Abstract
Aim:
A growing body of evidence suggests that α-synuclein accumulation may play an important role in the pathogenesis of Parkinson's disease. The aim of this study was to investigate the roles of the proteasome and autophagy pathways in the clearance of wild-type and mutant α-synuclein in PC12 cells.
Methods:
PC12 cells overexpressing either wild-type or A30P mutant α-synuclein were treated with the proteasome inhibitor epoxomicin, the macroautophagy inhibitor 3-MA and the macroautophagy activator rapamycin alone or in combination. The cell viability was assessed using MTT assay. Immunofluorescence and Western blot analysis were used to detect the level of α-synuclein, LAMP-2A, E1 activase, and E2 ligase in the cells. Chymotrypsin-like proteasomal activity was measured using a commercial kit.
Results:
When the proteasome and macroautophagy in the wild-type and mutant cells were inhibited with epoxomicin and 3-MA, respectively, the cell viability was significantly decreased, and the α-synuclein level was increased. Both epoxomicin and 3-MA activated the chaperone-mediated autophagy (CMA) by increasing the level of the CMA-limiting enzyme LAMP-2A. Furthermore, 3-MA or epoxomicin significantly decreased chymotrypsin-like proteasomal activity. 3-MA or epoxomicin did not change E1 activase expression in either mutant or wild-type cells, but increased E2 ligase expression, especially when used together. Macroautophagy inducer rapamycin increased the cell viability and reduced epoxomicin-induced α-synuclein accumulation. Interestingly, CMA was also activated by rapamycin.
Conclusion:
Our results demonstrate the existence of complex crosstalk between different forms of autophagy and between autophagy and the proteasome pathway in the clearance of α-synuclein in PC12 cells.
Keywords: autophagy, proteasome, α-synuclein, lysosome-associated membrane protein type 2A, Parkinson's disease
Introduction
Parkinson's disease (PD) is a common degenerative disorder of the central nervous system and a condition associated with protein aggregate formation in brain tissue1. Gene mutations and structural changes of α-synuclein are etiological factors in PD, and disordered metabolism of the α-synuclein protein is also associated with PD2,3,4,5,6,7.
The ubiquitin-proteasome system (UPS) plays an important role in the degradation of abnormal intracellular proteins. These degenerate, misfolded, and abnormal accumulations are degraded by the UPS8,9. Functional impairment of the UPS causes protein accumulation, which further inhibits UPS activity and results in a vicious cycle8,9, leading to neuronal impairment or death. Wild-type and mutant α-synuclein are both degraded by the UPS, and some components of ubiquitin and the proteasome are also found in Lewy bodies10,11,12.
As a lysosome-based degradation pathway present in all eukaryotic cells, autophagy also participates in α-synuclein metabolism and PD pathology10,13. Wild-type α-synuclein is degraded mainly by chaperone-mediated autophagy (CMA). The selectivity of CMA is conferred by recognition of a pentapeptide amino acid motif in CMA substrates by a cytosolic chaperone [heat shock cognate [hsc] protein 70 kDa]14. The mutant forms of α-synuclein bind strongly to the CMA receptors on lysosomes but are not translocated into the lysosomal lumen, and this failure to translocate impairs the degradation of CMA protein substrates, including α-synuclein itself. Consequently, despite having a high affinity for the CMA receptor, pathogenic α-synuclein mutations are poorly degraded by CMA5,8. The clearance of mutant α-synuclein is strongly dependent on macroautophagy, which is generally referred to as autophagy5,10,15. Therefore, the relationship between α-synuclein and proteasome/autophagy functions, their roles in the formation of Lewy bodies, and their roles in the degeneration of dopaminergic cells are all important.
We set out to ascertain the differences between the degradation of α-synuclein by (a) the proteasomal pathway and (b) autophagy in different cell types. Furthermore, we assessed the relationship between the proteasomal pathway and autophagy.
Materials and methods
Cell culture
PC12 cells were transfected with the recombinant vectors pEGFP-C3-SNCA using liposome to construct cell lines overexpressing human wild-type and A30P α-synuclein. Transfected PC12 cells were further screened with the G418 aminoglycoside antibiotic selection agent (Mediatech, Olando, FL, USA) and were obtained by a limiting dilution assay. Stably transfected A30P and wild-type α-synuclein PC12 cell lines were grown in RPMI-1640 (GIBCO, USA) containing 10% calf serum and stored at 37 °C in 5% CO2 (details in Ref 15)16.
Treatment with autophagy/proteasome drugs
After adherence, cells were cultured in either control medium (GIBCO, USA) or medium that contained 10 mmol/L 3-methyladenine (3-MA, Sigma, USA) or 0.2 μg/mL rapamycin (Sigma, St Louis, USA) or 100 nmol/L epoxomicin (Sigma, USA). As described previously, 3-MA was dissolved in water, and rapamycin and epoxomicin were solvated in DMSO.
MTT assay
Six wells were prepared for each group in parallel. Cells were plated into 96-well plates (1×104 cells per well) in 100 μL complete culture medium. After overnight culture, the medium was replaced with either drug-free medium or drugs dissolved in complete medium. The cells were cultured for the times indicated, and then the MTT solution (20 μL) was added. Six hours later, the supernatant was discarded and DMSO (150 μL/well) was added. Optical density was read using a multiwell scanning spectrophotometer (ELISA reader).
Immunofluorescence
Cells were seeded on coverslips placed in 24-well dishes, then incubated with autophagy/proteasome drugs for 24 h. Cells were fixed with 4% paraformaldehyde (Sigma) for 15 min, washed with PBS, and permeabilized with 0.1% Triton-X-100 (Sigma) for 15 min. Next, the cells were incubated with an anti-α-synuclein antibody (Sigma; diluted 1:250) for 2 h, washed, and then incubated for 1 h with FITC-conjugated anti-rabbit secondary antibodies (Beyotime Institute of Biotechnology, Haimen, China; diluted 1:500). Finally, slides were mounted in Citifluor (Citifluor Ltd, London, UK) with 3 μg/mL 4′,6-diamidino-2-phenylindole (DAPI, Sigma, St Louis, USA). Cells were visualized using a LEICA TCS ST2 confocal microscope.
Western blot analysis
Cells were seeded at 1.5×108 cells/mL, treated with different drugs as described above, and cultured for 24 h. Cell pellets were collected and lysed in 1×lysis buffer before electrophoresis on SDS-PAGE and transfer to nitrocellulose membrane. The blots were probed with anti-β-actin (Santa Cruz Biotechnology, Santa Cruz, USA), anti-α-synuclein (Sigma), and anti-Lamp-2a (Abcam, Cambridge, UK) antibodies, as well as antibodies against the E1 and E2 enzymes; and signals were detected using an enhanced chemiluminescence system (Pierce, Rockford, lL, USA) or the TMB stabilized substrate for HRP (Promega, Madison, WI, USA). Membrane-bound proteins from three independent experiments were quantitatively analyzed with an ImageJ software analyzer system.
Chymotrypsin-like proteasomal activity assays
Cells were cultured in 96-well plates, and drug treatment was performed as above. At 24 h after drug treatment, cells were measured with a chymotrypsin-like proteasomal activity determination kit (Promega, Madison, WI, USA) with a multisection microplate reader according to the manufacturer's instructions.
Statistical analysis
Normally distributed data are shown as the mean±SD and were analyzed by one-way ANOVA using SPSS (Statistical Product and Service Solutions) 12.0 statistical software. Multiple comparisons among groups were performed using SNK post hoc analysis.
Results
Effects of the proteasome pathway or autophagy on cell viability
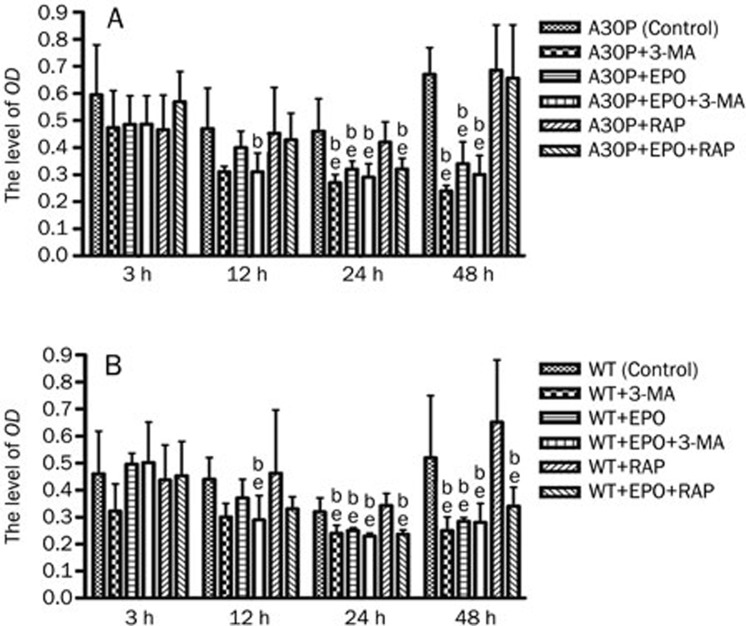
We measured the viability of A30P and wild-type (WT) cells at 3, 12, 24, and 48 h in culture. Cell viability decreased 24 h after treatment with the macroautophagy inhibitor 3-MA and/or the proteasome inhibitor epoxomicin; 3-MA alone was the most effective (Figure 1). Cell viability was significantly greater in 24 h cultures treated with the macroautophagy stimulator rapamycin than that in any other group (Figure 1). Rapamycin also improved A30P cell viability, which decreased in response to epoxomicin treatment.
Figure 1.
Analysis of cell proliferation after drug treatments. (A) Stably transfected A30P α-synuclein PC12 cells were treated with epoxomicin and/or 3-MA and rapamycin for 3, 12, 24, or 48 h before assessing cell proliferation by MTT assay. (B) PC12 cells expressing wild-type α-synuclein were also assessed after treatment. EPO, epoxomicin; RAP, rapamycin. Mean±SD. bP<0.05 vs control. eP<0.05 vs RAP.
Effects of inhibition of the proteasome or autophagy on α-synuclein accumulation
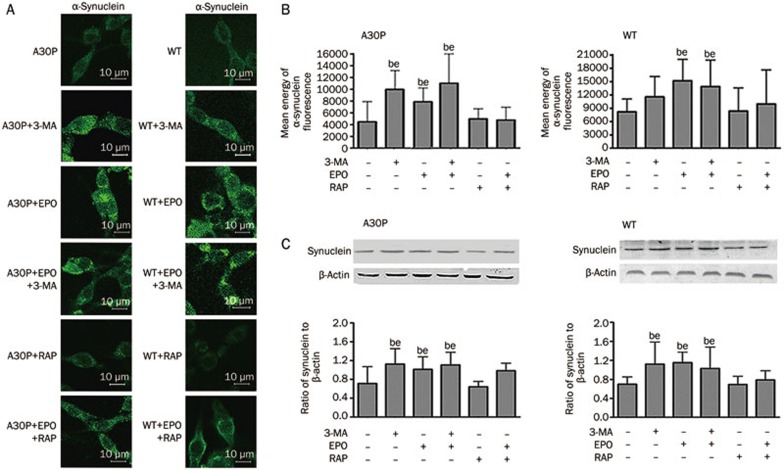
Conflicting results have emerged from previous studies of the effects of proteasomal inhibition on α-synuclein metabolism. When we incubated cells with the proteasome inhibitor epoxomicin and/or the macroautophagy inhibitor 3-MA for 24 h, we found an increase in α-synuclein levels in both wild-type and mutant cell lines. Western blot analysis showed that cells incubated with a combination of the proteasome inhibitor epoxomicin and the autophagy activator rapamycin had less α-synuclein accumulation than cells exposed to epoxomicin alone (Figure 2C). Furthermore, confocal microscopy images showed an accumulation of α-synuclein in cells treated either with epoxomicin alone or with epoxomicin in combination with 3-MA (Figure 2A, 2B).
Figure 2.
(A) Cells were imaged by confocal microscopy. A30P and wild-type cells treated with the indicated drugs were permeabilized and immunostained with anti-α-synuclein followed by FITC-labeled secondary antibodies for imaging. (B) The immunofluorescence mean energy of cells was analyzed. At least 50 cells in each group were randomly selected using the same settings. The data were normally distributed and statistically analyzed using one-way ANOVA. (C) Western blot analysis of total exogenous human α-synuclein in A30P and wild-type PC12 cell lines. Mean±SD. bP<0.05 vs control. eP<0.05 vs RAP.
Activation of CMA in cells with inhibition of the proteasome or macroautophagy
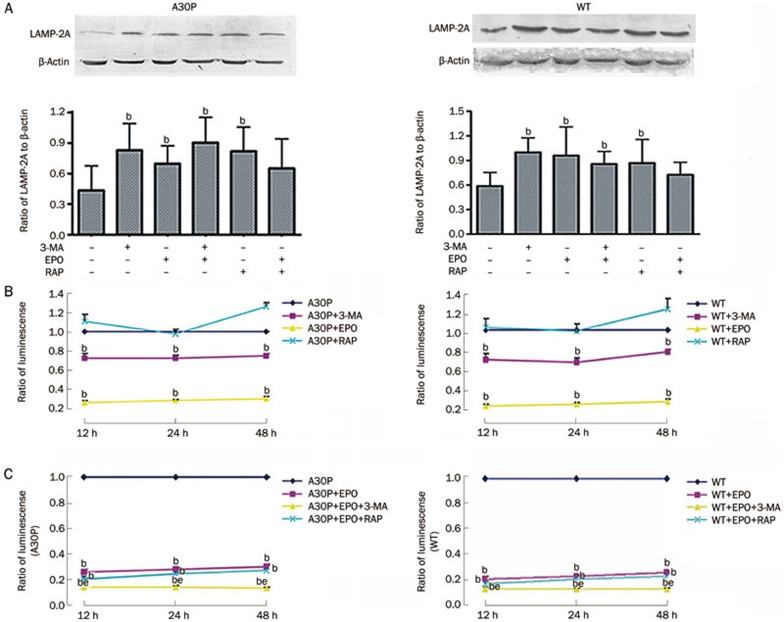
Binding of substrate proteins to lysosome-associated membrane protein type 2A (LAMP-2A) is a limiting step for CMA. Levels of LAMP-2A at the lysosomal membrane are tightly controlled and constitute a regulatory mechanism for CMA16,17. So, we investigated LAMP-2A levels by immunoblotting and confirmed that the increase in LAMP-2A was related to the inhibition of either the proteasome or macroautophagy (Figure 3A). Interestingly, compared with the control, cells grown in medium with rapamycin also had an increased LAMP-2A/actin ratio (Figure 3A).
Figure 3.
The crosstalk among different autophagy types and between autophagy and the proteasome pathway. (A) LAMP-2A was determined by immunoblotting the same cells used in Figure 2. bP<0.05 vs control. (B) Chymotrypsin-like proteasomal activity following 3-MA, epoxomicin (EPO) or rapamycin (RAP) treatment for the indicated times in A30P and wild-type cells (ratio of chymotrypsin-like proteasomal activity between cells incubated in experimental vs control medium [A30P or wild-type]). (C) Chymotrypsin-like proteasomal activity after EPO, EPO+3-MA, or EPO+RAP treatment. Mean±SD. bP<0.05 vs control. eP<0.05 vs EPO.
Macroautophagy inhibitor 3-MA decreased chymotrypsin-like proteasomal activity
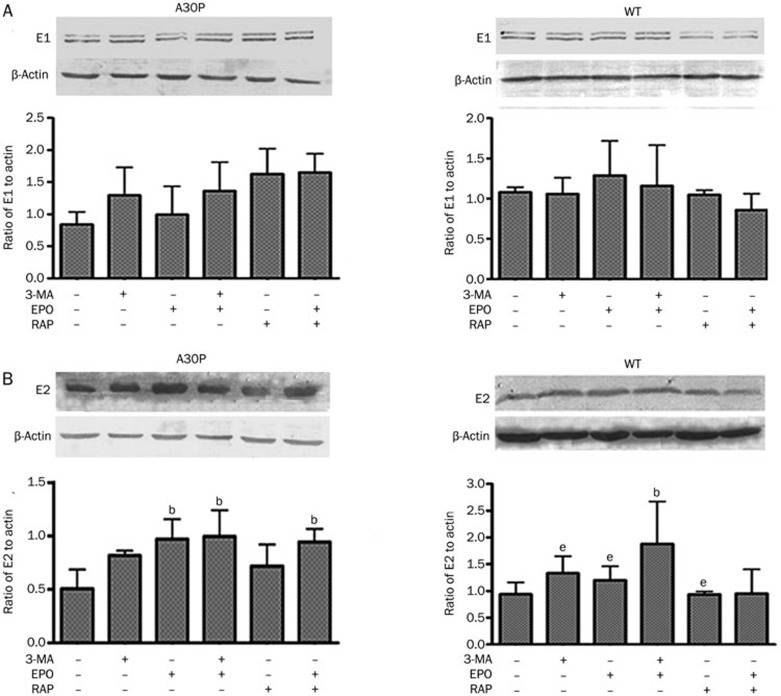
The macroautophagy inhibitor 3-MA decreased chymotrypsin-like proteasomal activity compared with the control, and rapamycin tended to increase chymotrypsin-like proteasomal activity (Figure 3B). To further verify the effects of macroautophagy inhibition or stimulation on the proteasome pathway, we combined epoxomicin with 3-MA or rapamycin. Again, proteasomal activity was significantly lower in 3-MA together with epoxomicin than in epoxomicin alone (Figure 3C). Ubiquitin-like processes of the proteasome pathway also require E1-like and E2-like enzymes18. We analyzed E1 activase and E2 ligase expression in the proteasome pathway by Western blot. E1 enzyme expression did not change in either mutant or wild-type cells (Figure 4A). 3-MA and epoxomicin increased E2 enzyme expression, especially when used together (Figure 4B). We hypothesized that the E2 enzyme is likely to be one of the interaction points between autophagy and the proteasome pathway. Thus, the macroautophagy inhibitor 3-MA inhibits chymotrypsin-like proteasomal activity.
Figure 4.
(A) Western blot analysis of E1 enzyme in PC12 cell lines overexpressing A30P or wild-type α-synuclein. (B) E2 enzyme expression following indicated drug treatments (24 h). Mean±SD. bP<0.05 vs control. eP<0.05 vs EPO+3-MA.
Discussion
PD is a common neurodegenerative disease affecting old population and has brought a heavy burden to family and society. So far, there is no cure for PD, although some neuroprotective treatment and replacement therapy can provide some relief from its symptoms. It is possible that there's key targets in regulating autophagic degration of α-synuclein and the drugs activating autophagy may become promising treatments for PD.
Our studies have revealed that inhibiting the macroautophagy and proteasome pathways decreased cell viability of PC12 cells overexpressing either A30P mutant or wild-type α-synuclein, while stimulating macroautophagy increased cell viability. Previous studies indicated that cells with impaired CMA but compensatory activation of macroautophagy can accommodate nutritional stress without changing their viability19,20. However, activation of macroautophagy in cells with proteasome inhibition10,21 may not be enough to maintain viability under normal conditions. Our results indicated that macroautophagy inducer rapamycin could not relieve the decrease in cell viability caused by the proteasome inhibitor epoxomicin in PC12 cells overexpressing A30P mutant and wild-type α-synuclein. As demonstrated by the highly effective impact of 3-MA alone, macroautophagy does play a role in maintaining cell viability; however, it alone is insufficient for viability.
Some groups have reported that proteasomal inhibition causes an accumulation of α-synuclein, inclusion formation, and increased cell death12,22,23,24, whereas others have suggested that α-synuclein is not a proteasome substrate25,26. The effects of proteasomal inhibition on α-synuclein metabolism have been investigated in a number of studies. Our studies have suggested that inhibiting the proteasome pathway and macroautophagy decreased the clearance of α-synuclein. Now it has been widely accepted that wild-type α-synuclein containing five peptide motif KFERQ was degradated by both CMA and macroautophagy, while A30P mutant α-synuclein was only degradated by macroautophagy because of lack the specific five peptide motif. Although there was no statistical significance, our data showed that macroautophagy inhibitors did induce relatively higher levels of mutant α-synuclein than the proteasome inhibitor. However, we found the opposite result in PC12 cells overexpressing wild-type α-synuclein. This finding provided evidence to the view that mutant α-synuclein is mainly cleared by macroautophagy, whereas wild-type α-synuclein is mainly degraded by the CMA pathway5,10.
It is interesting that the combination of 3-MA and epoxomicin did not produce greater cell death and more α-synuclein accumulation than either 3-MA or epoxomicin alone. Many recent studies suggest that compensatory activation of macroautophagy is induced by proteasomal inhibition10,21. Our previous results27 also support this idea. The compensatory activation of macroautophagy caused by proteasomal inhibition may help in part to weaken the effects of the combination of 3-MA and epoxomicin in cell viability and α-synuclein clearance.
In order to understand the crosstalk between the autophagy and the proteasome pathway involved in the degradation of α-synuclein, we detected the proteasome activity (chymotrypsin-like proteasomal activity) and the level of the CMA-limiting enzyme LAMP-2A after intervening macroautophagy and the proteasome pathway. We found that inhibiting the proteasome pathway enhanced expression of LAMP-2A. This finding may be explained by the fact that proteasomal inhibition induces oxidative stress and abnormal protein accumulation, and CMA is activated as a compensation strategy during mild oxidative stress and exposure to toxic compounds28. In addition, we found that macroautophagy inhibition reduced proteasomal activity and increased LAMP-2A. This may be because inhibiting macroautophagy results in α-synuclein accumulation, which further inhibits UPS activity, and then increases the level of LAMP-2A. This may be another reason that macroautophagy inhibition leads to cell injury and abnormal protein accumulation. It is noted that although LAMP-2A increased after macroautophagy and/or proteasome inhibitor treatment, A30P and wild-type α-synuclein still accumulated in the cytoplasm. These results indicate that the compensatory activation of CMA in PC12 cells overexpressing wild-type α-synuclein could not rescue wild-type α-synuclein accumulation caused by a macroautophagy and/or proteasome inhibitor and that the compensatory activation of CMA in PC12 cells overexpressing A30P α-synuclein has no effect on clearance of A30P α-synuclein.
Previous studies revealed that the macroautophagy inducer rapamycin promoted the clearance of all forms of α-synuclein10,12,15. In our study, rapamycin decreased the cell death and α-synuclein accumulation caused by proteasome impairment. Interestingly, rapamycin also upregulated the level of LAMP-2A, and LAMP-2A increased in line with the increase of macroautophagy. The results suggest that rapamycin also activates CMA accompanied with the induction of macroautophagy. Macroautophagy and CMA probably share the same regulatory pathway which could be regulated by rapamycin. This may be another mechanism by which rapamycin decreases the accumulation of α-synuclein and so has therapeutic potential for PD.
Furthermore, our results indicated that the expression of E2 ligase changed with inhibition of the proteasome and macrophagy and that E2 ligase may be one of the interaction points between autophagy and the proteasome pathway. However, further studies are required to assess whether there is an intersection between autophagy and UPS and, if so, the nature of the mechanisms involved.
In summary, our results demonstrate the existence of complex crosstalk between different forms of autophagy and between autophagy and the proteasome pathway in the clearance of α-synuclein in PC12 cells. Further studies are required to explore the exact mechanisms of their interactions. Rapamycin is a potential therapeutic agent for the treatment of PD. Its mechanisms are likely to include upregulating macroautophagy and upregulating CMA.
Author contribution
Prof Chun-feng LIU designed the research; Fang YANG and Ya-ping YANG performed the research; Cheng-jie MAO, Ling LIU, and Hui-fen ZHENG analyzed the data; Fang YANG and Ya-ping YANG wrote the paper; and Prof Li-fang HU and Chun-feng LIU revised the paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No 81171213) and the Natural Science Foundation of the Jiangsu Province of China (No BK2010228). We are grateful to Professor IC Bruce for critical reading of the manuscript.
References
- Ozansoy M, Basak AN. The central theme of Parkinson's disease: alpha-synuclein. Mol Neurobiol. 2013;47:460–52. doi: 10.1007/s12035-012-8369-3. [DOI] [PubMed] [Google Scholar]
- Kahle PJ, Haass C, Kretzschmar HA, Neumann M. Structure/function of alpha-synuclein in health and disease: rational development of animal models for Parkinson's and related diseases. J Neurochem. 2002;82:449–57. doi: 10.1046/j.1471-4159.2002.01020.x. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–7. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, et al. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson's disease. Nat Genet. 1998;18:106–8. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–5. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- Feany MB, Bender WW. A Drosophila model of Parkinson's disease. Nature. 2000;404:394–8. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- Masliah E, Rockenstein E, Veinbergs I, Mallory M, Hashimoto M, Takeda A, et al. Dopaminergic loss and inclusion body formation in alpha-synuclein mice: implications for neurodegenerative disorders. Science. 2000;287:1265–9. doi: 10.1126/science.287.5456.1265. [DOI] [PubMed] [Google Scholar]
- Chung KK, Dawson VL, Dawson TM. The role of the ubiquitin-proteasomal pathway in Parkinson's disease and other neurodegenerative disorders. Trends Neurosci. 2001;24:S7–14. doi: 10.1016/s0166-2236(00)01998-6. [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–5. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–13. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- Li K, Ito H, Tanaka K, Hirano A. Immunocytochemical co-localization of the proteasome in ubiquitinated structures in neurodegenerative diseases and the elderly. J Neuropathol Exp Neurol. 1997;56:125–31. doi: 10.1097/00005072-199702000-00002. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–17. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Engelender S. alpha-Synuclein fate: proteasome or autophagy. Autophagy. 2012;8:418–20. doi: 10.4161/auto.19085. [DOI] [PubMed] [Google Scholar]
- Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–9. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–42. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- Qian JJ, Cheng YB, Yang YP, Mao CJ, Qin ZH, Li K, et al. Differential effects of overexpression of wild-type and mutant human alpha-synuclein on MPP+-induced neurotoxicity in PC12 cells. Neurosci Lett. 2008;435:142–6. doi: 10.1016/j.neulet.2008.02.021. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–18. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Polo RA, Boya P, Pauleau AL, Jalil A, Larochette N, Souquere S, et al. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci. 2005;118:3091–102. doi: 10.1242/jcs.02447. [DOI] [PubMed] [Google Scholar]
- Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2006;103:5805–10. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X, Song W, Zhang W, Chen L, Xi Z, Xin Z, et al. Mitochondrially localized EGFR is subjected to autophagic regulation and implicated in cell survival. Autophagy. 2008;4:641–9. doi: 10.4161/auto.5971. [DOI] [PubMed] [Google Scholar]
- Bennett MC, Bishop JF, Leng Y, Chock PB, Chase TN, Mouradian MM. Degradation of alpha-synuclein by proteasome. J Biol Chem. 1999;274:33855–8. doi: 10.1074/jbc.274.48.33855. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Mytilineou C, Jnobaptiste R, Yabut J, Shashidharan P, Jennert P, et al. Impairment of the ubiquitin-proteasome system causes dopaminergic cell death and inclusion body formation in ventral mesencephalic cultures. J Neurochem. 2002;81:301–6. doi: 10.1046/j.1471-4159.2002.00821.x. [DOI] [PubMed] [Google Scholar]
- Tofaris GK, Layfield R, Spillantini MG. Alpha-synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Lett. 2001;509:22–6. doi: 10.1016/s0014-5793(01)03115-5. [DOI] [PubMed] [Google Scholar]
- Rideout HJ, Larsen KE, Sulzer D, Stefanis L. Proteasomal inhibition leads to formation of ubiquitin/alpha-synuclein-immunoreactive inclusions in PC12 cells. J Neurochem. 2001;78:899–908. doi: 10.1046/j.1471-4159.2001.00474.x. [DOI] [PubMed] [Google Scholar]
- Ancolio K, Alves da Costa C, Ueda K, Checler F. Alpha-synuclein and the Parkinson's disease-related mutant Ala53Thr-alpha-synuclein do not undergo proteasomal degradation in HEK293 and neuronal cells. Neurosci Lett. 2000;285:79–82. doi: 10.1016/s0304-3940(00)01049-1. [DOI] [PubMed] [Google Scholar]
- Yang F, Yang YP, Mao CJ, Cao BY, Cai ZL, Shi JJ, et al. Role of autophagy and proteasome degradation pathways in apoptosis of PC12 cells overexpressing human alpha-synuclein. Neurosci Lett. 2009;454:203–8. doi: 10.1016/j.neulet.2009.03.027. [DOI] [PubMed] [Google Scholar]
- Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–40. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]