Abstract
Aim:
Ubiquitin-proteasome system (UPS) and autophagosome-lysosome pathway (ALP) are the most important machineries responsible for protein degradation in Parkinson's disease (PD). The aim of this study is to investigate the adaptive alterations in autophagy upon proteasome inhibition in dopaminergic neurons in vitro and in vivo.
Methods:
Human dopaminergic neuroblastoma SH-SY5Y cells were treated with the proteasome inhibitor lactacystin (5 μmol/L) for 5, 12, or 24 h. The expression of autophagy-related proteins in the cells was detected with immunoblotting. UPS-impaired mouse model of PD was established by microinjection of lactacystin (2 μg) into the left hemisphere of C57BL/6 mice that were sacrificed 2 or 4 weeks later. The midbrain tissues were dissected to assess alterations in autophagy using immunofluorescence, immunoblotting and electron microscopy assays.
Results:
Both in SH-SY5Y cells and in the midbrain of UPS-impaired mouse model of PD, treatment with lactacystin significantly increased the expression levels of LC3-I/II and Beclin 1, and reduced the levels of p-mTOR, mTOR and p62/SQSTM1. Furthermore, lactacystin treatment in UPS-impaired mouse model of PD caused significant loss of TH-positive neurons in the substantia nigra, and dramatically increased the number of autophagosomes in the left TH-positive neurons.
Conclusion:
Inhibition of UPS by lactacystin in dopaminergic neurons activates another protein degradation system, the ALP, which includes both the mTOR signaling pathway and Beclin 1-associated pathway.
Keywords: proteasome inhibitor, lactacystin, ubiquitin-proteasome system, dopaminergic neuron, Parkinson's disease, autophagy-lysosomal pathway, autophagosome, mTOR, Beclin 1
Introduction
Parkinson's disease (PD) produces both motor and non-motor symptoms that substantially degrade a patient's quality of life. Clinically, PD affects motor function because of the loss of midbrain dopamine (DA) neurons. Biochemical and imaging data suggest a greater than 70% decrease in DA neurons at the time PD is diagnosed. Neuronal death continues as the disease progresses, and eventually, nearly all of these DA neurons are lost. Pathologically, PD is characterized by the accumulation of protein aggregates (called Lewy bodies) in the substantia nigra (SN) of the midbrain1. The accumulation of misfolded proteins and the formation of protein aggregates are closely related to cell dysfunction and death in individuals with PD. Extensive molecular and cellular changes occur that indicate that the impairment of the proteolytic system and protein aggregation are central to the pathogenesis of PD2,3. Preventing aggregation or disaggregating misfolded proteins might provide therapeutic benefit by slowing or preventing the progression of PD4.
The ubiquitin-proteasome system (UPS) is an important degradation system that selectively degrades short-lived intracellular proteins5,6. Several studies have converged to show that the failure of the UPS to degrade misfolded proteins plays an important role in the pathogenesis of both familial and sporadic PD, as well as other neurodegenerative diseases7,8. The autophagy-lysosomal pathway (ALP) is another important protein degradation system4,9. The ALP is a less selective multistep process involving the formation of double-membrane structures known as autophagosomes (AVs) followed by the fusion of these AVs with lysosomes to form autophagolysosomes, the contents of which are digested4,10,11. Generally, the ALP is primarily responsible for controlling the homeostasis of long-lived intracellular proteins and organelles. ALP dysfunction has been implicated in a number of neurodegenerative disorders, including PD10. Greater accumulation of AVs has been observed in postmortem patient brains compared with brains from normal controls12. Because autophagy is generally considered to be a bulk, less selective pathway, we thus hypothesized that if an aggregated substrate cannot be efficiently cleared by the proteasome, the activity of lysosomal systems, especially autophagy, is expected to increase as a compensatory mechanism to degrade potentially toxic proteins.
Previous studies have shown that the treatment of neuronal cell lines with low doses of proteasome inhibitors can activate autophagy13. Several studies have also shown that the proteasome inhibitor MG132 can enhance autophagy, accelerating the degradation of aggregated poly-ubiquitinated proteins in murine embryonic fibroblasts and DU145 prostate cancer cells14,15. It has remained unclear, however, how these aggregates are efficiently collected and delivered to autophagosomes16. In our previous work, we attempted to generate a UPS-impaired mouse model of PD by administering the proteasome inhibitor lactacystin directly into the medial forebrain bundle (MFB) of mice17. This UPS impairment model recapitulates many features of PD; however, it is not known whether UPS inhibition can alter autophagy in this model. In this study, we used both in vitro cell culture and in vivo animal models to assess the changes in autophagy activity in DA neurons and the correlation of these changes with DA neuron degeneration.
Materials and methods
Cell culture and treatment
SH-SY5Y cells (Cell Bank of Chinese Academy of Sciences, Shanghai, China) were maintained in DMEM medium containing 10% FBS and 1% penicillin/streptomycin at 37 °C in a humidified 5% CO2 incubator. The proteasome inhibitor lactacystin (Calbiochem, San Diego, California) was prepared in dimethylsulfoxide (DMSO) at a stock concentration of 1 mmol/L and then added to the cultures to a final concentration of 5 μmol/L for 5, 12, or 24 h. The drug vehicle (0.1% DMSO) was used as the treatment control. The total protein was then isolated from the cells at the indicated time points.
Animals and proteasome inhibitor treatment
C57BL/6 mice (male, 12 weeks old) were housed under conditions of constant temperature and controlled lighting (12 h/12 h light/dark cycle) and microinjected with a proteasome inhibitor, lactacystin, directly into the MFB according to a previously described protocol6,18. Briefly, the mice were anesthetized before being placed in a Kopf stereotaxic frame, and they received unilateral infusion of either lactacystin (2 μg in 2 μL of 0.1% DMSO) in the left hemisphere or vehicle (2 μL of 0.1% DMSO) on the right side. Lactacystin was freshly prepared, snap frozen and stored at −20 °C before use. The stereotaxic coordinates are as follows: anteroposterior (AP) −1.3 mm, lateromedial (ML) ±1.2 mm, and ventral from the dura (DV) −5.1 mm from the bregma. After treatment for 2 weeks or 4 weeks, half of the mice were sacrificed by anesthesia followed by transcardial perfusion with ice-cold 0.1 mol/L PBS and decapitated. The brain tissues were then fixed with 4% paraformaldehyde overnight, dehydrated with 20% sucrose and 30% sucrose, embedded in Tissue-Tek optimal cutting temperature compound (OCT, Sakura Finetek Inc) and finally snap frozen and stored at −80 °C for histological analysis. For the remaining mice, the midbrain tissues were immediately micro-dissected on ice for total protein isolation. Animal care and procedures were performed in accordance with the Laboratory Animal Care Guidelines approved by the Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences.
Immunofluorescence analysis
To perform the immunofluorescence analysis, the OCT-embedded brain tissues were sectioned with a Leica cryostat. Serial sections of the midbrain were cut to a 12 μm thickness and mounted on Poly-D-Lysine-coated slides. The slides were then incubated in blocking buffer, followed by an overnight incubation at 4 °C with the primary antibodies against LC3 (1:500; Sigma) and TH (1:2000; Sigma). The sections were then washed in PBS and incubated with a secondary antibody conjugated with Alexa Fluor-488/555 (1:400; Invitrogen), followed by staining the nuclei with Hoechst 33342. Finally, the sections were visualized under a fluorescence microscope (Olympus IX81, Japan) or a confocal microscope (Leica SP5, Germany).
Immunoblotting analysis
The collected mouse midbrain tissues or cultured SH-SY5Y cells were sonicated in ice-cold RIPA lysis buffer (Beyotime) supplemented with 1% PMSF according to the manufacturer's protocol. Aliquots containing 40 μg of total protein were separated on a 7.5% or 12% SDS gel and transferred onto 0.45 μm or 0.22 μm PVDF membranes. The nonspecific sites were blocked in 5% nonfat milk for 1 h at room temperature. The membranes were then incubated overnight in the presence of the primary antibodies against the following proteins: LC3 (1:1000; CST), p62 (1:30 000; MBL), Beclin 1 (1:1000; CST), p-mTOR (1:1000; CST) and mTOR (1:1000; CST). A mouse anti-β-actin antibody (1:30 000; Sigma) was used to demonstrate equal protein loading. The secondary antibodies conjugated with horseradish peroxidase were used. The detection of the proteins followed the manufacturer's instructions for the SuperSignal Detection Kit (Pierce, USA).
Electron microscopy
SNc tissues (approximately 1 mm3) were punched from the fixed midbrain blocks using the Palkovits method. The tissue punches were fixed in 2.5% glutaraldehyde in 100 mmol/L PBS and cut into 50 μm thick sections using a vibratome. The sections were postfixed with 1% OsO4, dehydrated, embedded in Durcupan (ACM; 14040) on a microscope slide and coverslipped. These sections were further cut into 70 nm-thick sections using a Reichert ultramicrotome. These ultra-thin sections were then stained with uranyl acetate and lead citrate and evaluated by electron microscopy (Philips CM120, Holland).
Statistical analysis
All values are presented as the mean±SEM values. Statistical significance was determined using Student's t test. The results were considered significant when P<0.05.
Results
Proteasome inhibition enhances autophagy in vitro
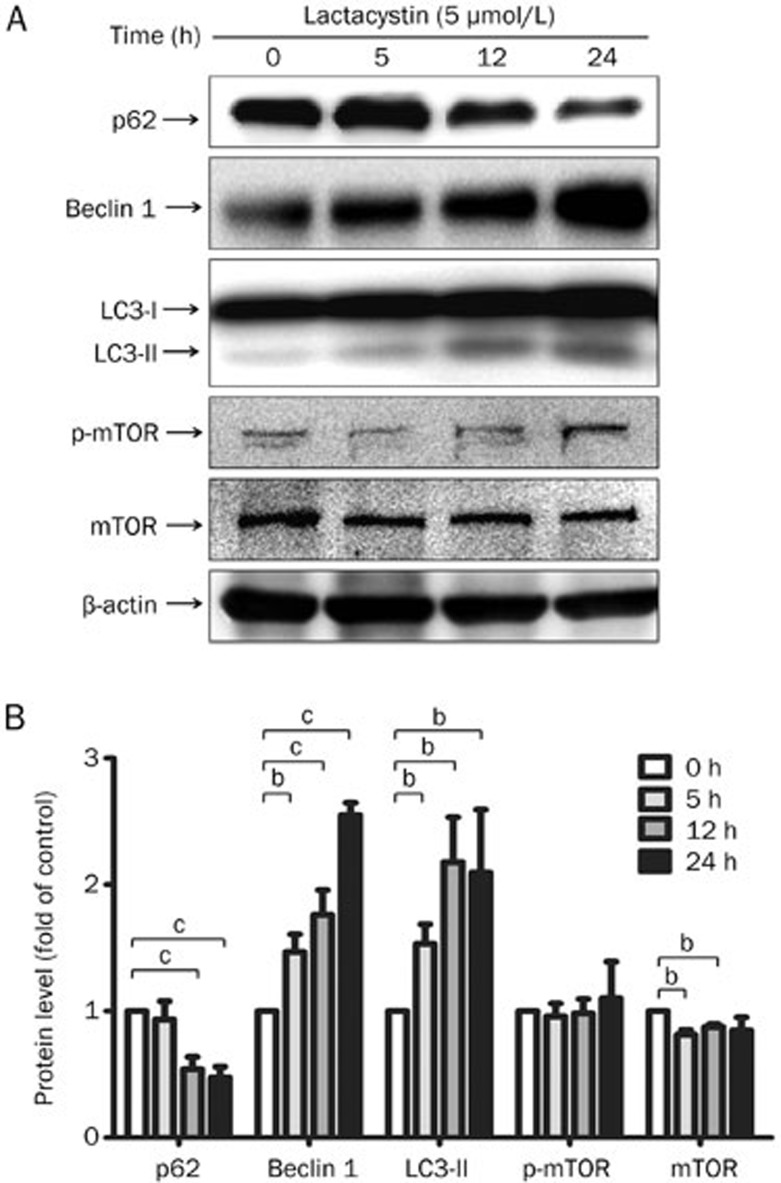
To investigate whether the inhibition of the proteasome enhances autophagy in vitro, we incubated human neuroblastoma SH-SY5Y cells with 5 μmol/L lactacystin, a specific inhibitor of the 20S proteasome, for 5, 12, or 24 h. We found that proteasome inhibition by lactacystin caused extensive neuronal death, as previously reported14,15. We then harvested the total protein from the remaining cells at the desired time. To evaluate the augmentation of diverse autophagy pathways, we assessed the expression of proteins associated with the mTOR signaling pathway, including mammalian target of rapamycin (mTOR) and phosphorylated mTOR (p-mTOR), and the Beclin signaling pathway, including Beclin 1, in addition to the autophagy markers LC3-I/II and p62/SQSTM1. The immunoblotting analysis revealed that treatment with lactacystin led to time-dependent increases in the level of LC3-II, a lipidated form of cleaved LC3 that is incorporated into the limiting membrane of AVs, and Beclin 1, a positive regulator involved in the formation of AVs and the initiation of autophagy (Figure 1). The polyubiquitin-binding protein p62/SQSTM1, which is found in cellular inclusion bodies together with polyubiquitinated proteins and in cytosolic protein aggregates, can bind directly to LC3-A and LC3-B, thereby facilitating the degradation of p62/SQSTM1-positive bodies containing polyubiquitinated proteins by autophagy19. We therefore also detected the protein level of p62/SQSTM1 and found, as we expected, that it was decreased after lactacystin treatment (Figure 1). We also found that mTOR expression was significantly down-regulated at 5 and 12 h after treatment, Whereas the p-mTOR level was not affected though it appeared a slight while not siginificant increase 24 h after lactacystin insult (Figure 1). Together, these results indicate that proteasome inhibition enhances autophagy in an in vitro cell model, most likely through regulating the protein expression levels of autophagy-related genes.
Figure 1.
Proteasome inhibition induces autophagy enhancement in the cell culture. (A) The human DAergic neuroblastoma SH-SY5Y cells were treated with lactacystin at a concentration of 5 μmol/L for 5, 12, and 24 h respectively. The protein expression of LC3-I/II, Beclin 1 and p62/SQSTM1 were determined by immunoblotting. (B) The density of the bands was analyzed by Image J software and expressed as fold of control. Mean±SEM. n=5. bP<0.05, cP<0.01.
Proteasome inhibition enhances autophagy in the UPS impairment mouse model
To determine whether proteasome inhibition leads to autophagy enhancement in vivo, we established a UPS impairment animal model by microinjecting mice with lactacystin. This mouse model exhibits many behavioral, biochemical and pathological features resembling those associated with the progressive nigral DA neuron dysfunction observed in PD, which is characterized by the loss of TH-positive neurons and the reduction of the striatal DA levels. Because this modelmimics certain aspects of PD, it has potential uses in the study of the role of autophagy in PD. The progressive PD-like symptoms of this model start to appear after one week and continue to be observed for up to 3 months17.
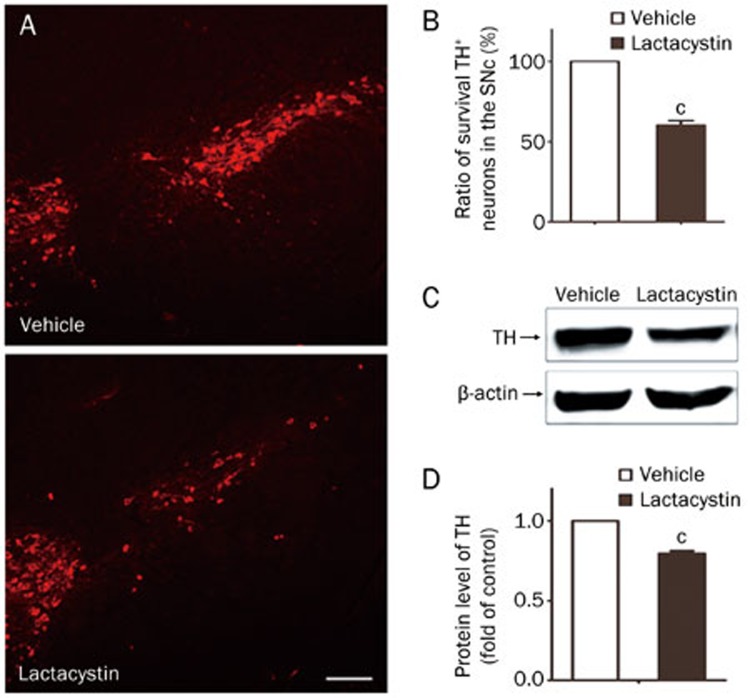
After the administration of 2 μg lactacystin into the MFB of C57BL/6 mice for 2–4 weeks, the mice were examined by immunostaining and immunoblotting assays. The number of TH-positive cells in the SN was determined by immunofluorescence staining with an anti-TH antibody (Figure 2A). Our results showed that lactacystin administration to the MFB caused a nearly 40% loss of TH-positive neurons in the SN after 4 weeks compared with the number of TH-positive neurons in the control (Figure 2B) (P<0.01). We also determined the TH protein level by immunoblotting and found that the protein expression of TH was significantly decreased after 4 weeks in the midbrain tissues (Figure 2C, 2D). Overall, these results suggest that lactacystin-induced UPS impairment caused marked nigral DA neuron degeneration.
Figure 2.
Loss of TH-positive neurons in the SN and decrease of TH protein in the midbrain of the UPS impaired mouse model. (A) TH-positive neurons in the SN were determined by immunofluorescence assay with anti-TH antibody 4 weeks after lactacystin injection. Scale bar, 200 μm. (B) The number of TH-positive neurons in the SNpc was quantified stereologically. (C, D) The protein level of TH in the 2 weeks and 4 weeks midbrain tissues of injected mice was determined by immunoblotting. The density of the bands was analyzed by Image J software and expressed as fold of control. Mean±SEM. n=5. cP<0.01.
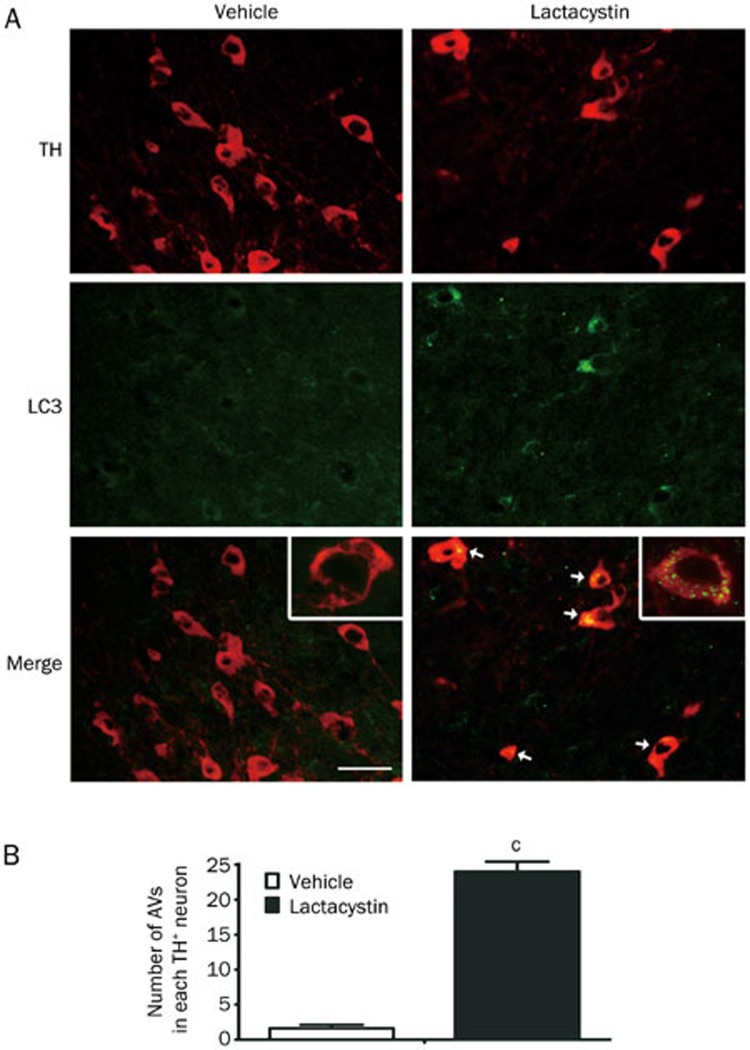
To investigate the changes in autophagy in the UPS impairment animal model, we employed double immunofluorescence staining with anti-LC3B and anti-TH antibodies to visualize the formation of AVs in DA neurons in the brain sections. The results showed that lactacystin treatment significantly increased the number and size of LC3-labeled AV aggregates in TH-positive neurons (Figure 3). Very few TH-negative neurons on the lactacystin injected side expressed LC3 and harbored LC3 vesicles in the cytoplasm, suggesting that changes in autophagy alteration were primarily limited to the DA neurons.
Figure 3.
Aggregation of autophagosomes (AVs) in DA neurons after proteasome inhibition. (A) AVs were visualized by double immunofluorescent staining with anti-LC3B and TH antibodies. The LC3-labled TH+ neurons are indicated by white arrows. Insets are magnified neurons. Scale bar, 100 μm. (B) The number of AVs in each TH+ neuron is counted. Mean±SEM. n=5. cP<0.01.
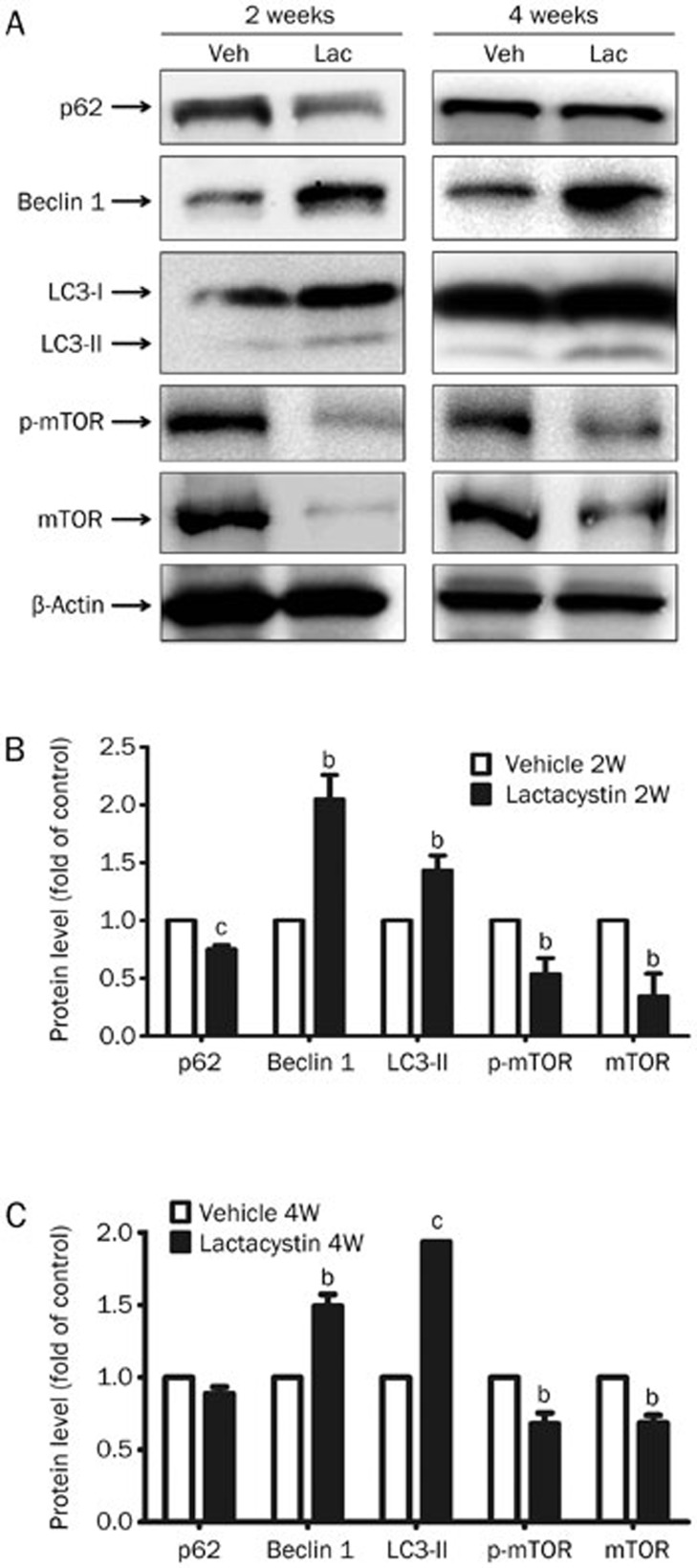
To detect the levels of markers in different autophagy pathways, we performed immunoblotting to examine the expression levels of proteins in the midbrains of lactacystin-injected mice, and we found that the protein levels of mTOR and p-mTOR were greatly down-regulated, whereas the levels of the autophagy markers LC3-II and Beclin 1 were accordingly elevated in the midbrain tissues of mice 2 weeks and 4 weeks after lactacystin injection (Figure 4). We also found that p62/SQSTM1 expression was greatly decreased 2 weeks after lactacystin injection, but the change in p62/SQSTM1 expression had partially recovered at 4 weeks (Figure 4). This phenomenon was most likely due to the complicated transcriptional regulation of p62/SQSTM1 by autophagy. Taken together, the results of the immunostaining and immunoblotting assays demonstrate that the ALP pathway is indeed activated by UPS dysfunction in vivo.
Figure 4.
UPS impairment induces autophagy augmentation in the animal model. (A) The protein expression of autophagy associated genes in the 2 weeks and 4 weeks midbrain tissues of injected mice including p62/SQSTM1, Beclin 1, LC3-I/II, p-mTOR, and mTOR was detected by immunoblotting. The density of the bands of 2 weeks (B) or 4 weeks (C) midbrain tissues was analyzed by Image J software and expressed as fold of control. Mean±SEM. n=5. bP<0.05, cP<0.01.
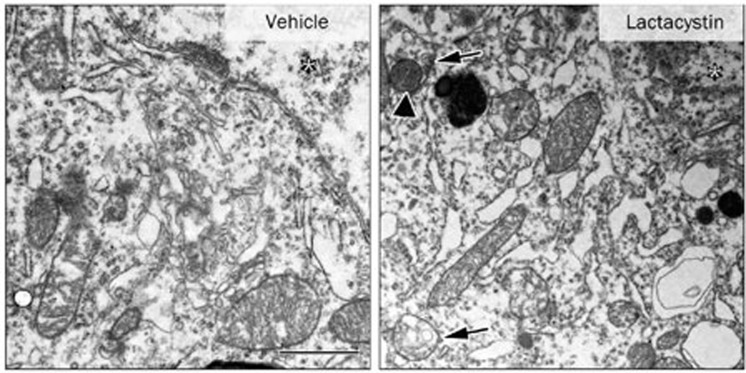
Although a number of other techniques have been used to evaluate autophagy, electron microscopy remains the gold standard. Autophagosomes (AVs) are characterized by double-membrane structures that engulf intracellular organelles and are accompanied by multilamellar bodies in the cytoplasm. In the electron microscopy images, we also observed many double-membrane structures harboring swollen mitochondria in the lactacystin-treated midbrain neurons (Figure 5), indicating that enhanced AV formation and autophagy alterations are induced by UPS impairment.
Figure 5.
Autophagosomes (AVs) were visualized under electron microscopy. Images shown are formation of AVs including swollen mitochondria (arrowhead) and AVs (arrows) induced by lactacystin injection. The asterisk represents the nucleus. Scale bar, 1 μm.
Discussion
Lewy bodies, which contain abundant intracytoplasmic misfolded protein aggregates, are the pathological hallmark of PD. The proteolytic systems responsible for protein degradation include the UPS, which is the primary non-lysosomal degradation system for ubiquitinated proteins, and the ALP, a lysosome-mediated degradation pathway20. Previously, the UPS and autophagy pathways have been viewed as complementary but parallel degradation systems that do not interact21,22. This view has been challenged by two recent studies that demonstrated an interaction between the UPS and the ALP: impairment of the UPS induced autophagy in vitro, and in turn, the conditional knockout of autophagy in the mouse brain gave rise to neurodegeneration with a ubiquitin-positive pathology23,24,25,26. In our study, we examined the functional relationship between these two systems and hypothesized that UPS dysfunction would lead to ALP activation in DA neurons both in vitro and in a UPS-impaired mouse model.
There have been many models used to evaluate the role of autophagy in the etiopathogenesis of PD, such as tissue-specific Atg5-deficient mice26, Atg7-deficient mice27, and the rapamycin-mediated mouse model28. Although these models have their own advantages, they also have limitations with respect to recapitulating either the neuropathology or clinical phenotype of PD. Obviously, models with nigral UPS impairment have advantages in modeling the two cardinal pathological features of PD, nigral degeneration and the aberrant protein degradation that regulates autophagy. Specifically, this mouse model has many behavioral, biochemical and pathological features resembling those due to the progressive nigral DA neuron dysfunction observed in PD, which is characterized by the loss of TH-positive neurons and a reduction in striatal DA levels17. We therefore established a UPS-impaired mouse model of PD by administering lactacystin directly into the MFB of mice, thereby mimicking the progressive neuron degeneration of PD. We observed the abundant accumulation of AVs by electron microscopy, immunoblotting and immunofluorescence staining in the DA neurons under conditions of UPS dysfunction. The immunoblotting results also indicated that lactacystin enhanced autophagy in a manner closely related to both the mTOR signaling pathway and the Beclin 1- associated pathway, which are considered to be the two core regulators of autophagy. The inhibition of the mTOR pathway by rapamycin can induce the activity of the autophagy machinery. Malagelada et al28 have found that rapamycin can protect neurons by sparing the phosphorylation of Akt in the mTOR signaling pathway to promote neuron survival. These authors also found that the administration of rapamycin to inhibit mTOR signaling in the MPTP-treated mouse model of PD greatly alleviated the loss of DA neurons.
It is still debated whether autophagy enhancement is a cell defense mechanism that acts as a compensatory degradation system in response to UPS impairment or a cell-killing program11,29. Some recent studies have revealed that autophagy might have a dual role in cell survival that depends on the cellular environment. At first, the activation of autophagy may be able to protect cells by degrading intracellular macromolecules and organelles to provide energy for cell functions and attenuate further cell injury by facilitating the removal of damaged organelles and aggregated proteins. However, prolonged or vigorous autophagy enhancement may lead to the excessive elimination of the affected cells through apoptosis19,30,31. In a previous study, Du et al32found that p53 can mediate the proteasome inhibition-induced activation of autophagy, which, in turn, may partially block p53 as well as its downstream mitochondria-dependent apoptotic pathways. Moreover, rapamycin treatment can protect VM neurons from lactacystin-mediated cell death by down-regulating p53-related apoptotic pathways and enhancing the degradation of misfolded proteins32. Pandey et al33 have also shown that autophagy acts as a compensatory degradation system that is induced in response to mutations affecting the proteasome when the UPS is impaired in a Drosophila melanogaster model of the neurodegenerative disease spinobulbar muscular atrophy. They also demonstrated that histone deacetylase 6 (HDAC6), a microtubule-associated deacetylase, is an essential mechanistic link in this compensatory interaction. In addition, the expression of HDAC6 was sufficient to rescue in vivo degeneration in an autophagy-dependent manner, suggesting that the impairment of autophagy might be a predisposing factor for neurodegeneration33. Because we know that autophagy is not strictly a parallel degradation system and that there may be a compensatory interaction between these systems, the molecular link must be investigated further. This link is of importance in delving into the functions of autophagy during neuron degeneration.
In conclusion, our findings indicate that lactacystin-induced UPS dysfunction can activate another protein degradation system, the ALP, including the mTOR signaling pathway and the beclin 1-associated pathway. Thus, in vitro and in vivo models may be of great help in investigating the role of autophagy in DA neuron degeneration associated with PD and in discovering novel therapies for PD.
Author contribution
Wei-dong LE designed this study; Xiao-jie ZHANG and Wei-dong LE supervised the research; Yu-fei SHEN and Yu TANG performed the experiments; Yu TANG analyzed the data; Yu TANG and Yu-fei SHEN wrote the manuscript; and Kai-Xing HUANG and Wei-dong LE edited the manuscript.
Acknowledgments
This study was supported by funding from the National Natural Sciences Foundation of China (No 81171201) and the National Basic Research Program of China (No 2011CB510003).
References
- Braak H, Del Tredici K, Rüb U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- Lansbury PT, Lashuel HA. Acentury-ole debateon protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–9. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL. Molecularpathways of neurodegeneration in Parkinson disease. Science. 2003;302:819–22. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- Pan T, Kondo S, Le W, Jankovic J. The role of autophagy-lysosome pathway in neurodegeneration associated with Parkinson disease. Brain. 2008;131:1969–78. doi: 10.1093/brain/awm318. [DOI] [PubMed] [Google Scholar]
- Bové J, Zhou C, Jackson-Lewis V, Taylor J, Chu Y, Rideout HJ, et al. Proteasomal inhibition and Parkinson disease modeling. Ann Neurol. 2006;60:260–4. doi: 10.1002/ana.20937. [DOI] [PubMed] [Google Scholar]
- Zhu W, Xie W, Pan T, Xu P, Fridkin M, Zheng H, et al. Prevention and restoration of lactacystin-induced nigrostriatal dopamine neuron degeneration bynovel brain permeable iron chelators. FASEB J. 2007;21:3835–44. doi: 10.1096/fj.07-8386com. [DOI] [PubMed] [Google Scholar]
- Olanow CW, McNaught KS. Ubiquitin-proteasome system and Parkinson's disease. Mov Disord. 2006;21:1806–23. doi: 10.1002/mds.21013. [DOI] [PubMed] [Google Scholar]
- Sawada H, Kohno R, Kihara T, Izumi Y, Sakka N, Ibi M, et al. Proteasome mediates dopaminergic neuronal degeneration, and its inhibition causes alpha-synuclein inclusions. J Biol Chem. 2004;279:10710–9. doi: 10.1074/jbc.M308434200. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–7. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Martinez VM, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–61. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- McCray BA, Taylor JP. The role of autophagy in age-related neurodegeneration. Neurosignals. 2008;16:75–84. doi: 10.1159/000109761. [DOI] [PubMed] [Google Scholar]
- Marino G, Lopez OC. Autophagy: molecular mechanisms, physiological functions and relevance in human pathology. Cell Mol Life Sci. 2004;61:1439–54. doi: 10.1007/s00018-004-4012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Dimayuga E, Martin S, Bruce-Keller AJ, Nukala V, Cuervo AM, et al. Characterization of chronic low-level proteasome inhibition on neural homeostasis. J Neurochem. 2003;86:489–97. doi: 10.1046/j.1471-4159.2003.01885.x. [DOI] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–24. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortun J, Verrier JD, Go JC, Madorsky I, Dunn WA, Notterpek L. The formation of peripheral myelin protein 22 aggregates is hindered by the enhancement of autophagy and expression of cytoplasmic chaperones. Neurobiol Dis. 2007;25:252–65. doi: 10.1016/j.nbd.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Sekito T, Ohsumi Y. Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr Top Microbiol Immunol. 2004;279:73–84. doi: 10.1007/978-3-642-18930-2_5. [DOI] [PubMed] [Google Scholar]
- Xie W, Li X, Li C, Zhu W, Jankovic J, Le W. Proteasome inhibition modeling nigral neurondegeneration in Parkinson's disease. J Neurochem. 2010;115:188–99. doi: 10.1111/j.1471-4159.2010.06914.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Xie W, Qu S, Pan T, Wang X, Le W. Neuroprotection by iron chelator against proteasome inhibitor-induced nigral degeneration. Biochem Biophys Res Commun. 2005;333:544–9. doi: 10.1016/j.bbrc.2005.05.150. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Finley D, Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutants. Cell. 1984;37:57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–90. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–92. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- Rideout HJ, Lang RI, Stefanis L. Involvement of macroautophagy in the dissolution of neuronal inclusions. Int J Biochem Cell Biol. 2004;36:2551–62. doi: 10.1016/j.biocel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki MR, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Masaaki K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Malagelada C, Jin ZH, Jackson LV, Przedborski S, Greene LA. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson's disease. J Neurosci. 2010;30:1166–75. doi: 10.1523/JNEUROSCI.3944-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F, Di BS, Nardacci R, Fuoco C, Corazzari M, Giunta L, et al. A novel role for autophagy in neurodevelopment. Autophagy. 2007;3:506–8. doi: 10.4161/auto.4616. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Du Y, Yang D, Li L, Luo G, Li T, Fan X, et al. An insight into the mechanistic role of p53-mediated autophagy induction in response to proteasomal inhibition-induced neurotoxicity. Autophagy. 2009;5:663–75. doi: 10.4161/auto.5.5.8377. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–63. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]