Abstract
As autophagy is involved in cell growth, survival, development and death, impaired autophagic flux has been linked to a variety of human pathophysiological processes, including neurodegeneration, cancer, myopathy, cardiovascular and immune-mediated disorders. There is a growing need to identify and quantify the status of autophagic flux in different pathological conditions. Given that autophagy is a highly dynamic and complex process that is regulated at multiple steps, it is often assessed accurately. This perspective review article will focus on the autophagic flux defects in different human disorders and update the current methods of monitoring autophagic flux. This knowledge is essential for developing autophagy-related therapeutics for treating the diseases.
Keywords: autophagy, autophagic flux, autophagosome-lysosome fusion, neurodegenerative disease, cancer, myopathy, cardiovascular disease, immune-mediated disorder
Introduction
Macroautophagy, herein referred to as autophagy, is a sequential process through which eukaryotic cells degrade long-lived proteins, misfolded proteins and impaired cytoplasmic organelles1. Autophagy is considered to be a dynamic process that comprises three sequential steps: formation of autophagosomes, the fusion of autophagosomes with lysosomes and degradation2. The entrapped cargo materials within autolysosomes are degraded by hydrolases, the most important component of the cathepsin family of proteases3. Autophagy is tightly associated with a wide range of physiological processes, such as embryonic development and the establishment of immunologic self-tolerance4. Moreover, emerging evidence suggests that the dysregulation of autophagy may contribute to a broad spectrum of human diseases, including cancer, cardiovascular disease, muscular disease and neurodegenerative disease4,5.
The increasing significance attached to autophagy in development and disease has generated a growing need to accurately identify and quantify autophagy in live cells, animals, and patients. To date, the principal methods used to monitor autophagic activation are the detection of LC3 processing by Western blot analysis and the detection of autophagosome formation by fluorescence and electron microscopy6,7. One critical point that must be kept in mind is that autophagy is a highly dynamic, multi-step process. The accumulation of autophagosomes could indicate either autophagic activation or a blockage of downstream steps in autophagy, such as inefficient fusion or decreased lysosomal degradation4,8. Therefore, the mere detection of the number of autophagosomes and the presence of LC3 processing is insufficient for an overall evaluation of the entire autophagic system.
The term “autophagic flux” is used to represent the dynamic process of autophagy. In detail, autophagic flux refers to the whole process of autophagy, including autophagosome formation, maturation, fusion with lysosomes, subsequent breakdown and the release of macromolecules back into the cytosol (Figure 1). In this review, we will summarize the relationship between autophagic flux defects and human diseases and discuss the latest methods to monitor autophagic flux in mammalian cells.
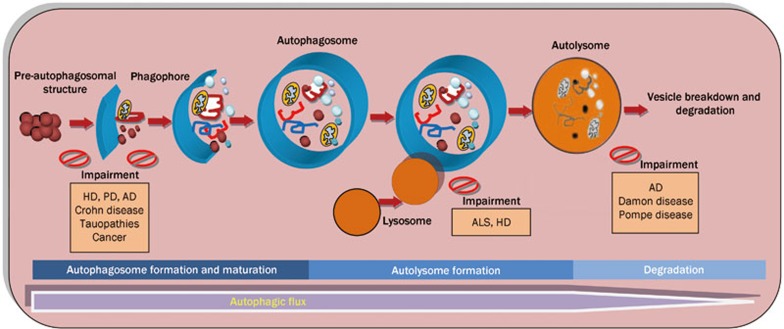
Figure 1.
The process of autophagic flux and the putative steps of autophagic dysfunction in human diseases. A normal autophagic flux includes the autophagosome formation and maturation step, the autolysosome formation step and the degradation step. The possible defects that could lead to autophagic dysfunction in different human diseases are depicted: (1) autophagy induction and autophagosome formation; (2) inefficient autophagosome-lysosome fusion; (3) inefficient degradation of the autophagic vesicles in the lysosome. Examples of diseases for which alterations in each autophagic step have been described are shown. AD, Alzheimer's disease; ALS, Amyotrophic lateral sclerosis; HD, Huntington's disease; PD, Parkinson's disease.
Autophagic flux defects contribute to human diseases
Consistently, impaired autophagic progress is involved in a growing list of pathologies, including neurodegeneration, cancer, myopathy, cardiovascular diseases and immune-mediated disorders (Figure 1).
Autophagic flux defects in neurodegenerative diseases
Autophagy is essential for neuronal homeostasis, and its dysfunction has been directly linked to a growing number of neurodegenerative diseases8. Early reports have demonstrated the neuronal accumulation of autophagosomes in diverse neurodegenerative disorders, including Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD) and amyotrophic lateral sclerosis (ALS)9,10. Currently, many studies are dedicated to revealing the detailed mechanisms of the autophagic flux defects in those disorders to help identify possible therapeutic targets8,11. Impairment in the induction of autophagy may result in an autophagic flux defect that causes misfolded protein aggregation. The recruitment of a complex composed of three proteins (Vps15, Vps34, and Beclin-1) is important to the phagophore formation induction process12. It is now conceivable that changes in the enzymes of this complex could partially underlie autophagic dysfunction in neurodegenerative diseases13.
In addition to defects in the induction process, genetic or functional alterations may occur in the autophagosome maturation, autolysosome formation, or autophagosome clearance processes14. For example, when components of the dynein motor machinery, such as dynein, dynactin or tubulin deacetylases, undergo changes, autophagosome-lysosome fusion is impaired, leading to decreased autophagic clearance of protein aggregates and enhanced neurotoxicity in animal models of HD and ALS15,16,17. In addition, a recent report indicates that rapamycin (a classic autophagic activator) treatment causes further accumulation of autophagic vacuoles but fails to reduce the mutant SOD1 aggregates in ALS transgenic mice, indicating the possibility of abnormal autophagic flux in ALS18,19. Furthermore, it has been described that alterations in the lysosomal function, such as reduced lysosomal acidification or decreased activity of lysosomal hydrolases, underlie the failure of autophagic clearance (observed as increased autophagosome accumulation)8,11. A primary defect in lysosome acidification and proteolysis caused by mutations in presenilin-1 has recently been identified in AD, suggesting that defective lysosomal proteolysis may represent a basis for pathogenic protein accumulations and neuronal cell death in AD and other neurodegenerative diseases20.
Autophagic flux defects in muscular and cardiovascular diseases
Under most circumstances, defective autophagic flux plays an important role in the pathogenesis of inherited cardiovascular diseases (such as Danon disease and Pompe disease). Danon disease (a lysosomal glycogen storage disease) is characterized by cardiomyopathy, skeletal myopathy and variable mental retardation21. Lysosomal-associated membrane protein 2 (LAMP-2) deficiency is considered to be caused by genetic mutations and to lead to the extensive accumulation of autophagosomes in the muscles of mice and patients with Danon disease22. The impairment of autophagic flux by defective lysosomal function has been observed in an inherited disorder of skeletal and cardiac muscle, Pompe disease, which is caused by the deficiency of the glycogen-degrading lysosomal enzyme acid alpha-glucosidase23.
Autophagic flux defects in cancer
The relationship between autophagy and tumorigenesis has been intensively explored, but whether autophagy plays a role of pro- or anti-tumorigenesis in tumor development and cancer therapy is still controversial24. Several genetic links have been demonstrated between autophagic defects and cancer, implying that autophagy is a part of tumor suppressor pathways25. For example, partial deletion of the Beclin 1 gene is found in a high percentage of human breast, ovarian, and prostate cancers. A large number of human breast, ovarian, and brain cancers are characterized by decreased expression of Beclin 126. Furthermore, the autophagy related gene, Atg5, has been reported to suppress tumor genesis in a mouse xenograft model27. More interestingly, the expression of both Beclin 1 and UVRAG has been shown to repress the growth of human cancer cell xenografts28. Thus, although the molecular mechanisms are poorly understood, it is reasonable to postulate the importance of autophagy in tumor suppression.
Many studies have reported that autophagy may exert an anti-cancer function via its effects in maintaining the stability of the intracellular environment and its crucial roles in protein and organelle quality control to prevent cancer cell damage24. More importantly, the dual roles of autophagy in cell survival and cell death might explain the complicated effects of autophagy in the tumor cell response to anticancer therapies. Many anticancer agents have been reported to induce autophagy in tumor cell lines, and specific inhibition of autophagy with siRNAs targeted against ATG genes has been reported to accelerate tumor cell death, indicating that inhibition of autophagy is beneficial in cancer therapy29. However, autophagy has also been implicated in promoting chemoresistance in cancer cells, attenuating the therapeutic efficacy of chemotherapy30. Further in vivo studies are needed to determine the underlying mechanisms of autophagic flux defects in tumor suppression.
Autophagic flux defects in immune-mediated diseases
Recent studies indicate that autophagy acts as an immune effector that mediates pathogen clearance31. The roles of autophagy, included antigen presentation, promotion of lymphocyte homeostasis and survival, and regulation of cytokine production, bridge both the innate and adaptive immune systems32. A recent discovery suggests that a defect in autophagy caused by an ATG16L variant may contribute to the pathogenesis of Crohn's disease, a chronic inflammatory disorder of the bowel33. In vivo studies using ATG16L or other ATG gene knockout mice may help elucidate the potential relationships between impaired autophagy flux and Crohn's disease or other immune-mediated diseases32.
Methods for monitoring autophagic flux
Although the quantification of autophagosomes and autolysosomes by electron microscopy can help to identify the status of autophagic flux, it does not provide direct evidence about the autophagic substrate degradation in lysosomes. Therefore, electron microscopy is not recommended as a classical method for a “autophagic flux” assay34 (Table 1).
Table 1. Current methods for monitoring autophagosomes and autophagic flux.
| |
|
|
Results |
|||
|---|---|---|---|---|---|---|
| Assays | Mehods | Basal | Induction | Early step suppression | Late step suppression | |
| Autophagosomes | Number of LC3 positive puncta | IHC | → | ↑↑ | ↓ | ↑↑ |
| LC3-II level | WB | → | ↑↑ | ↓ | ↑↑ | |
| Morphology | AP/AL detection | TEM | AP → | AP ↑ | AP ↓ | AP ↑↑ |
| AL → | AL ↑ | AL ↓ | AL ↓ | |||
| Autophagic flux | LC3 turnover | WB | → | ↑↑ | ↓ | ↓ |
| P62 level | IHC/WB | → | ↓ | ↑ | ↑ | |
| mCherry-GFP-LC3 | IHC/FM | Red → | Red ↑ | Red ↓ | Red ↓ | |
| Yellow → | Yellow ↑ | Yellow ↓ | Yellow ↑↑ | |||
| Long lived protein degradation | IR | → | ↑↑ | ↓ | ↓ | |
AP, Autophagosome; AL, Autolysosome; IHC, Immunohistochemistry; IR, Isotope release; FM, Flow cytometry; TEM, Transmission electron microscopy; WB, Western blotting; Early step suppression refers to impairments in autophagosome formation or maturation; Late step suppression refers to impairments in the fusion and degradation steps.
Assay based on dynamic LC3 turnover
Perhaps the most experimentally straightforward method for monitoring autophagy is the detection of LC3 protein processing35. LC3 proteins are specifically cleaved at the C terminus by Atg4 to become LC3-I, which then conjugates to phosphatidylethanolamine to form LC3-II35. Based on the observation that LC3-II is degraded in autolysosomes, the level of LC3-II or GFP-LC3-II is widely used as a marker for monitoring the autophagic process6. However, an increased level of LC3-II or an accumulation of GFP-LC3 puncta is not always indicative of autophagy induction and may represent a blockade in autophagosome maturation36.
Autophagic flux can be detected by LC3-II turnover using Western blot analysis in the presence and absence of lysosomal degradation inhibitors, such as pepstatin A, E64d, bafilomycin A1, chloroquine and NH4Cl37,38. If autophagic flux is occurring, the level of LC3-II will be increased in the presence of the a lysosomal degradation inhibitor because the transit of LC3-II through the autophagic pathway will be blocked39. Using a tandem fusion of LC3 to the acid-insensitive mCherry (or other red fluorescent protein such as RFP) and the acid-sensitive GFP, a novel autophagic flux report system has been developed to analyze autophagosome maturation and degradation40,41. In this system, LC3 is fused to both GFP and mCherry and forms an mCherry-GFP-LC3 vector. At first, both GFP and mCherry are detected in autophagosomes, which appear as yellow puncta40. Once autophagosomes fuse with lysosomes, the green fluorescence is lost because of the degradation of GFP by acid lysosomal proteases, resulting in LC3 emitting only red fluorescence40. The dynamic switch from yellow to red fluorescence indicates a functional autophagic flux process.
Assay of degradation of p62
P62, also known as SQSTM1/sequestome 1, serves as a link between LC3 and ubiquitinated substrates and is efficiently degraded by autophagy42. Thus, the level of p62 proteins can be used to monitor autophagic flux. For example, autophagic suppression correlates with an increased p62 level, and similarly, autophagic activation correlates with a decreased p62 level43. Although the measurement of the cellular p62 level appears to correlate well with other markers of autophagic flux, this assay has some potential experimental pitfalls. Firstly, p62 is degraded by both the autophagy and ubiquitin-proteasome system, and its level may be increased when the proteasome is inhibited44. Secondly, besides LC3, p62 contains domains that interact with several signaling molecules, indicating that p62 may have other functions with regard to its role in autophagy45. Finally, p62 can be transcriptionally upregulated under certain conditions46. Given these potential pitfalls, to monitor autophagic flux it is recommended that the measurement of the p62 level be performed in combination with other methods such as LC3-II turnover.
Assay of degradation of long-lived proteins
Long-lived protein degradation assays represent a traditional and well-established method for evaluating autophagic flux47. The general strategy is first to culture cells with isotope-labeled amino acids (usually [14C]-leucine, [14C]-valine or [35S]-methionine) for a sufficient length of time to label long-lived proteins (the autophagic substrates). This step is followed by a second incubation without isotope-labeled amino acids to wash out the short-lived cellular proteins (the proteasome substrates)48. Next, the time-dependent, trichloroacetic acid-soluble radioactivity is measured, which is the most quantitative index of autophagic flux48. To further ensure that the measured radioactivity is the result of degradation of proteins by the autophagic pathway or another proteolytic pathway, it is recommended to compare the radioactive assay between samples with or without an autophagy inhibitor (such as 3-methyladenine, 3-MA) and with or without isotope-labeled amino acids.
Conclusions
There is no doubt that research on autophagy has been expanded dramatically, and a growing number of studies have pinpointed a causal relationship between autophagic flux defects and several diseases; thus, it is critical to set a standard method to measure autophagic flux. However, currently there is no single “gold standard” to measure autophagic flux. It is recommended to use a combination of several assays to accurately measure the status and functions of autophagic flux. Clearly, we need to put more effort into defining the autophagic signaling pathways, understanding the molecular role of Atg proteins, and identifying the regulatory mechanisms of autophagosome-lysosome fusion. With that research focus, we will be able to establish a better methodology for monitoring autophagic flux, find more specific autophagic activators or inhibitors, and develop more ideal animal models with the goal of uncovering the pathologic role of the autophagic pathway in human diseases such as neurodegeneration, cancer, infection, and cardiovasculopathy. For detailed information, please see the autophagy-related databases, including the Human Autophagy-dedicated Database (HADb; http://www.autophagy.lu/) and the Autophagy Database (http://tp-apg.genes.nig.ac.jp/autophagy/).
Acknowledgments
This review was supported by the National Natural Science Foundation of China (No 81000541 and No 81171201) and the National Basic Research Program of China (No 2011CB510003).
References
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF. Autopahgy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Adeli K, Agholome L, et al. Guidelines for the use and interpretation of assays for monitoring autophagy Autophagy 201281–100.22082964 [Google Scholar]
- Rubinsztein DC. The role of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nature. 2010;13:805–11. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–12. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- Sarkar S. Role of autophagy in neurodegenerative diseases. Curr Sci. 2011;101:1–6. [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickford F, Masliah E, Britschqi M, Lucin K, Narasimhan R, Jaeger PA, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–9. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravilumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O'Kane CJ, et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005;37:771–6. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- Webb JL, Ravikumar B, Rubinstztein DC. Microtubule disruption inhibits autophagosome-lysosome fusion: implications for studying the roles of aggresomes in polyglutamine diseases. Int J Biochem Cell Biol. 2004;36:2541–50. doi: 10.1016/j.biocel.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–92. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct. 2008;33:109–22. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- Zhang XJ, Li L, Chen S, Yang D, Wang Y, Zhang X, et al. Rapamycin treatment augments motor neuron degeneration in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7:412–25. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang XJ, Song L, Le W. Autophagy dysregulation in amyotrophic lateral sclerosis. Brain Pathol. 2012;22:110–6. doi: 10.1111/j.1750-3639.2011.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–58. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicdan MC, Nishino I. Autophagy in lysosomal myopathies. Brain Pathol. 2012;22:82–8. doi: 10.1111/j.1750-3639.2011.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–10. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Ahearn M, Roberts A, Mattaliano RJ, Zaal K, Ralston E, et al. Autophagy and mistargeting of therapeutic enzyme in skeletal muscle in Pompe disease. Mol Ther. 2006;14:831–9. doi: 10.1016/j.ymthe.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Zhao L, Kuang M, Zhang B, Liang Z, Yi T, et al. Autophagy in tumorigenesis and cancer therapy: Dr Jekyll or Mr Hyde. Cancer Lett. 2012;323:115–27. doi: 10.1016/j.canlet.2012.02.017. [DOI] [PubMed] [Google Scholar]
- Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–7. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- Miracco C, Cosci E, Oliveri G, Luzi P, Pacenti L, Monciatti I, et al. Protein and RNA expression of autophagy gene Beclin 1 in human brain tumours. Int J Oncol. 2007;30:429–36. [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–32. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, et al. Autophagic and tumour suppressor activity of a novel Beclin-1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Amaravadi RK, Yu D, Lum JJ, Bui T, Christonphorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy: an emerging immunological paradigm. J Immunol. 2012;189:15–20. doi: 10.4049/jimmunol.1102108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–77. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey DC, Parkes M. Genome-wide association scanning highlights two autophagy genes, ATG16L and IRGM, as being significantly associated with Crohn's disese. Autophagy. 2007;3:649–51. doi: 10.4161/auto.5075. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem. 2006;281:36303–16. doi: 10.1074/jbc.M607031200. [DOI] [PubMed] [Google Scholar]
- Ju JS, Varadhachary AD, Miller SE, Weihl CC. Quantitation of “autophagic flux” in mature skeletal muscle. Autophagy. 2010;6:929–35. doi: 10.4161/auto.6.7.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes. Autophagy. 2008;4:849–50. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tendem fluorescent-tagged LC3. Autophagy. 2007;3:452–60. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–5. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Bardag-Gorce F, Francis T, Nan L, Li J, He Lue Y, French BA, et al. Modifications in P62 occur due to proteasome inhibition in alcoholic liver disease. Life Sci. 2005;77:2594–602. doi: 10.1016/j.lfs.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–4. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaso K, Yoshimoto Y, Nakano T, Takeshima T, Fukuhara Y, Yasui K, et al. Transcriptional activation of p62/A170/ZIP during the formation of the aggregates: possible mechanisms and the role of Lewy body formation in Parkinson's disease. Brain Res. 2004;1021:42–51. doi: 10.1016/j.brainres.2004.03.029. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–7. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Bauvy C, Meijer AJ, Codogno P. Assaying of autophagic protein degradation. Methods Enzymol. 2009;452:47–61. doi: 10.1016/S0076-6879(08)03604-5. [DOI] [PubMed] [Google Scholar]