MH2 domain of I-Smads is responsible for their association with Peli1, which has non-redundant binding sequences for Smad6 and Smad7, suggesting potentially simultaneous interaction of Peli1 with both I-Smad proteins.89 Indeed, induction of both I-Smads can suppress NF-κB activity more effectively than any one of the two. These findings shed light on a novel function of Peli1 in NF-κB regulation, although additional studies are required for better understanding the underlying molecular mechanism and physiological relevance of the Peli1/I-Smad interplay.
Conclusions and outstanding questions
Since the initial characterization of mammalian Peli proteins, significant progress has been made in understanding the regulation and biological functions of this family of evolutionarily conserved signaling factors. In particular, the characterization of Peli members as RING finger E3s has provided critical clues to their signaling functions. Since Peli possesses both K63 and K48 types of ubiquitin ligase activity, the function of Peli may be more complex than what was initially proposed. Indeed, recent Peli1 gene-targeting studies have revealed critical, but opposing, functions in the regulation of innate immunity and T-cell activation. One future task is to characterize additional physiological or pathological functions of Peli and to examine how the different functions of Peli are regulated. Since Peli members share extensive sequence homology, it is important to examine whether the different Peli proteins have functional redundancies. Another important issue to be addressed is the potential association of Peli with human diseases. Although this possibility is suggested by the current findings, it is yet to be examined by genome-wide association studies.
Understanding how Peli is regulated in vivo represents another challenge for future studies. Nevertheless, an important clue has been provided by the recent observations that IRAKs and TBK1/IKKε mediate the inducible phosphorylation of Peli1 by IL-1R and TLR, respectively. It appears that signal-induced Peli phosphorylation may serve as a molecular trigger for its activation. A number of outstanding questions remain to be addressed in this area. For example, it is unclear how Peli1 phosphorylation stimulates its E3 ubiquitin ligase activity. It remains to be tested whether phosphorylation promotes the binding of Peli1 by specific E2s. Future studies should also examine whether phosphorylation similarly regulates the E3 ligase activity of other Peli family members in vivo and whether this involves the same or different protein kinases. Another outstanding issue to be further addressed is regarding the IRAK1/Peli mutual regulation model previously proposed based on in vitro studies. Recent studies support the involvement of IRAK1 in the in vivo activation of Peli1, but the inverse function of Peli in IRAK1 regulation remains to be speculative. Notwithstanding, accumulative findings have established Peli as a new family of RING finger E3s with important roles in the regulation of both innate immune receptor signaling and T-cell tolerance.
Acknowledgments
The research of S-CS is supported by grants from the US National Institutes of Health (AI057555, AI064639 and GM84459) and the GS Hogan Gastrointestinal Cancer Fund.
References
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol. 2008;8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Moynagh PN. The Pellino family: IRAK E3 ligases with emerging roles in innate immune signalling. Trends Immunol. 2009;30:33–42. doi: 10.1016/j.it.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Schauvliege R, Janssens S, Beyaert R. Pellino proteins: novel players in TLR and IL-1R signalling. J Cell Mol Med. 2007;11:453–61. doi: 10.1111/j.1582-4934.2007.00040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Jin W, Sun SC. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat Immunol. 2009;10:1089–1095. doi: 10.1038/ni.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Jin W, Chang JH, Xiao Y, Brittain GC, Yu J, et al. The ubiquitin ligase Peli1 negatively regulates T cell activation and prevents autoimmunity. Nat Immunol. 2011;12:1002–1009. doi: 10.1038/ni.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H, Peggie M, Campbell DG, Vandermoere F, Carrick E, Cohen P. Identification of the phosphorylation sites on the E3 ubiquitin ligase Pellino that are critical for activation by IRAK1 and IRAK4. Proc Natl Acad Sci USA. 2009;106:4584–4590. doi: 10.1073/pnas.0900774106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H, Liu XY, Dai L, Goh ET, Chan AT, Xi J, et al. The role of TBK1 and IKKεpsilon in the expression and activation of Pellino 1. Biochem J. 2011;434:537–548. doi: 10.1042/BJ20101421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh ET, Arthur JS, Cheung PC, Akira S, Toth R, Cohen P.Identification of the protein kinases that activate the E3 ubiquitin ligase Pellino 1 in the innate immune system Biochem Jin press; 2011 [DOI] [PubMed]
- Sun SC, Ley SC. New insights into NF-kappaB regulation and function. Trends Immunol. 2008;29:469–478. doi: 10.1016/j.it.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen ZJ. Expanding role of ubiquitination in NF-κB signaling. Cell Res. 2011;21:6–21. doi: 10.1038/cr.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. NF-κB in immunobiology. Cell Res. 2011;21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- Sun SC. Non-canonical NF-κB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue JI, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Bhoj V, Seth RB. Ubiquitin, TAK1 and IKK: is there a connection. Cell Death Differ. 2006;13:687–692. doi: 10.1038/sj.cdd.4401869. [DOI] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA. Rip1 mediates the TRIF-dependent Toll-like receptor 3- and 4-induced NF-kappaB activation but does not contribute to interferon regulatory factor 3 activation. J Biol Chem. 2005;280:36560–36566. doi: 10.1074/jbc.M506831200. [DOI] [PubMed] [Google Scholar]
- Häcker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- Oganesyan G, Saha SK, Guo B, He JQ, Shahangian A, Zarnegar B, et al. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- Chuang TH, Ulevitch RJ. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat Immunol. 2004;5:495–502. doi: 10.1038/ni1066. [DOI] [PubMed] [Google Scholar]
- Bachmaier K, Toya S, Gao X, Triantafillou T, Garrean S, Park GY, et al. E3 ubiquitin ligase Cblb regulates the acute inflammatory response underlying lung injury. Nat Med. 2007;13:920–926. doi: 10.1038/nm1607. [DOI] [PubMed] [Google Scholar]
- Tseng PH, Matsuzawa A, Zhang W, Mino T, Vignali DA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol. 2010;11:70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Wang C, Zhu X, Tang S, Shi L, Cao X, et al. E3 ubiquitin ligase CHIP facilitates Toll-like receptor signaling by recruiting and polyubiquitinating Src and atypical PKCzeta. J Exp Med. 2011;208:2099–2112. doi: 10.1084/jem.20102667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa A, Tseng PH, Vallabhapurapu S, Luo JL, Zhang W, Wang H, et al. Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science. 2008;321:663–668. doi: 10.1126/science.1157340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultmark D. Drosophila immunity: paths and patterns. Curr Opin Immunol. 2003;15:12–19. doi: 10.1016/s0952-7915(02)00005-5. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- Boman HG, Steiner H. Humoral immunity in Cecropia pupae. Curr Top Microbiol Immunol. 1981;94:75–91. doi: 10.1007/978-3-642-68120-2_2. [DOI] [PubMed] [Google Scholar]
- Sun SC, Faye I. Cecropia immunoresponsive factor, an insect immunoresponsive factor with DNA-binding properties similar to nuclear factor κB. Eur J Biochem. 1992;204:885–892. doi: 10.1111/j.1432-1033.1992.tb16708.x. [DOI] [PubMed] [Google Scholar]
- Sun SC, Faye I. Affinity purification and characterization of CIF, a Cecropia DNA-binding protein involved in the induction of the immune genes. Comp Biochem Physiol. 1992;103B:225–233. doi: 10.1016/0305-0491(92)90436-u. [DOI] [PubMed] [Google Scholar]
- Ip YT, Reach M, Engstrom Y, Kadalayil L, Cai H, Gonzalez-Crespo S, et al. Dif, a dorsal-related gene that mediates an immune response in Drosophila. . Cell. 1993;75:753–763. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- Towb P, Sun H, Wasserman SA. Tube Is an IRAK-4 homolog in a Toll pathway adapted for development and immunity. J Innate Immun. 2009;1:309–321. doi: 10.1159/000200773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans J, Bergmann A, Haffter P, Nusslein-Volhard C. Activation of the kinase Pelle by Tube in the dorsoventral signal transduction pathway of Drosophila embryo. Nature. 1994;372:563–566. doi: 10.1038/372563a0. [DOI] [PubMed] [Google Scholar]
- Grosshans J, Schnorrer F, Nüsslein-Volhard C. Oligomerisation of Tube and Pelle leads to nuclear localisation of dorsal. Mech Dev. 1999;81:127–138. doi: 10.1016/s0925-4773(98)00236-6. [DOI] [PubMed] [Google Scholar]
- Haghayeghi A, Sarac A, Czerniecki S, Grosshans J, Schock F. Pellino enhances innate immunity in Drosophila. . Mech Dev. 2010;127:301–307. doi: 10.1016/j.mod.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Rich T, Allen RL, Lucas AM, Stewart A, Trowsdale J. Pellino-related sequences from Caenorhabditis elegans and Homo sapiens. Immunogenetics. 2000;52:145–149. doi: 10.1007/s002510000249. [DOI] [PubMed] [Google Scholar]
- Resch K, Jockusch H, Schmitt-John T. Assignment of homologous genes, Peli1/PELI1 and Peli2/PELI2, for the Pelle adaptor protein Pellino to mouse chromosomes 11 and 14 and human chromosomes 2p13.3 and 14q21, respectively, by physical and radiation hybrid mapping. Cytogenet Cell Genet. 2001;92:172–17. doi: 10.1159/000056895. [DOI] [PubMed] [Google Scholar]
- Yu KY, Kwon HJ, Norman DA, Vig E, Goebl MG, Harrington MA. Cutting edge: mouse pellino-2 modulates IL-1 and lipopolysaccharide signaling. J Immunol. 2002;169:4075–4078. doi: 10.4049/jimmunol.169.8.4075. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Johnson HJ, Nie H, Qin J, Bird TA, Li X. Pellino 1 is required for interleukin-1 (IL-1)-mediated signaling through its interaction with the IL-1 receptor-associated kinase 4 (IRAK4)-IRAK-tumor necrosis factor receptor-associated factor 6 (TRAF6) complex. J Biol Chem. 2003;278:10952–10956. doi: 10.1074/jbc.M212112200. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Hernandez LD, Galan JE, Janeway CA, Jr, Medzhitov R, Flavell RA. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- Strelow A, Kollewe C, Wesche H. Characterization of Pellino2, a substrate of IRAK1 and IRAK4. FEBS Lett. 2003;547:157–161. doi: 10.1016/s0014-5793(03)00697-5. [DOI] [PubMed] [Google Scholar]
- Jensen LE, Whitehead AS. Pellino2 activates the mitogen activated protein kinase pathway. FEBS Lett. 2003;545:199–202. doi: 10.1016/s0014-5793(03)00533-7. [DOI] [PubMed] [Google Scholar]
- Jensen LE, Whitehead AS. Pellino3, a novel member of the Pellino protein family, promotes activation of c-Jun and Elk-1 and may act as a scaffolding protein. J Immunol. 2003;171:1500–1506. doi: 10.4049/jimmunol.171.3.1500. [DOI] [PubMed] [Google Scholar]
- Butler MP, Hanly JA, Moynagh PN. Pellino3 is a novel upstream regulator of p38 MAPK and activates CREB in a p38-dependent manner. J Biol Chem. 2005;280:27759–27768. doi: 10.1074/jbc.M500756200. [DOI] [PubMed] [Google Scholar]
- Xiao H, Qian W, Staschke K, Qian Y, Cui G, Deng L, et al. Pellino 3b negatively regulates interleukin-1-induced TAK1-dependent NF kappaB activation. J Biol Chem. 2008;283:14654–14664. doi: 10.1074/jbc.M706931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauvliege R, Janssens S, Beyaert R. Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: a role as novel RING E3-ubiquitin-ligases. FEBS Lett. 2006;580:4697–4702. doi: 10.1016/j.febslet.2006.07.046. [DOI] [PubMed] [Google Scholar]
- Freemont PS. The RING finger. A novel protein sequence motif related to the zinc finger. Ann NY Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- Butler MP, Hanly JA, Moynagh PN. Kinase-active interleukin-1 receptor-associated kinases promote polyubiquitination and degradation of the Pellino family: direct evidence for PELLINO proteins being ubiquitin-protein isopeptide ligases. J Biol Chem. 2007;282:29729–29737. doi: 10.1074/jbc.M704558200. [DOI] [PubMed] [Google Scholar]
- Ordureau A, Smith H, Windheim M, Peggie M, Carrick E, Morrice N, et al. The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem J. 2008;409:43–52. doi: 10.1042/BJ20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier ME, Du Pasquier L, Degnan BM. The genome of the sponge Amphimedon queenslandica provides new perspectives into the origin of Toll-like and interleukin 1 receptor pathways. Evol Dev. 2010;12:519–533. doi: 10.1111/j.1525-142X.2010.00436.x. [DOI] [PubMed] [Google Scholar]
- Lin CC, Huoh YS, Schmitz KR, Jensen LE, Ferguson KM. Pellino proteins contain a cryptic FHA domain that mediates interaction with phosphorylated IRAK1. Structure. 2008;16:1806–1816. doi: 10.1016/j.str.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, et al. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell. 2000;6:1169–1182. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Kee Y, Huibregtse JM. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem Biophys Res Commun. 2007;354:329–333. doi: 10.1016/j.bbrc.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder PM. Down-regulating destruction: phosphorylation regulates the E3 ubiquitin ligase Nedd4-2. Sci Signal. 2009;2:pe41. doi: 10.1126/scisignal.279pe41. [DOI] [PubMed] [Google Scholar]
- Meek DW, Hupp TR. The regulation of MDM2 by multisite phosphorylation—opportunities for molecular-based intervention to target tumours. Semin Cancer Biol. 2010;20:19–28. doi: 10.1016/j.semcancer.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Kim JH, Sung KS, Jung SM, Lee YS, Kwon JY, Choi CY, et al. Pellino-1, an adaptor protein of interleukin-1 receptor/toll-like receptor signaling, is sumoylated by Ubc9. Mol Cells. 2011;31:85–89. doi: 10.1007/s10059-011-0006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T, Sun H. Crosstalk between the SUMO and ubiquitin pathways. Ernst Schering Found Symp Proc. 2008;1:1–16. doi: 10.1007/2789_2008_098. [DOI] [PubMed] [Google Scholar]
- Weighardt H, Jusek G, Mages J, Lang R, Hoebe K, Beutler B, et al. Identification of a TLR4- and TRIF-dependent activation program of dendritic cells. Eur J Immunol. 2004;34:558–564. doi: 10.1002/eji.200324714. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, et al. IKKεpsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Marquez RT, Wendlandt E, Galle CS, Keck K, McCaffrey AP. MicroRNA-21 is upregulated during the proliferative phase of liver regeneration, targets Pellino-1, and inhibits NF-kappaB signaling. Am J Physiol Gastrointest Liver Physiol. 2010;298:G535–G541. doi: 10.1152/ajpgi.00338.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Radu C, Diab A, Tsen MF, Hussain R, Cowdery JS, et al. IL-1 receptor-associated kinase 1 regulates susceptibility to organ-specific autoimmunity. J Immunol. 2003;170:2833–2842. doi: 10.4049/jimmunol.170.6.2833. [DOI] [PubMed] [Google Scholar]
- Kawagoe T, Sato S, Matsushita K, Kato H, Matsui K, Kumagai Y, et al. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat Immunol. 2008;9:684–691. doi: 10.1038/ni.1606. [DOI] [PubMed] [Google Scholar]
- Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, et al. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- Goodnow CC, Sprent J, Fazekas de St Groth B, Vinuesa CG. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature. 2005;435:590–597. doi: 10.1038/nature03724. [DOI] [PubMed] [Google Scholar]
- Tzachanis D, Lafuente EM, Li L, Boussiotis VA. Intrinsic and extrinsic regulation of T lymphocyte quiescence. Leuk Lymphoma. 2004;45:1959–1967. doi: 10.1080/1042819042000219494. [DOI] [PubMed] [Google Scholar]
- von Boehmer H. Mechanisms of suppression by suppressor T cells. Nat Immunol. 2005;6:338–344. doi: 10.1038/ni1180. [DOI] [PubMed] [Google Scholar]
- Wells AD. New insights into the molecular basis of T cell anergy: anergy factors, avoidance sensors, and epigenetic imprinting. J Immunol. 2009;182:7331–7341. doi: 10.4049/jimmunol.0803917. [DOI] [PubMed] [Google Scholar]
- Wan YY. Regulatory T cells: immune suppression and beyond. Cell Mol Immunol. 2010;7:204–210. doi: 10.1038/cmi.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggirwar SB, Harhaj EW, Sun SC. Regulation of the interleukin-2 CD28 responsive element by NF-ATp and various NF-κB/Rel transcription factors. Mol Cell Biol. 1997;17:2605–2614. doi: 10.1128/mcb.17.5.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köntgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoural immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- Liou HC, Jin Z, Tumang J, Andjelic S, Smith KA, Liou ML. c-Rel is crucial for lymphocyte proliferation but dispensable for T cell effector function. Int Immunol. 1999;11:361–371. doi: 10.1093/intimm/11.3.361. [DOI] [PubMed] [Google Scholar]
- Hilliard BA, Mason N, Xu L, Sun J, Lamhamedi-Cherradi SE, Liou HC, et al. Critical roles of c-Rel in autoimmune inflammation and helper T cell differentiation. J Clinic Invest. 2002;110:843–850. doi: 10.1172/JCI15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason NJ, Liou HC, Hunter CA. T cell-intrinsic expression of c-Rel regulates Th1 cell responses essential for resistance to Toxoplasma gondii. J Immunol. 2004;172:3704–3711. doi: 10.4049/jimmunol.172.6.3704. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Liou HC, Sen R. c-Rel-dependent priming of naive T cells by inflammatory cytokines. Immunity. 2005;23:445–458. doi: 10.1016/j.immuni.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Chen G, Hardy K, Bunting K, Daley S, Ma L, Shannon MF. Regulation of the IL-21 gene by the NF-kappaB transcription factor c-Rel. J Immunol. 2010;185:2350–2359. doi: 10.4049/jimmunol.1000317. [DOI] [PubMed] [Google Scholar]
- Deenick EK, Po L, Chapatte L, Murakami K, Lu YC, Elford AR, et al. c-Rel phenocopies PKCtheta but not Bcl-10 in regulating CD8+ T-cell activation versus tolerance. Eur J Immunol. 2010;40:867–877. doi: 10.1002/eji.200939445. [DOI] [PubMed] [Google Scholar]
- Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19:71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KC, Lee YS, Lim S, Choi HK, Lee CH, Lee EK, et al. Smad6 negatively regulates interleukin 1-receptor-Toll-like receptor signaling through direct interaction with the adaptor Pellino-1. Nat Immunol. 2006;7:1057–1065. doi: 10.1038/ni1383. [DOI] [PubMed] [Google Scholar]
- Lee YS, Kim JH, Kim ST, Kwon JY, Hong S, Kim SJ, et al. Smad7 and Smad6 bind to discrete regions of Pellino-1 via their MH2 domains to mediate TGF-beta1-induced negative regulation of IL-1R/TLR signaling. Biochem Biophys Res Commun. 2010;393:836–843. doi: 10.1016/j.bbrc.2010.02.094. [DOI] [PubMed] [Google Scholar]
- Letterio JJ, Roberts AB. Regulation of immune responses by TGF-beta. Annu Rev Immunol. 1998;16:137–161. doi: 10.1146/annurev.immunol.16.1.137. [DOI] [PubMed] [Google Scholar]
- Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- Itoh S, Itoh F, Goumans MJ, Ten Dijke P. Signaling of transforming growth factor-beta family members through Smad proteins. Eur J Biochem. 2000;267:6954–6967. doi: 10.1046/j.1432-1327.2000.01828.x. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Souchelnytskyi S, Heldin CH. Smad regulation in TGF-beta signal transduction. J Cell Sci. 2001;114:4359–4369. doi: 10.1242/jcs.114.24.4359. [DOI] [PubMed] [Google Scholar]
- Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, et al. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389:622–666. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, et al. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, et al. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- Ulloa L, Doody J, Massague J. Inhibition of transforming growth factor-beta/SMAD signalling by the interferon-gamma/STAT pathway. Nature. 1999;397:710–713. doi: 10.1038/17826. [DOI] [PubMed] [Google Scholar]
- Bitzer M, von Gersdorff G, Liang D, Dominguez-Rosales A, Beg AA, Rojkind M, et al. A mechanism of suppression of TGF-beta/SMAD signaling by NF-kappa B/RelA. Genes Dev. 2000;14:187–197. [PMC free article] [PubMed] [Google Scholar]