Abstract
Aim:
To study the molecular mechanism underlying the effect of aristolochic acid (AA), a major active component of plants from the Aristolochiaceae family using microarray analysis.
Methods:
Human kidney (HK-2) cells were treated with AA (0, 10, 30, and 90 μmol/L) for 24 h, and the cell viability was measured by a 3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide assay. Complementary DNA microarrays were used to investigate the gene expression pattern of HK-2 cells exposed to AA in triplicate. A quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) assay was used to verify the microarray data for selected nuclear factor kappa B (NF-κB)-regulated genes. Furthermore, the subcellular localization of NF-κB p65 was visualized by immunofluorescence confocal microscopy in HK-2 cells. The NF-κB activity was examined by a luciferase reporter assay in HK-2/NF-κB transgenic cells.
Results:
AA exhibited a dose-dependent cytotoxic effect in HK-2 cells and induced alterations in the gene expression profiles related to the DNA damage response, DNA repair, macromolecule metabolic process, carbohydrate metabolic process, DNA metabolic process, apoptosis, cell cycle, and transcription. In addition, 9 biological pathways associated with immunomodulatory functions were down-regulated in AA-treated HK-2 cells. A network analysis revealed that NF-κB played a central role in the network topology. Among NF-κB-regulated genes, 8 differentially expressed genes were verified by qRT-PCR. The inhibition of NF-κB activity by AA was further confirmed by immunofluorescence confocal microscopy and by NF-κB luciferase reporter assay.
Conclusion:
Our data revealed that AA could suppress NF-κB activity in normal human cells, perhaps partially accounting for the reported anti-inflammatory effects of some plants from the genus Aristolochia.
Keywords: aristolochic acid, microarray analysis, nuclear factor-kappa B, human kidney HK-2 cells, confocal microscopy, luciferase reporter assay
Introduction
Aristolochic acid (AA), a major active component of plants from the Aristolochiaceae family, was first detected in Aristolochia clematitis in 19431. Many plant species of the genus Aristolochia have been used worldwide for centuries in folk medicine. For example, there are many formulas containing various species of the genus Aristolochia that are commonly used in traditional medicine in China, Japan, and Singapore2. Beside those used in East Asia, several species of the genus Aristolochia have been used to regulate menstruation, induce labor, expel parasites, relieve pain, and treat arthritis, cancer, diarrhea, and snake bites in India, West Africa, the Mediterranean, and South America 3, 4, 5.
Pharmacological studies have demonstrated that aristolochic acid I (AAI) and aristolochic acid II (AAII) are the major active components of plants in the Aristolochiaceae family1, 6. It has been shown that AA protects against infections and inflammation in several biological systems, including humans. AAI inhibits the growth of bacteria, including Escherichia coli, Pseudomonas aeruginosa, Streptococcus faecalis, Staphylococcus aureus, and Staphylococcus epidermidis5. AA can block H2O2-induced platelet aggregation and suppress hydroxyl radical-induced platelet activation through the arachidonic acid pathway7, 8. AA was found to possess anti-inflammation effects as demonstrated by its ability to inhibit phospholipase A2 (PLA2) when administered by intramuscular or intraperitoneal injection9, 10. Furthermore, AA was also reported to inhibit Group I PLA2 in humans with sepsis11. From in vitro studies, AA has been shown to suppress phospholipohydration of PLA2 derived from human synovial fluid, cobra venom, porcine pancreas, and human platelets12. The anti-inflammatory activities of AA in different models of inflammation have promoted its use in many countries in herbal formulations for arthritis, rheumatism, gout and chronic inflammatory skin diseases13, 14. Moreover, double-blind studies in healthy volunteers show that AA increased the phagocytic activity of peripheral granulocytes after treatment with AA 0.9 mg/d for three to ten consecutive days15.
Plants of the genus Aristolochia were used as therapeutic drugs until cases of rapidly progressive renal failure were reported in Belgium16 in 1993, which were found in 1994 to be caused by the inadvertent replacement of Stephania tetrandra by Aristolochia fangchi17. High cumulative doses of AA have been reported to be associated with nephropathy in humans18. Aristolochic acid nephropathy complicated with urothelial malignancy has been described, and the cumulative dose (>200 g) of ingested Aristolochia was found to be a significant risk factor for urothelial carcinoma19, 20. Moreover, there is evidence that AA is nephrotoxic and carcinogenic in animals including humans2, 6. However, herbs of the genus Aristolochia administrated at low cumulative doses (<30 g of raw drugs) did not increase the risks for chronic kidney disease in a large Chinese cohort study of 200 000 subjects21.
In this paper, the effects of AA and its underlying molecular mechanism were examined in normal human kidney cells (HK-2) through microarray analysis. Results from these analyses showed that NF-κB is an important modulator of gene expression in AA-treated normal human cells. Furthermore, AA-induced NF-κB inhibition was confirmed by immunofluorescence confocal microscopy in HK-2 cells and by a luciferase reporter assay in HK-2/NF-κB transgenic cells.
Materials and methods
Cell culture and reagents
HK-2 cells, derived from an immortalized proximal tubule epithelial cell line from the normal adult human kidney, were purchased from the Bioresource Collection and Research Center (Hsinchu, Taiwan). HK-2 cells were grown in keratinocyte serum-free basal medium (Gibco) supplemented with 5 ng/mL of recombinant epidermal growth factor and 50 μg/mL of bovine pituitary extract without antibiotics in 5% CO2 at 37 °C. Aristolochic acid sodium salt, a mixture of AAI (41%) and AAII (56%) (Sigma, St Louis, MO) was dissolved in double-distilled water. HK-2 cells were seeded in 96-well plates and incubated for 24 h before the AA treatment. Various concentrations of AA (10, 30, or 90 μmol/L) were added to HK-2 cells for 24 h. The control cells received equal amounts of water only.
3-[4,5-dimethylthiazol-2yl]-2,5-diphenyltetrazolium bromide (MTT) assay
After the AA treatment for 24 h, 5 mg/mL MTT was added to the cells for 4 h. The purple formazan crystals were solubilized in 150 μL acidic isopropanol (0.1 mol/L HCl), and the absorbance was measured at a test wavelength of 570 nm and a reference wavelength of 650 nm using a microplate reader. The relative survival rate was calculated using the following equation: survival rate (%)=(absorbance of AA-treated cells/ absorbance of untreated control cells)×100%. The data are expressed as the mean±SD of three independent experiments.
Total RNA extraction
Total RNA was extracted from HK-2 cells grown in 75 cm2 flasks following treatment with AA (0, 10, 30, or 90 μmol/L) (n=3) using an RNeasy Mini kit (Qiagen, Valencia, CA). The total RNA was quantified using a Beckman DU800 spectrophotometer (Beckman Coulter, Fullerton, CA). Samples with A260/A280 ratios greater than 1.8 were further evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA samples with an RNA integrity number greater than 8.0 were used for the subsequent microarray analysis.
Microarray analysis
The microarray analysis was performed as previously described22. Briefly, fluorescence-labeled RNA targets were prepared from 5 μg of total RNA samples using a MessageAmpTM RNA kit (Ambion, Austin, TX) and Cy5 dye (Amersham Pharmacia, Piscataway, NJ). Triplicate fluorescent targets were hybridized to the Human Whole Genome OneArrayTM (Phalanx Biotech Group, Hsinchu, Taiwan) and scanned using an Axon 4000 scanner (Molecular Devices, Sunnyvale, CA). The Cy5 fluorescent intensity of each spot was analyzed using the Genepix 4.1 software (Molecular Devices). The signal intensity of each spot was corrected by subtracting the background signals in the surrounding area. Control probes were filtered out to measure the background-corrected signal intensity of each spot. We have submitted the original microarray data to Gene Expression Omnibus (GEO), series number GSE18243. Two samples and one series are included (samples=mock, treatment; series=mock+treatment). The sample numbers are GSM455880 (treatment) and GSM455881 (mock). The spots were normalized using the Limma package of the R program23. The normalized data were tested for differential expression using the Gene Expression Pattern Analysis Suite v3.124. The 'GeneSetTest' function in the Limma package was used to test which biological pathways were affected by AA in HK-2 cells. This function computes a P-value to test the hypothesis that the selected genes in a pathway tend to be differentially expressed. The score for each pathway following the AA treatment was defined as follows: score=-log (2P), if P-value≤0.5; or score=log [2(1−P)], if P-value>0.5. There were 352 pathways used in this analysis, and they were extracted from ArrayTrack (http://www.fda.gov/nctr/science/centers/toxicoinformatics/ArrayTrack/), which included the KEGG pathways (http://www.genome.jp/kegg/pathway.html) and PathArt pathways (Jubilant Biosys). Furthermore, the interaction network for genes with fold changes >2.0 and false discovery rates <0.05 was constructed using BiblioSphere Pathway Edition software (Genomatix Applications, http://www.genomatix.de/index.html) based on knowledgebase analyses25. Finally, the Cytoscape software was used to visualize the interaction network of AA-regulated genes, including genes related to NF-κB26.
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
The expression levels of NF-κB-related genes were further validated by qRT-PCR. RNA samples were reverse-transcribed for 120 min at 37 °C with High Capacity cDNA Reverse Transcription Kit according to the standard protocol of the supplier (Applied Biosystems). Quantitative RT-PCR was performed using 1 μL of cDNA and 2×SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). The reaction conditions were as follows: 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Each assay was run on an Applied Biosystems 7300 Real-time PCR system in triplicate. Fold changes were calculated using the comparative CT method. The primer sets for selected genes are shown in Table 1.
Table 1. Primer sequences of selected genes for qRT-PCR.
| Gene | Primer sequence |
|---|---|
| CCL20 | Sense: 5′-GCTCCTGGCTGCTTTGATGT-3′ |
| Antisense: 5′-GAATACGGTCTGTGTATCCAAGACA-3′ | |
| CD68 | Sense: 5′-TCTGCCCACCCAGAACCA-3′ |
| Antisense: 5′-CACAGGGCTGGGAACCATT-3′ | |
| CYP1A1 | Sense: 5′-ATGGGCAAGCGGAAGTGTAT-3′ |
| Antisense: 5′-CCAGTGGCACGCTGAATTC-3′ | |
| IGFBP3 | Sense: 5′-CAGCGCTACAAAGTTGACTACGA-3′ |
| Antisense: 5′-ATTTCTCTACGGCAGGGACCAT-3′ | |
| IL-8 | Sense: 5′-FCTTTCCACCCCAAATTTATCAAAG-3′ |
| Antisense: 5′-RAGAGCTCTCTTCCATCAGAAAGCT-3′ | |
| LTB | Sense: 5′-ACTTCTCTGGTGACCTTGTTGCT-3′ |
| Antisense: 5′-AGCTTCTGAAACCCCAGTCCTT-3′ | |
| SAA2 | Sense: 5′-CCGATCAGGCTGCCAATAAA-3′ |
| Antisense: 5′-GCAGAGTGAAGAGGAAGCTCAGT-3′ | |
| TNFRSF9 | Sense: 5′-TGCGAGAGAGCCAGGACACT-3′ |
| Antisense: 5′-GAAACGGAGCGTGAGGAAGA-3′ |
Immunofluorescence and confocal microscopy
HK-2 cells were fixed with acetone for 30 min at 4 °C and incubated with the p65 subunit of the NF-κB rabbit polyclonal antibody (Abcam-ab7970) overnight at 4 °C. Alexa Fluor® 488-conjugated secondary goat anti-rabbit antibody was added, and the reaction was incubated at room temperature for 1 h. After the last step for the NF-κB single staining, cell nuclei were labeled by propidium iodide for 15 min at room temperature. The fluorescence imaging was conducted using a laser scanning confocal microscope (TCS SP2 system, Leica) equipped with an argon laser adjusted to 488 nm excitation with a HeNe1 laser.
Transfection and luciferase assay
HK-2 cells were transiently transfected with 5 μg of NF-κB expression plasmid or empty vector (pcDNA 3.1) using the SuperFect® transfection reagent (Qiagen, Valencia, CA, USA). Cells were cultured in 24-well plates overnight and then treated with 10 μmol/L AA for 24 h. The cells were then washed with ice-cold PBS, lysed with 350 μL Triton lysis buffer (50 mmol/L Tris-HCl, 1% Triton X-100, 1 mmol/L dithiothreitol, pH 7.8) and collected with a cell scraper. The luciferase activity was measured as described previously22. The relative luciferase activity was calculated by dividing the relative luciferase units (RLU) of the experimental groups by the RLU of untreated NF-κB-transfected cells.
Statistical analysis
Data are presented as the mean±SD. Student's t-test was used for comparisons between the control and AA-treated groups. A value of P<0.05 was determined to be statistically significant.
Results
Cell viability in AA-treated HK-2 cells
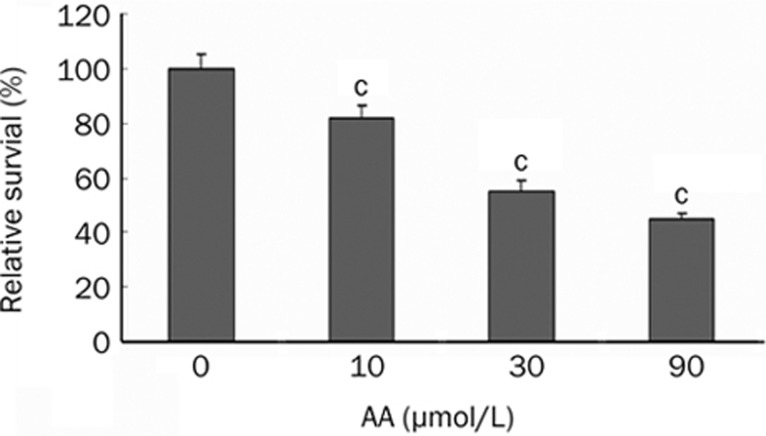
The cell viability (using the MTT assay) was measured after exposing HK-2 cells to various concentrations of AA (10, 30, or 90 μmol/L). AA exhibited cytotoxicity in a dose-dependent manner. The relative survival rates of cells after treatment with 10, 30, and 90 μmol/L AA were 81.9%±4.6%, 55.2%±3.7%, and 45.2%±1.9%, respectively. These results represent the average of three independent experiments (Figure 1).
Figure 1.
The cytotoxicity of various doses of AA in HK-2 cells. HK-2 cells were treated with 0, 10, 30, and 90 μmol/L of AA. The cell viability was measured by MTT assay. Values are mean±SD of three independent experiments. cP<0.01 vs control group.
Cluster analysis of the gene expression profiles in AA-treated HK-2 cells
An unsupervised analysis was used to predict significant differences among the gene expression levels in HK-2 cells in response to 10, 30, and 90 μmol/L of AA treatment. As shown in Table 2, DNA repair, the response to the DNA damage stimulus, the macromolecule metabolic process, and the carbohydrate metabolic process were shown to be significantly regulated in cells in all AA treatment groups.
Table 2. Biological processes significantly regulated by AA in HK-2 cells.
| Biological processes | Genes | 10 ×mol/L _P | 30 ×mol/L_P | 90 ×mol/L_P |
|---|---|---|---|---|
| DNA repair | 166 | 3.15×10−7 | 7.70×10−5 | 1.75×10−7 |
| Response to DNA damage stimulus | 122 | 5.40×10−7 | 2.00×10−4 | 2.20×10−6 |
| Macromolecule metabolic process | 202 | 4.07×10−6 | 3.00×10−4 | 3.18×10−6 |
| Carbohydrate metabolic process | 205 | 4.90×10−5 | 4.00×10−4 | 6.17×10−5 |
| DNA metabolic process | 236 | 1.00×10−4 | 3.70×10−3 | 7.43×10−7 |
| Apoptosis | 295 | 4.00×10−4 | 1.60×10−3 | 1.19×10−5 |
| Cell cycle | 169 | 2.40×10−3 | 3.04×10−2 | 8.30×10−6 |
| Transcription | 148 | 5.40×10−3 | 3.80×10−3 | 2.20×10−5 |
The biological pathways affected by AA in HK-2 cells were examined by 'GeneSetTest' function in the Limma package. This function computes a P-value to test the hypothesis that the selected genes in a pathway tend to be differentially expressed. “P” indicated P values.
Pathway analysis of gene expression profiles in AA-treated HK-2 cells
The GeneSetTest function was used to investigate which biological pathways could be downregulated in HK-2 cells in response to treatment with 10, 30, or 90 μmol/L of AA. In this study, pathways with P values <0.001 (scores >2.7 or <-2.7) in all AA treatment groups were considered differentially regulated. The pathway analysis revealed that nine pathways were dysregulated following all AA treatments (Table 3). A negative score was indicative of an AA-induced downregulation at all doses. Most of the AA-regulated pathways were associated with immunomodulatory functions, indicating that AA might participate in the regulation of immune genes.
Table 3. Estimates of various pathways regulated by different doses of AA.
| Pathway | 10 μmol/L_P | 30 μmol/L_P | 90 μmol/L_P | 10 μmol/L_s | 30 μmol/L_s | 90 μmol/L_s |
|---|---|---|---|---|---|---|
| Cytokine-cytokine receptor interaction | <2.20×10−16 | <2.20×10−16 | <2.20×10−16 | <−15.66 | <−15.66 | <−15.66 |
| TNF signaling pathway | 2.00×10−7 | 5.70×10−5 | <2.20×10−16 | −6.40 | −3.94 | <−15.66 |
| Interleukin signaling pathway | 4.05×10−5 | 1.37×10−4 | <2.20×10−16 | −4.09 | −3.56 | <−15.66 |
| Hematopoietic cell lineage | 3.45×10−5 | 3.10×10−6 | 4.30×10−5 | −4.16 | −5.21 | −4.07 |
| Cell adhesion molecules (CAMs) | 3.75×10−4 | 7.20×10−6 | 4.40×10−6 | −3.13 | −4.84 | −5.06 |
| Insulin signaling pathway | 3.13×10−4 | 2.00×10−6 | 6.84×10−5 | −3.20 | −5.40 | −3.86 |
| IFN signaling pathway | 5.44×10−5 | 4.03×10−4 | 2.80×10−6 | −3.96 | −3.09 | −5.25 |
| Long-term depression | 5.73×10−4 | 3.24×10−5 | 1.95×10−5 | −2.94 | −4.19 | −4.41 |
| IL-2 signaling pathway | 3.04×10−4 | 3.19×10−5 | 3.64×10−4 | −3.22 | −4.20 | −3.14 |
Pathways with P values <0.001 (scores >2.7 or <−2.7) calculated by “GeneSetTest” function in all AA treatments were considered differentially regulated. Pathway analysis revealed that 9 pathways were regulated in all AA treatments. The minus sign of scores in each pathway meant that all AA treatments downregulated every pathway. The results of this study were in triplicate. “P” indicated P values and “s” indicated score.
The gene interaction network regulated by AA treatment
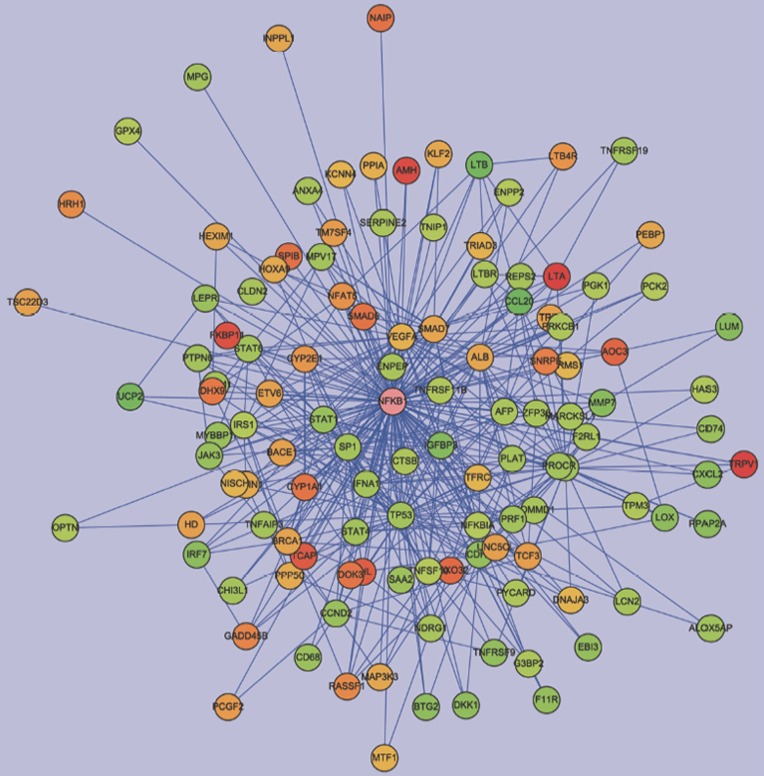
To explore the pharmacological and molecular mechanisms of AA, we had first chosen the subtoxic dose of AA (10 μmol/L) to interpret the gene interaction network. Genes with fold changes >2.0 and false discovery rates <0.05 after the 10 μmol/L AA treatment were further selected to construct the interaction network using the Pathway Edition software. The connections between NF-κB and the 10μmol/L AA-regulated genes were noted following the analysis using the Cytoscape software. Nodes for regulated genes and NFKB1 were color-coded according to their log2 expression values (Figure 2). As shown in Figure 2, most genes were connected with NF-κB, suggesting that NF-κB played a central role in the network. The NF-κB-connected genes with fold changes >2.0 and false discovery rates <0.05 after the treatments of 10, 30, and 90 μmol/L AA are listed in Table 4. The upregulated genes were AOC3, CYP1A1, CYP2E1, DHX9, LTB4R, RASSF1, SPIB, TCF3, TRPV1, and VHL. The downregulated genes were CCL20, CCND2, CD68, CD74, CDH1, CXCL2, EBI3, IGFBP3, IKIP, IL8, LTB, MMP7, PPAP2A, SAA2, STAT1, TNFRSF9, and UCP2.
Figure 2.
Network analysis of expression profiles of 10 μmol/L AA in HK-2 cells. We constructed the interaction network by BiblioSphere Pathway Edition software which was selected the genes with fold changes >2.0 and false discovery rates <0.05 after 10 μmol/L AA treatment. Intensity of nodes for regulated genes and NFKB1 are color-coded according to their log2 expression values in gradient with red and green indicating the degree of gene upregulation or downregulation, respectively. As shown, NF-κB played a central role in the network.
Table 4. Gene list of NF-κB connected significantly regulated genes in AA- treated HK-2 cells.
| Symbol |
Description |
10 μmol/L AA |
30 μmol/L AA |
90 μmol/L AA |
|||
|---|---|---|---|---|---|---|---|
| FC±SD | P value | FC±SD | P value | FC±SD | P value | ||
| AOC3 | Amine oxidase, copper containing (vascular adhesion protein 1) | 3 3.8±0.53 | 1.54×10−2 | 10.16±1.55 | 5.94×10−3 | 10.25±1.66 | 4.46×10−3 |
| CCL20 | Chemokine (C-C motif) ligand 20 | −11.46±2.20 | 2.15×10−3 | −11.4±1.04 | 4.31×10−4 | −17.4±4.26 | 5.29×10−4 |
| CCND2 | Cyclin D2 | −2.24±0.13 | 8.18×10−3 | −2.89±0.16 | 2.70×10−3 | −5.71±0.84 | 1.35×10−3 |
| CD68 | CD68 antigen | −2.16±0.34 | 2.24×10−2 | 1.21±0.24 | 2.98×10−1 | 1.46±0.22 | 4.76×10−2 |
| CD74 | CD74 molecule, major histocompatibility complex, class II invariant chain | −2.18±0.50 | 4.56×10−2 | −2.33±0.69 | 2.54×10−2 | −2.37±0.57 | 1.54×10−2 |
| CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | −2.42±0.67 | 3.31×10−2 | −3.97±0.72 | 6.01×10−3 | −4.78±0.94 | 3.42×10−3 |
| CXCL2 | Chemokine (C-X-C motif) ligand 2 | −2.71±0.47 | 1.30×10−2 | −3.07±0.50 | 4.42×10−3 | −2.74±0.20 | 1.99×10−3 |
| CYP1A1 | Cytochrome P450, family 1, subfamily A, polypeptide 1 | 3.21±0.82 | 2.17×10−2 | 4.9±1.43 | 1.43×10−2 | 4.85±1.24 | 4.53×10−3 |
| CYP2E1 | Cytochrome P450, family 2, subfamily E, polypeptide 1 | 2.15±0.33 | 1.77×10−2 | 4.21±0.78 | 7.35×10−3 | 4.3±0.68 | 2.02×10−3 |
| DHX9 | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | 3.05±0.65 | 1.78×10−2 | 2.45±0.66 | 3.27×10−2 | 4.59±0.92 | 1.49×10−3 |
| EBI3 | Epstein-Barr virus induced gene 3 | −2.37±0.22 | 1.66×10−2 | −2.8±0.40 | 6.80×10−3 | −1.77±0.31 | 1.73×10−2 |
| IGFBP3 | Insulin-like growth factor binding protein 3 | −3.19±0.41 | 1.09×10−2 | −3.36±0.92 | 7.05×10−3 | −3.77±1.06 | 3.95×10−3 |
| IKIP | IKK interacting protein | −1.39±0.04 | 4.49×10−3 | −1.8±0.17 | 2.51×10−2 | −5.2±0.06 | 1.94×10−5 |
| IL8 | Interleukin 8 | −2.04±0.29 | 1.94×10−2 | −1.35±0.23 | 7.96×10−2 | −6.66±0.66 | 1.40×10−3 |
| LTB | Lymphotoxin beta (TNF superfamily, member 3) | −5.15±1.69 | 1.88×10−2 | −9.95±2.42 | 5.90×10−3 | −5.07±1.12 | 4.87×10−3 |
| LTB4R | Leukotriene B4 receptor | 2.21±0.55 | 3.37×10−2 | 2.74±0.64 | 5.75×10−3 | 4.8±1.16 | 2.00×10−3 |
| MMP7 | Matrix metallopeptidase 7 (matrilysin, uterine) | −2.97±0.23 | 4.19×10−3 | −2.93±0.35 | 2.70×10−3 | −4.23±0.22 | 8.69×10−4 |
| PPAP2A | Phosphatidic acid phosphatase type 2A | −2.81±0.89 | 4.39×10−2 | −2.51±0.44 | 1.83×10−2 | −3.21±0.61 | 1.05×10−2 |
| RASSF1 | Ras association (RalGDS/AF-6) domain family 1 | 2.49±0.34 | 1.35×10−2 | 4.24±0.84 | 1.19×10−2 | 4.81±1.38 | 2.51×10−2 |
| SAA2 | Serum amyloid A2 | −2.08±0.34 | 2.25×10−2 | −4.48±0.77 | 3.66×10−3 | −7.18±0.86 | 1.86×10−3 |
| SPIB | Spi-B transcription factor (Spi-1/PU.1 related) | 3.34±0.80 | 2.12×10−2 | 6.82±1.67 | 6.33×10−3 | 6.95±1.86 | 6.94×10−3 |
| STAT1 | Signal transducer and activator of transcription 1, 91kDa | −2.28±0.24 | 1.88×10−2 | −3.34±0.63 | 6.61×10−3 | −4.21±0.42 | 2.86×10−3 |
| TCF3 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | 2.01±0.14 | 8.79×10−3 | 2.24±0.33 | 1.65×10−2 | 1.61±0.11 | 3.94×10−3 |
| TNFRSF9 | Tumor necrosis factor receptor superfamily, member 9 | −2.45±0.37 | 9.97×10−3 | −3.76±0.16 | 8.99×10−4 | −6.05±0.58 | 6.01×10−4 |
| TRPV1 | Transient receptor potential cation channel, subfamily V, member 1 | 7.62±1.46 | 8.18×10−3 | 10.86±1.96 | 1.53×10−3 | 13.12±2.94 | 4.50×10−3 |
| UCP2 | Uncoupling protein 2 (mitochondrial, proton carrier) | −4.75±0.63 | 1.35×10−2 | −5.04±1.08 | 6.01×10−3 | −2.91±0.51 | 6.07×10−3 |
| VHL | Von Hippel-Lindau tumor suppressor | 4.19±0.54 | 7.12×10−3 | 2.49±0.62 | 3.96×10−2 | 4.12±0.71 | 5.83×10−3 |
aThe statistical analysis was performed by t-statistics. Genes with P<0.05 are listed.
Validation of NF-κB-related gene expression changes by qRT-PCR
Eight NF-κB-regulated genes (downregulated: CCL20, CD68, IGFBP3, IL8, TNFRSF9, LTB, and SAA2; upregulated: CYP1A1) were chosen for qRT-PCR verification. The expression levels of NF-κB-related genes that were validated by qRT-PCR showed correlations in the general trends for the microar ray analysis (Table 5).
Table 5. Expression levels of selective NF-κB regulated genes by qRT-PCR in AA-treated HK-2 cells.
| Gene |
Fold changes |
|
|---|---|---|
| 10 μmol/L AA | 30 μmol/L AA | |
| CCL20 | −9.09 | −9.09 |
| CD68 | −1.45 | 1.01 |
| CYP1A1 | 1.79 | 3.83 |
| IGFBP3 | −4.55 | −3.57 |
| IL8 | −1.69 | −2.17 |
| LTB | −4.17 | −4.17 |
| SAA2 | −2.38 | −3.23 |
| TNFRSF9 | −2.22 | −2.08 |
The data represent the mean fold changes in triplicates.
Effect of AA on NF-κB localization and activity in HK-2 cells
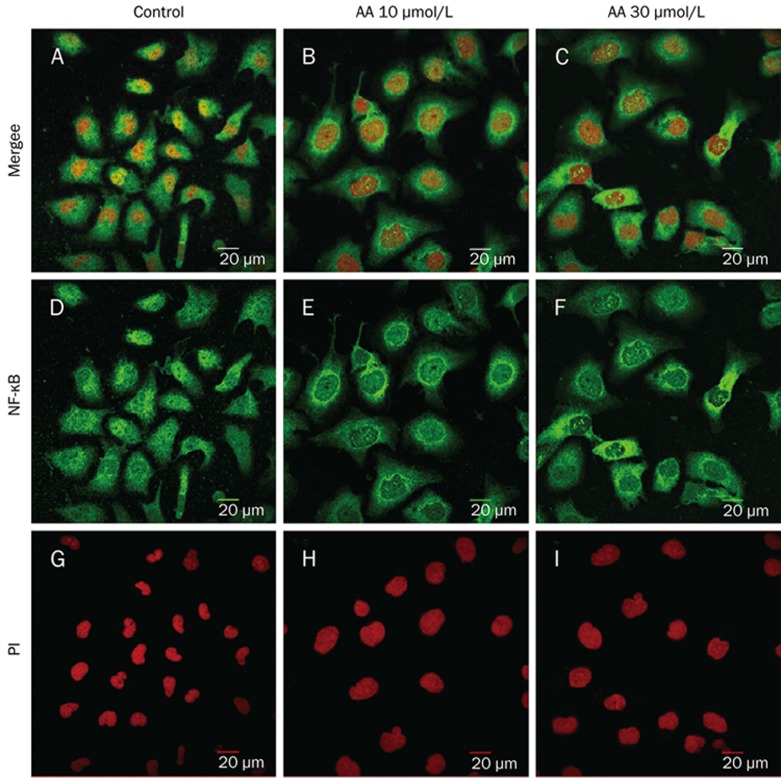
The cellular localization of NF-κB was visualized by confocal microscopy after staining with an antibody against NF-κB p65 (green) (Figure 3D–3F) and propidium iodide (nuclear DNA, red) (Figure 3G–3I). The nuclear distribution of NF-κB in AA (10 μmol/L and 30 μmol/L)-treated cells was less apparent than in normal HK-2 cells. By contrast, the cytoplasmic distribution of NF-κB in AA (10 μmol/L and 30 μmol/L)-treated cells was increased compared with the distribution in normal HK-2 cells (Figure 3A–3F).
Figure 3.
Cellular localization of the p65 subunit of NF-κB before and after AA treatment in HK-2 cells. NF-κB localization was visualized in panel D–F (green) and PI was used for nuclear staining (red) (panel G–I) by immunofluorescence and confocal microscopy. The nuclear distribution of NF-κB in AA-treated cells was less apparent than that in normal HK-2 cells (A–F).
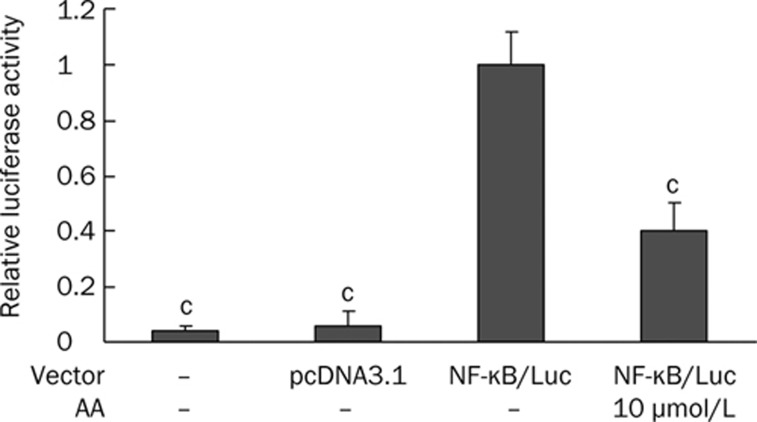
To further confirm that AA was able to affect the activity of NF-κB, transgenic cells carrying the NF-κB luciferase gene were used to estimate the NF-κB activity after AA treatment. Because the cell viability of HK-2 cells after treatment with 10 μmol/L of AA was 81.9%±4.6%, whereas that after 30 μmol/L of AA was 55.2%±3.7%, we chose the subtoxic dose of AA (10 μmol/L) to examine the effect of AA on the NF-κB activity by a luciferase assay. The HK-2 cells transfected with empty vector behaved like non-transfected cells. Both non-transfected and empty vector-transfected control groups showed a significantly much lower luciferase activity compared to cells transfected with the NF-κB luciferase reporter gene (Figure 4). NF-κB transgenic HK-2 cells were treated with 10 μmol/L AA, and the luciferase activity was analyzed. The results from these analyses indicated that AA significantly inhibited the luciferase activity in these cells compared with the untreated cells (Figure 4). These data revealed that 10 μmol/L AA can significantly suppress the NF-κB activity in HK-2 cells.
Figure 4.
The inhibitory effect of 10 μmol/L AA on NF-κB activity in HK-2 cells. HK-2 cells were transfected with NF-κB luciferase gene and were subsequently incubated with 10 μmol/L AA for 24 h. Experiments were done in triplicate and all results of luciferase assay expressed as relative luciferase units (RLU). Relative luciferase activity was calculated by dividing RLU of non-transfected cells, empty vector (pcDNA 3.1)-transfected cells, and AA-treated NF-κB-transfected cells by the RLU of untreated NF-κB-transfected cells. Error bars represent mean±SD of relative luciferase activity. cP<0.01 vs untreated NF-κB-transfected cells.
Discussion
Microarray analysis has become a popular and useful tool to study the effects of agents on gene expression in cells, tissues, and organs; however, only a few studies have applied DNA microarray analysis to investigate the effects of AA. In this in vitro study on the gene expression profiles of AA-treated (HK-2) cells, the normal human proximal tubular cell line was used, in contrast with the human colorectal cancer cell line (HCT 116) used by Simoes27. Our results showed that HK-2 cells were more sensitive to AA-induced cytotoxicity than HCT 116 cells. The IC50 values of AA after 24 h treatment in HK-2 cells and HCT 116 cells were 30 μmol/L and 100 μmol/L, respectively. Moreover, the effect of AA on gene expression was more profound in HK-2 cells than in HCT 116 cells27. A previous report indicated no significant difference in TP53 expression in P53-WT HCT 116 cells after a 24 h exposure to AA (100 μmol/L), whereas our study showed a marked downregulation of TP53 gene expression (fold change -1.92 after qRT-PCR confirmation) in 10 μmol/L AA-treated HK-2 cells (data not shown). In an in vivo study exploring the gene expression profiles of AA, rats were exposed to AA to distinguish the carcinogenic effects on target (kidney) and non-target (liver) tissues28. The microarray analysis after the AA treatment showed that there were more significantly altered genes and biological processes in the kidney compared to the liver. These data suggest that the analysis of gene expression profiles can be used to define the different responses of the kidney and liver to AA. Another experiment using gene expression profiling was conducted to study AA-induced renal tumorigenesis in the short-term AA-treated Eker rats (heterozygous for a mutation in the tuberous sclerosis 2 [Tsc2] tumor suppresser gene) and wild-type rats29. The results indicated that the gene expression profiles of AA-treated Eker and wild-type rats were Tsc2-independent.
Our finding revealed that AA exhibited cytotoxicity in a dose-dependent manner. These results were similar to an earlier study by Guo et al30 demonstrating that AAI, AAIa, and AAII inhibited the growth of HK-2 cells in a concentration-dependent and time-dependent manner. They also found that AAI, AAIa and AAII could affect the cell cycle and even induce cell apoptosis. Moreover, by microarray analysis, we noticed that genes involved in DNA repair, response to DNA damage stimulus, apoptosis, and cell cycle were significantly regulated by different concentrations of AA (Table 2). The pathway analysis showed that most of the AA-regulated pathways were associated with immunomodulatory functions, indicating that AA might participate in the regulation of immune genes. Furthermore, it has been noted that NF-κB is downregulated following AA exposure, and that it plays a central role in the gene networks regulated by AA (Figure 2). NF-κB is a nuclear transcription factor consisting of heterodimers including Rel A (p65), Rel B, c-Rel, p50, and p52. NF-κB activity can be induced by infection, pathogenic microorganisms, carcinogens, necrotic cell products, and inflammatory cytokines. When stimulated, activated NF-κB is translocated from the cytoplasm to the nucleus, where it binds to specific DNA sequences in the promoter regions of genes and initiates the expression of genes that encode proteins to control stress responses, cell adhesion, proliferation, and apoptosis31. After NF-κB is activated, it may serve as a transcription factor, and a small change in the gene expression of NF-κB may lead to significant changes in the expression levels of NF-κB-regulated downstream genes. In our study, the mRNA expression level of NF-κB in 10, 30, and 90 μmol/L AA-treated cells was 1.24±0.14, 1.04±0.24, and 1.20±0.23, respectively.
Constitutive NF-κB activity can induce the overexpression of proinflammatory genes, which can lead to acute or chronic inflammatory disease, such as rheumatoid arthritis, inflammatory bowel disease, and atherosclerosis32. Similarly, the upregulated expression of NF-κB has been found in pancreatic cancer, hepatocellular carcinoma, and colorectal cancer33. Recent research strongly suggests a relationship between chronic inflammation, NF-κB activation, and cancer33, 34. Because NF-κB target genes promote tumor cell proliferation, survival, migration and angiogenesis, the inhibition of NF-κB was thought to be useful as an anticancer therapy in clinical studies. However, the inhibition of NF-κB has been found to promote squamous cell carcinomas in epidermal skin35, 36. Although plants of the genus Aristolochia have been used as an anti-inflammatory (eg, for arthritis), analgesic, diuretic, and anticancer agents in the past, it was interesting to note that more recently, AA has been reported to be nephrotoxic and carcinogenic. These different properties of AA are similar to the dual effects of NF-κB. In our study, the results from immunofluorescence confocal microscopy and a luciferase reporter assay revealed that NF-κB activity in HK-2 cells was inhibited by AA. The nuclear localization of NF-κB in AA (10 μmol/L and 30 μmol/L)-treated cells was less apparent than in normal HK-2 cells. It was suggested that AA decreased the nuclear staining of NF-κB by the inhibition of NF-κB translocation from the cytoplasm to the nucleus. Concurrently, our study showed that AA significantly inhibited NF-κB activity in HK-2/NF-κB transgenic cells compared with the control cells.
Among the list of genes significantly regulated by the 10, 30, and 90 μmol/L AA treatments, chemokine ligand 20 (CCL20) was the most markedly downregulated. CCL20 is a macrophage inflammatory protein that plays a role in B-cell adhesion to the inflamed endothelium37. Interleukin 8 (IL-8) was another downregulated gene identified in our study. The gene expression of IL-8 can be induced by IL-1 and TNF and simultaneously regulated by NF-κB38. Insulin-like growth factor binding protein 3 (IGFBP3) was also shown to be downregulated following AA treatment in this study. IGFBP-3 can inhibit the proliferative function and anti-apoptotic functions of insulin-like growth factor (IGF)39. Recently, it has been shown that the regulation of IGFBP-3 involves the phosphatidylinositol-3 kinase (PI3K) and NF-κB pathways39, 40. Another downregulated gene identified in this study was CD68, which is a transmembrane glycoprotein highly expressed in human monocytes and tissue macrophages. CD68 is also a member of the scavenger receptor family and typically functions to clear cellular debris, promote phagocytosis, and mediate the recruitment and activation of macrophages. Based on immunohistochemistry, CD68 is histiocytic (similar to the T-lymphocytic marker CD3) and is associated with multicentric reticulohistiocytosis41, rheumatoid arthritis42, Crohn's disease43, and vasculitic neuropathies44. Besides the genes mentioned above, the gene expression of other NF-κB downregulated genes, including SAA2, LTB, TNFRSF9, was confirmed by real-time qRT-PCR. Most of these NF-κB downregulated genes are involved the inflammatory response. In summary, the downregulation of NF-κB regulated genes by AA may partly explain the use of Chinese herbs of Aristolochiaceae to treat inflammatory diseases such as arthritis in the past.
In conclusion, AA exhibited a dose-dependent cytotoxic effect in HK-2 cells and induced alterations in gene expression profiles related to DNA damage response, stress response, and other pathways. In addition, we demonstrated that AA treatment downregulated nine biological pathways, most of which were associated with immunomodulatory functions in HK-2 cells, and NF-κB plays a central role in the network topology. The relationship between AA and NF-κB was further confirmed by real-time qRT-PCR, confocal microscopy in AA-treated HK-2 cells and the luciferase reporter assay in HK-2/ NF-κB transgenic cells. Our study reveal that AA can suppress NF-κB activity in human cells, and these results may partly explain the toxicological and pharmacological effects of the genus Aristolochia.
Author contribution
Jaung-geng LIN conceived and coordinated the study; Su-yin CHIANG, Tin-yun HO, and Chien-yun HSIANG designed the study and supervised the data collection; Ya-yin CHEN, Hsiu-ching WU, and Shung-te KAO performed the experiments; Tin-yun HO and Chien-yun HSIANG contributed analytical tools; and Ya-yin CHEN analyzed the data and wrote the manuscript.
Acknowledgments
We thank Mr Wei-shuen SHEN, Miss Zih-syuan WANG, Miss Wan-yu TIEN, Miss Wei-pin HUANG, and Miss Zhao-ying DING for their technical assistance. This work was supported by grants from the National Research Program for Genomic Medicine, the National Science and Technology Program for Agricultural Biotechnology, the National Science Council, the Committee on Chinese Medicine and Pharmacy of the Department of Health (CCMP 96-RD-201 and CCMP 97-RD-201), and China Medical University (CMU95-051).
References
- Rosenmund H, Reichstein T. Zur Kenntnis der Aristolochiasäure. Pharm Acta Helv. 1943;18:243–61. [Google Scholar]
- Debelle FD, Vanherweghem JL, Nortier JL. Aristolochic acid nephropathy: a worldwide problem. Kidney Int. 2008;74:158–69. doi: 10.1038/ki.2008.129. [DOI] [PubMed] [Google Scholar]
- Zhang G, Shimokawa S, Mochizuki M, Kumamoto T, Nakanishi W, Watanabe T, et al. Chemical constituents of Aristolochia constricta: antispasmodic effects of its constituents in guinea-pig ileum and isolation of a diterpeno-lignan hybrid. J Nat Prod. 2008;71:1167–72. doi: 10.1021/np800041t. [DOI] [PubMed] [Google Scholar]
- Messiano GB, Vieira L, Machado MB, Lopes LM, de Bortoli SA, Zukerman-Schpector J. Evaluation of insecticidal activity of diterpenes and lignans from Aristolochia malmeana against Anticarsia gemmatalis. J Agric Food Chem. 2008;56:2655–9. doi: 10.1021/jf703594z. [DOI] [PubMed] [Google Scholar]
- Hinou J, Demetzos C, Harvala C, Roussakis C. Cytotoxic and antimicrobial principles from the roots of Aristolochia longa. Int J Crude Drug Res. 1990;28:149–51. [Google Scholar]
- Arlt VM, Stiborova M, Schmeiser HH. Aristolochic acid as a probable human cancer hazard in herbal remedies: a review. Mutagenesis. 2002;17:265–77. doi: 10.1093/mutage/17.4.265. [DOI] [PubMed] [Google Scholar]
- Loiko EN, Samal AB, Shulyakovskaya SM. H2O2-induced platelet aggregation and increase in intracellular Ca2+ concentration are blocked by inhibitors of intracellular signaling. Biochemistry (Mosc) 2003;68:1210–6. doi: 10.1023/b:biry.0000009135.85883.65. [DOI] [PubMed] [Google Scholar]
- Iuliano L, Pedersen JZ, Pratico D, Rotilio G, Violi F. Role of hydroxyl radicals in the activation of human platelets. Eur J Biochem. 1994;221:695–704. doi: 10.1111/j.1432-1033.1994.tb18782.x. [DOI] [PubMed] [Google Scholar]
- Marshall LA, Chang JY, Calhoun W, Yu J, Carlson RP. Preliminary studies on phospholipase A2-induced mouse paw edema as a model to evaluate antiinflammatory agents. J Cell Biochem. 1989;40:147–55. doi: 10.1002/jcb.240400203. [DOI] [PubMed] [Google Scholar]
- Vishwanath BS, Fawzy AA, Franson RC. Edema-inducing activity of phospholipase A2 purified from human synovial fluid and inhibition by aristolochic acid. Inflammation. 1988;12:549–61. doi: 10.1007/BF00914317. [DOI] [PubMed] [Google Scholar]
- Lindahl M, Tagesson C. Selective inhibition of group II phospholipase A2 by quercetin. Inflammation. 1993;17:573–82. doi: 10.1007/BF00914195. [DOI] [PubMed] [Google Scholar]
- Rosenthal MD, Vishwanath BS, Franson RC. Effects of aristolochic acid on phospholipase A2 activity and arachidonate metabolism of human neutrophils. Biochim Biophys Acta. 1989;1001:1–8. doi: 10.1016/0005-2760(89)90299-3. [DOI] [PubMed] [Google Scholar]
- Moreno JJ. Effect of aristolochic acid on arachidonic acid cascade and in vivo models of inflammation. Immunopharmacology. 1993;26:1–9. doi: 10.1016/0162-3109(93)90061-t. [DOI] [PubMed] [Google Scholar]
- Shirwaikar A, Somashekar AP, Udupa AL, Udupa SL, Somashekar S. Wound healing studies of Aristolochia bracteolata Lam. with supportive action of antioxidant enzymes. Phytomedicine. 2003;10:558–62. doi: 10.1078/094471103322331548. [DOI] [PubMed] [Google Scholar]
- Kluthe R, Vogt A, Batsford S.Double blind study of the influence of aristolochic acid on granulocyte phagocytic activity Arzneimittelforschung 198232443–5.German. [PubMed] [Google Scholar]
- Vanherweghem JL, Depierreux M, Tielemans C, Abramowicz D, Dratwa M, Jadoul M, et al. Rapidly progressive interstitial renal fibrosis in young women: association with slimming regimen including Chinese herbs. Lancet. 1993;341:387–91. doi: 10.1016/0140-6736(93)92984-2. [DOI] [PubMed] [Google Scholar]
- Vanhaelen M, Vanhaelen-Fastre R, But P, Vanherweghem JL. Identification of aristolochic acid in Chinese herbs. Lancet. 1994;343:174. doi: 10.1016/s0140-6736(94)90964-4. [DOI] [PubMed] [Google Scholar]
- Martinez MC, Nortier J, Vereerstraeten P, Vanherweghem JL. Progression rate of Chinese herb nephropathy: impact of Aristolochia fangchi ingested dose. Nephrol Dial Transplant. 2002;17:408–12. doi: 10.1093/ndt/17.3.408. [DOI] [PubMed] [Google Scholar]
- Cosyns JP, Jadoul M, Squifflet JP, Van Cangh PJ, Van Ypersele de Strihou C. Urothelial malignancy in nephropathy due to Chinese herbs. Lancet. 1994;344:188. doi: 10.1016/s0140-6736(94)92786-3. [DOI] [PubMed] [Google Scholar]
- Nortier JL, Martinez MC, Schmeiser HH, Arlt VM, Bieler CA, Petein M, et al. Urothelial carcinoma associated with the use of a Chinese herb (Aristolochia fangchi) N Engl J Med. 2000;342:1686–92. doi: 10.1056/NEJM200006083422301. [DOI] [PubMed] [Google Scholar]
- Lai MN, Lai JN, Chen PC, Tseng WL, Chen YY, Hwang JS, et al. Increased risks of chronic kidney disease associated with prescribed Chinese herbal products suspected to contain aristolochic acid. Nephrology. 2009;14:227–34. doi: 10.1111/j.1440-1797.2008.01061.x. [DOI] [PubMed] [Google Scholar]
- Cheng WY, Hsiang CY, Bau DT, Chen JC, Shen WS, Li CC, et al. Microarray analysis of vanillin-regulated gene expression profile in human hepatocarcinoma cells. Pharmacol Res. 2007;56:474–82. doi: 10.1016/j.phrs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Smyth GK.editor. Limma: linear models for microarray data. New York: 2005. p 397–420.
- Montaner D, Tarraga J, Huerta-Cepas J, Burguet J, Vaquerizas JM, Conde L, et al. Next station in microarray data analysis: GEPAS. Nucleic Acids Res. 2006;34:W486–91. doi: 10.1093/nar/gkl197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert M, Scherf M, Epple A, Werner T. Multievidence microarray mining. Trends Genet. 2005;21:553–8. doi: 10.1016/j.tig.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes ML, Hockley SL, Schwerdtle T, da Costa GG, Schmeiser HH, Phillips DH, et al. Gene expression profiles modulated by the human carcinogen aristolochic acid I in human cancer cells and their dependence on TP53. Toxicol Appl Pharmacol. 2008;232:86–98. doi: 10.1016/j.taap.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Chen T, Guo L, Zhang L, Shi L, Fang H, Sun Y, et al. Gene expression profiles distinguish the carcinogenic effects of aristolochic acid in target (Kidney) and non-target (Liver) tissues in rats. BMC Bioinformatics. 2006;7:S20. doi: 10.1186/1471-2105-7-S2-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer K, Ellinger-Ziegelbauer H, Ahr HJ, Dietrich DR. Carcinogen-specific gene expression profiles in short-term treated Eker and wild-type rats indicative of pathways involved in renal tumorigenesis. Cancer Res. 2007;67:4052–68. doi: 10.1158/0008-5472.CAN-06-3587. [DOI] [PubMed] [Google Scholar]
- Guo YC, Lin ZX, Li H, Luo WH.The toxic effects of three Aristolochia compounds on HK-2 cell Carcinog Teratog & Mutagen 20061888–92.Chinese. [Google Scholar]
- Beinke S, Ley SC. Functions of NF-kappaB1 and NF-kappaB2 in immune cell biology. Biochem J. 2004;382:393–409. doi: 10.1042/BJ20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valledor AF, Ricote M. Nuclear receptor signaling in macrophages. Biochem Pharmacol. 2004;67:201–12. doi: 10.1016/j.bcp.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Rigas B. NF-kappaB, inflammation and pancreatic carcinogenesis: NF-kappaB as a chemoprevention target (review) Int J Oncol. 2006;29:185–92. [PubMed] [Google Scholar]
- Dobrovolskaia MA, Kozlov SV. Inflammation and cancer: when NF-kappaB amalgamates the perilous partnership. Curr Cancer Drug Targets. 2005;5:325–44. doi: 10.2174/1568009054629645. [DOI] [PubMed] [Google Scholar]
- Dajee M, Lazarov M, Zhang JY, Cai T, Green CL, Russell AJ, et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–43. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- van Hogerlinden M, Rozell BL, Ahrlund-Richter L, Toftgard R. Squamous cell carcinomas and increased apoptosis in skin with inhibited Rel/nuclear factor-kappaB signaling. Cancer Res. 1999;59:3299–303. [PubMed] [Google Scholar]
- Meissner A, Zilles O, Varona R, Jozefowski K, Ritter U, Marquez G, et al. CC chemokine ligand 20 partially controls adhesion of naive B cells to activated endothelial cells under shear stress. Blood. 2003;102:2724–7. doi: 10.1182/blood-2003-01-0007. [DOI] [PubMed] [Google Scholar]
- Mukaida N, Mahe Y, Matsushima K. Cooperative interaction of nuclear factor-kappa B- and cis-regulatory enhancer binding protein-like factor binding elements in activating the interleukin-8 gene by pro-inflammatory cytokines. J Biol Chem. 1990;265:21128–33. [PubMed] [Google Scholar]
- Gewirtz DA. Growth arrest and cell death in the breast tumor cell in response to ionizing radiation and chemotherapeutic agents which induce DNA damage. Breast Cancer Res Treat. 2000;62:223–35. doi: 10.1023/a:1006414422919. [DOI] [PubMed] [Google Scholar]
- Deng DX, Spin JM, Tsalenko A, Vailaya A, Ben-Dor A, Yakhini Z, et al. Molecular signatures determining coronary artery and saphenous vein smooth muscle cell phenotypes: distinct responses to stimuli. Arterioscler Thromb Vasc Biol. 2006;26:1058–65. doi: 10.1161/01.ATV.0000208185.16371.97. [DOI] [PubMed] [Google Scholar]
- Luz FB, Gaspar TAP, Kalil-Gaspar N, Ramos-e-Silva M. Multicentric reticulohistiocytosis. J Eur Acad Dermatol Venereol. 2001;15:524–31. doi: 10.1046/j.1468-3083.2001.00362.x. [DOI] [PubMed] [Google Scholar]
- Catalina-Fernandez I, Alvarez AC, Martin FC, Fernandez-Mera JJ, Saenz-Santamaria J. Cutaneous intralymphatic histiocytosis associated with rheumatoid arthritis: report of a case and review of the literature. Am J Dermatopathol. 2007;29:165–8. doi: 10.1097/01.dad.0000251824.09384.46. [DOI] [PubMed] [Google Scholar]
- Franchimont N, Reenaers C, Lambert C, Belaiche J, Bours V, Malaise M, et al. Increased expression of receptor activator of NF-kappaB ligand (RANKL), its receptor RANK and its decoy receptor osteoprotegerin in the colon of Crohn's disease patients. Clin Exp Immunol. 2004;138:491–8. doi: 10.1111/j.1365-2249.2004.02643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck KM, Bierhaus A, Erwin S, Kirchner A, Nawroth P, Schlotzer U, et al. Receptor for advanced glycation endproduct (RAGE)-mediated nuclear factor-kappaB activation in vasculitic neuropathy. Muscle Nerve. 2004;29:853–60. doi: 10.1002/mus.20039. [DOI] [PubMed] [Google Scholar]