Abstract
Melanoma differentiation-associated gene/interleukin-24 (mda-7/IL-24) is a cytokine that can activate monocytes and T helper 2 cells. The expression of mda-7/IL-24 gradually fades with the progression of melanoma, and it is undetectable at the metastatic stage. Ectopic expression of mda-7/IL-24 selectively suppresses growth and induces apoptosis in cancer cells with little harm to normal cells. However, the transcriptional regulation of the mda-7/IL-24 gene has not been extensively studied. In this study, we show that the expression of mda-7/IL-24 was upregulated by the histone deacetylase (HDAC) inhibitors trichostatin A (TSA) and sodium butyrate (NaBu), whereas it was downregulated by HDAC4. We also found that the histone acetylation level and the binding of the transcriptional factor Sp1 to the mad-7 promoter were reduced upon HDAC4 treatment. Moreover, the HDAC inhibitor TSA induced histone hyperacetylation and stimulated Sp1 binding to the mda-7/IL-24 promoter, which in turn enhanced the expression of mda-7/IL-24. Therefore, we conclude that histone acetylation modification plays an important role in the regulation of mda-7/IL-24 and that the transcription factor Sp1 participates in this process.
Keywords: HDAC4, HDAC inhibitors, histone acetylation, mda-7/IL-24, Sp1
Introduction
Melanoma differentiation-associated gene/interleukin-24 (mda-7/IL-24) was first identified as a cytokine gene that was induced by phytohemagglutinin or lipopolysaccharide in human peripheral blood mononuclear cells.1 A number of cytokine target genes have been shown to be upregulated by the adenovirus-mediated ectopic expression of mda-7/IL-24 (Ad.mda-7/IL-24); these genes include MCSF-1, CXC3, IFP53, PML and MyD88.2 There have been indications suggesting that mda-7/IL-24 can activate the interferon (IFN)-γ and NF-κB signaling pathways. The mda-7/IL-24 gene maps to the genomic cluster of IL-10-related genes including IL-10, IL-19, IL-20, IL-24 and IL-26,3 and mda-7/IL-24 was demonstrated to be a secreted glycosylated protein with 49 amino acid-long signal peptide that masked its cytokine identity. Treating peripheral blood mononuclear cells with purified secreted mda-7/IL-24 protein potently induced the secretion of IFN-γ, IL-6, tumor-necrosis factor-α, IL-1β and granulocyte/macrophage colony-stimulating factor, indicating that mda-7/IL-24 functions as a pro-Th1 cytokine. The cytokine activity of mda-7/IL-24 was inhibited by co-administration of IL-10.1 Ad-mda7 increased IL-6 and IFN-γ mRNA expression and induced the secretion of IFN-γ and IL-6 from melanoma cells.2 Microarray analyses of non-small cell lung cancer cells treated with Ad.mda7 further validated the cytokine-like activity of mda-7/IL-24. These data indicated that mda-7/IL-24 is part of a previously unrecognized immune-regulatory loop.
mda-7/IL-24 has also been identified as a tumor suppressor gene that is constitutively expressed in normal melanocytes2, 4, 5 to maintain the normal physiological function of cells and to induce terminal differentiation. The expression of mda-7/IL-24 gradually fades with the progression of the melanoma and is undetectable at the metastatic stage.4 It has been reported that ectopic expression of mda-7/IL-24 using an adenovirus vector (Ad.mda-7/IL-24) can selectively induce apoptosis in a broad range of tumor cells with little side effects in normal cells.4, 6, 7, 8 Furthermore, there is evidence that the absence of mda-7/IL-24 expression in cancer cells was not a result of an inactivating mutation in the gene, because no structural rearrangements were observed in a spectrum of cancer and normal cells.8, 9 The fact that mda-7/IL-24 can be induced in cancer cells5 suggests that there may be other regulatory mechanisms that govern the expression of mda-7/IL-24. Presumably, epigenetic changes such as histone-acetylation modification may play a role in mda-7/IL-24 regulation; however, this has not been studied.
The purpose of this study was to clarify whether histone acetylation is an epigenetic mechanism that participates in the transcriptional regulation of the mda-7/IL-24 gene. We demonstrated that the histone deacetylase (HDAC) inhibitors trichostatin A (TSA) and sodium butyrate (NaBu) were able to induce mda-7/IL-24 expression, whereas HDAC4 decreased the expression of mda-7/IL-24. We also investigated some of the molecular basis underlying this regulatory process.
Materials and methods
Plasmids
The luciferase reporter plasmid containing the mda-7/IL-24 promoter was obtained by subcloning the 1133-bp genomic DNA region upstream of the human mda-7/IL-24 gene into the pGL-3enhancer-luc vector using the primers 5′-GGA AGA TCT AGG TTA AGC CAT TCT CAG-3′ and 5′-CCC AAG CTT AGC CGT GGA AGT CAT T-3′. The expression plasmids for human HDACs were the gifts from Dr W. C. Greene (University of California, San Francisco, CA, USA). The Sp1 expression construct was a gift from Dr Robert Tjian (Department of Molecular and Cell Biology, University of California, Berkeley, CA, USA).
Cell culture
The A375 human malignant melanoma cell line was purchased from the Institute of Cell Biology, Shanghai, China. Cells were cultured in IMDM medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin, and the cells were kept in a humidified atmosphere of 5% CO2 at 37 °C.
Transient transfection and luciferase reporter assay
For transient transfection, cells were seeded in 24-well plates and were cultured for 18 h before being transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After transfection, cells were cultured for 24 h before harvesting, and after the cells were harvested, they were washed with PBS and then lysed in 30 µl lysis buffer. Reporter gene expression was measured and quantified using a dual luciferase reporter assay system (Promega, Madison, WI, USA). Relative luciferase activity was analyzed using a TD20/20 luminometer (Turner Designs, Sunnyvale, CA, USA). Firefly luciferase activity was normalized to the activity of the Renilla luciferase control. Extracts from at least three independent transfection experiments were assayed in triplicate. The results are shown as mean±SD.
RNA isolation and quantitative real-time PCR
Total RNA was extracted following the TaKaRa RNAiso Reagent manual and was reverse transcribed into cDNA using the ImProm-II Reverse Transcription System (A3802) supplied by Promega. The resulting cDNA was diluted fivefold with RNase-free water. Quantitative real-time PCR was performed on a Roche LightCycler System 480 using SYBR Green (TOYOBO, Osaka, Japan) as a double-strand DNA-specific fluorescent dye. β-actin was used to standardize the amount of mda-7/IL-24 mRNA. The amplification primers for the mda-7/IL-24 gene were 5′-GGCGGTTTCTGCTATTCC-3′ and 5′-AAGGGCGTGAAGTGTCCAG-3′. Data were analyzed using the 2−ΔΔCt method.10 All of the results represent the mean±SD of three independent experiments.
Western blotting
A375 cells were treated with 100 nM TSA or 1 mM NaBu, incubated for 24 h, and then lysed in lysis buffer (50 mM Tris-HCl, 1% Nonidet P-40, 150 mM NaCl, 1 mM EDTA and 1 mM PMSF). Cell lysates were separated by SDS–polyacrylamide gel electrophoresis in 15% gels, and then the proteins were transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA) and subjected to western blotting analysis with a rabbit polyclonal antibody against mda-7/IL-24 (Imgenex, San Diego, CA, USA) or a mouse monoclonal antibody against β-actin (Sigma, St Louis, MO, USA). The signals were visualized by using the chemiluminescent substrate method with the SuperSignal West Pico kit provided by Pierce Co. (Rockford, IL, USA).
Chromatin immunoprecipitation (ChIP)
ChIP assays were carried out using a kit supplied by Upstate Biotechnology (Lake Placid, NY, USA) following the manufacturer's protocol. Cells were plated at a density of 1×105 cells/ml in six-well plates and were cultured for 24 h. Cells were crosslinked in 2% formaldehyde for 10 min at 37 °C after transfection and were then lysed in SDS lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris, pH 8.1) containing protease inhibitors. The sonicated lysates were processed using a ChIP assay kit essentially as described by the manufacturer (Upstate Biotechnology). Antibodies against Ac-H3 and Ac-H4 were purchased from Upstate Biotechnology. An antibody against Sp1 was purchased from Santa Cruz. Immunoprecipitated chromatin was analyzed by quantitative PCR (ABI Prism 7000 Sequence Detection System Instrument) using SYBR green dye with primers specific to sequences at the mda-7/IL-24 promoter.
Statistical analysis
A Student's t-test was used to calculate the statistical significance of the experimental data. The level of significance was set at *P<0.05 and **P<0.01.
Results
HDAC inhibitors enhance the expression of mda-7/IL-24
To investigate whether histone-acetylation modification plays a role in regulation of mda-7/IL-24 expression, we treated A375 human malignant melanoma cells with the HDAC inhibitors TSA and NaBu. As shown in Figure 1a, the expression of mda-7/IL-24 mRNA was induced by treatment with TSA and NaBu. To further assess the effects of these HDAC inhibitors, we constructed a luciferase reporter plasmid containing the mda-7/IL-24 promoter for use in a reporter assay, and the results showed that the mda-7/IL-24 promoter was effective (Figure 1b). Moreover, TSA and NaBu increased the activity of the mda-7/IL-24 promoter-controlled luciferase reporter gene (Figure 1c). The results shown in Figure 1d demonstrate that the mda-7/IL-24 protein level was also upregulated upon TSA treatment. These results indicate that HDAC inhibitors were able to increase mda-7/IL-24 expression, suggesting that histone acetylation is a regulatory mechanism involved in mda-7/IL-24 gene expression.
Figure 1.
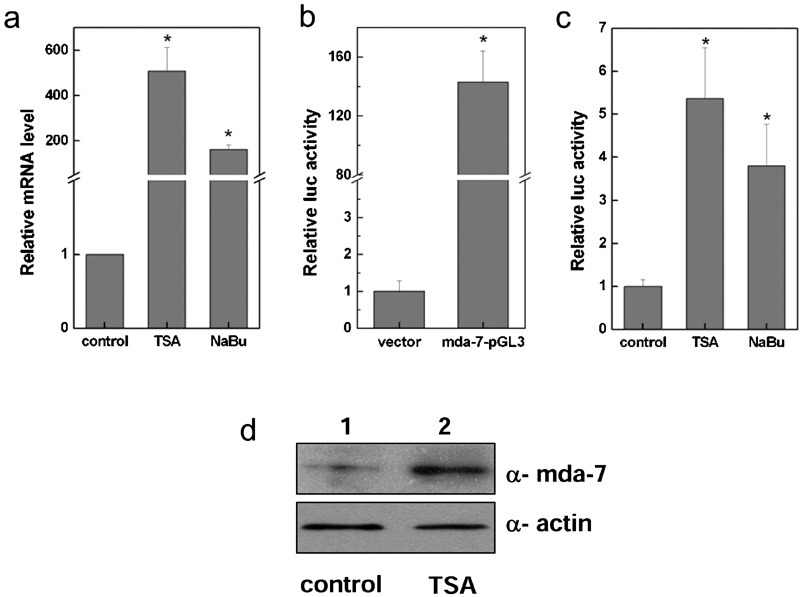
HDAC inhibitors enhanced the expression of mda-7/IL-24. A375 human malignant melanoma cells were treated with the HDAC inhibitors TSA and NaBu, and the expression of mda-7/IL-24 was examined by quantitative real-time PCR (a). A375 cells were transiently transfected for 24 h with a control vector (pGL3-enhancer) or with the mda-7/IL-24 promoter construct mda-7-pGL3 (mda-7/IL-24-pGL3-enhancer), and the expression of mda-7/IL-24 was measured using a reporter activity assay (b). A375 cells were treated with the HDAC inhibitors TSA and NaBu after transient transfection with the mda-7/IL-24 promoter, and the reporter activity was examined (c). A375 cells were collected 24 h after treatment with TSA, and the expression of mda-7/IL-24 was examined by western blotting. The protein bands were analyzed using Quantity One software (d). HDAC, histone deacetylase; mda-7/IL-24, melanoma differentiation-associated gene/interleukin-24; NaBu, sodium butyrate; TSA, trichostatin A.
Sp1 induced the expression of mda-7/IL-24, which was declined by HDAC4
To further explore the role of histone acetylation in mda-7/IL-24 regulation, we next intended to address the question of whether HDACs can inhibit mda-7/IL-24 expression. We co-transfected A375 cells with the mda-7/IL-24 promoter-luciferase reporter plasmid and with the six human HDACs (HDAC1–6), and we measured the luciferase activity assay. As can be seen in Figure 2a, among the six HDACs tested, HDAC4 was able to inhibit mda-7/IL-24 promoter activity, providing further evidence that HDAC inhibitors such as TSA may act on mda-7/IL-24 specifically by inhibiting HDAC4. There has been evidence that HDAC4 represses the p21 gene in cancer cells in an Sp1-dependent manner.11 Our results demonstrated that the transcription factor Sp1 upregulated the expression of mda-7/IL-24 (Figure 2b). To evaluate the effects of HDAC4 and Sp1 on mda-7/IL-24 expression, we co-transfected A375 cells with HDAC4 and Sp1 expression plasmids either alone or in combination, and the results in Figure 2c and d show that mda-7/IL-24 was significantly upregulated by Sp1, whereas co-transfection of Sp1 with HDAC4 decreased the Sp1-induced mda-7/IL-24 expression. These results indicate that the transcription factor Sp1 positively regulated mda-7/IL-24 expression, while HDAC4 inhibited mda-7/IL-24 activation.
Figure 2.
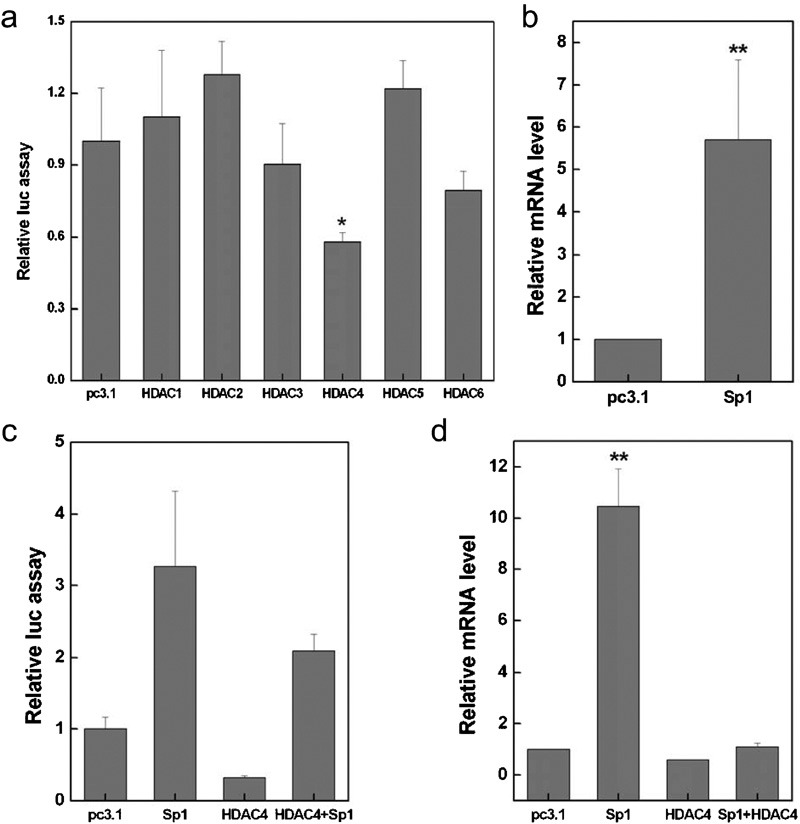
Sp1 induced mda-7/IL-24 expression and HDAC4 repressed mda-7/IL-24 expression. A375 cells were transiently transfected with pc3.1 (pcDNA3.1) or different HDAC constructs (HDACs-pcDNA3.1), and mda-7/IL-24 expression was evaluated using a reporter activity assay (a). A375 cells were transiently transfected with pc3.1 (pcDNA3.1) or with a Sp1 (Sp1-pcDNA3.1) expression plasmid, and quantitative real-time PCR was used to examine the expression of mda-7/IL-24 (b). A375 cells were co-transfected with Sp1 (Sp1-pcDNA3.1) and HDAC4 (HDAC4-pcDNA3.1) expression vectors, and mda-7/IL-24 expression was measured using reporter activity assay (c) and quantitative real-time PCR (d). HDAC, histone deacetylase; mda-7/IL-24, melanoma differentiation-associated gene/interleukin-24.
To further investigate the mechanism by which HDAC4 decreases the expression of mda-7/IL-24, we designed two primers to bind to the Sp1-binding regions in the mda-7/IL-24 promoter (Figure 3a) for ChIP assays. A375 cells were collected after transfection with pc3.1-vector or the Sp1 expression plasmid, and ChIP assays were performed using the antibody against Sp1. The ChIP results shown in Figure 3b revealed that Sp1 was enriched at the mda-7/IL-24 promoter upon Sp1 overexpression, while the binding of Sp1 was prominently reduced by HDAC co-transfection (Figure 3c). Moreover, the data in Figure 4 show that the acetylation levels of both histones H3 and H4 were decreased by HDAC4. The HDAC4 participated in this regulatory process by deacetylating histones at the mda-7/IL-24 promoter to form a closed chromatin structure, thus inhibiting the binding of the transcription factor Sp1.
Figure 3.
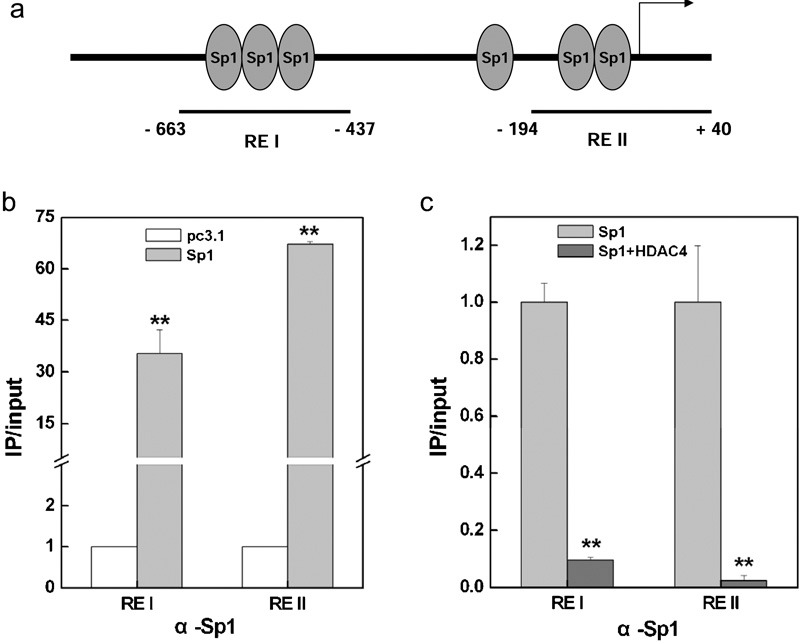
The binding of Sp1 at the mda-7/IL-24 promoter was inhibited by HDAC4. (a) Diagram showing the positions of the Sp1 binding sites at the mda-7/IL-24 promoter. The regions amplified by the PCR primers in the chromatin immunoprecipitation assay are indicated. ChIP and quantitative real-time PCR were used to evaluate the binding of Sp1 to the mda-7/IL-24 promoter after transfection with Sp1 expression plasmids (b). A375 cells were collected and precipitated with antibodies against Sp1 after transfection with HDAC4 expression plasmids, and ChIP assays were performed to examine the binding of Sp1 to the mda-7/IL-24 promoter (c). HDCA, histone deacetylase; ChIP, chromatin immunoprecipitation; mda-7/IL-24, melanoma differentiation-associated gene/interleukin-24.
Figure 4.
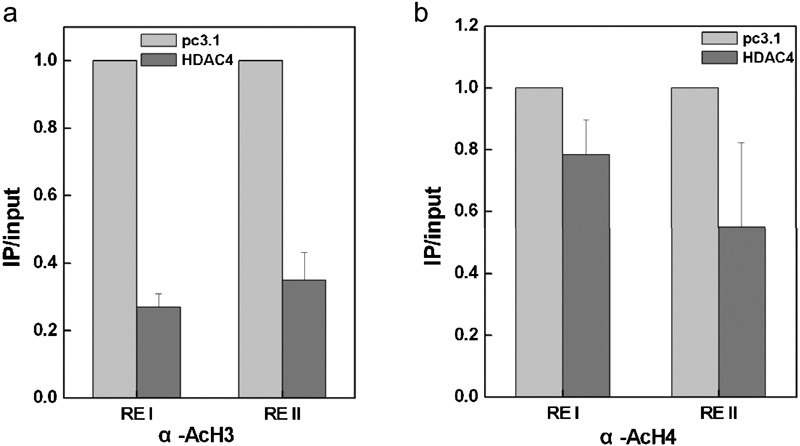
HDAC4 decreased the histone acetylation level at the mda-7/IL-24 promoter. A375 cells were transiently transfected with an expression plasmid for HDAC4 (HDAC4-pcDNA3.1), and the acetylation levels of histone H3 (a) and histone H4 (b) were examined by ChIP and quantitative real-time PCR. ChIP, chromatin immunoprecipitation; HDAC, histone deacetylase; mda-7/IL-24, melanoma differentiation-associated gene/interleukin-24.
The HDAC inhibitor TSA enhances the transcriptional activity of the mda-7/IL-24 promoter
The data presented above indicated that the HDAC inhibitors TSA and NaBu were able to induce mda-7/IL-24 expression, and Sp1 was a transcription factor for mda-7/IL-24 activation. We then asked the question of whether TSA affects the function of Sp1 in mda-7/IL-24 regulation. As shown in Figure 5a and b, TSA markedly upregulated mda-7/IL-24 expression by over 60-fold upon the Sp1 overexpression. Additionally, our ChIP assays revealed that TSA notably increased the acetylation level of histone H3 by fourfold at the RE I region and by 28-fold at the RE II region of the mda-7/IL-24 promoter (Figure 6a). Similarly, TSA enhanced the acetylation of histone H4 by 11-fold at the RE I region and by 43-fold at the RE II region of the mda-7/IL-24 promoter (Figure 6b). Furthermore, our data revealed that the binding efficiency of Sp1 on the mda-7/IL-24 promoter was prominently increased by treatment of the cells with TSA (Figure 6c). We concluded that HDAC inhibitors such as TSA can increase the histone acetylation level of the mda-7/IL-24 promoter, facilitating the binding of the transcription factor Sp1 to stimulate the transcription of the mda-7/IL-24 gene.
Figure 5.
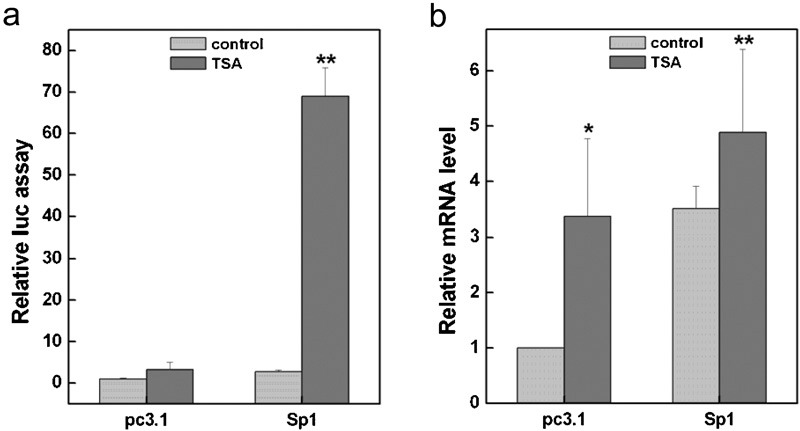
The HDAC inhibitor TSA enhanced the transcriptional activity of the mda-7/IL-24 gene. A375 cells were treated with TSA after transient transfection with pc3.1-vector (pcDNA3.1) or with a Sp1 (Sp1-pcDNA3.1) expression plasmid before analysis by a reporter assay (a) and quantitative real-time PCR (b). HDAC, histone deacetylase; TSA, trichostatin A.
Figure 6.
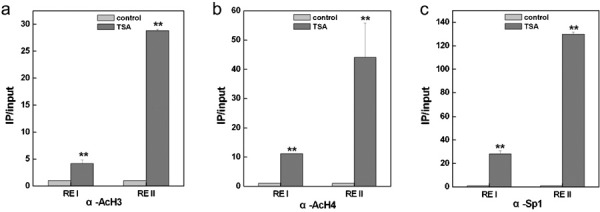
TSA increased the binding of Sp1 to the mda-7/IL-24 promoter. A375 cells were transiently transfected with pc3.1-vector or a Sp1 expression vector, and after treatment with TSA, the acetylation levels of histone H3 (a) and histone H4 (b) were examined by ChIP and quantitative real-time PCR. A375 cells were collected and precipitated with antibodies against Sp1 after treatment with TSA, and ChIP assays were performed to examine the binding of Sp1 to the mda-7/IL-24 promoter (c). ChIP, chromatin immunoprecipitation; mda-7/IL-24, melanoma differentiation-associated gene/interleukin-24; TSA, trichostatin A.
Discussion
The expression of the tumor suppressor gene mda-7/IL-24 declines in parallel with melanoma progression,4 and this process is not attributable to an inactivating mutation or to structural rearrangements,8, 9 suggesting that alternative regulatory mechanisms may be involved in the regulation of mda-7/IL-24. It has been well established that epigenetic modifications affect gene expression and phenotypes through mechanisms other than changes in the DNA sequence; these modifications include DNA methylation and histone modifications. Sequence analysis has shown that the GC content of the mda-7/IL-24 promoter is lower than 50%, and there are no typical CpG islands in the promoter of the gene. Moreover, we found that the treatment with the DNA methylation inhibitor 5-Aza had little effect on the expression of mda-7/IL-24 (data not shown in this report). Our results in Figure 1 show that mda-7/IL-24 expression can be induced by the HDAC inhibitors TSA and NaBu. Therefore, it is rational to postulate that histone modifications may play an important role in the regulation of mda-7/IL-24 expression.
There have been indications that the transcription factors activator protein 1 and CCAAT/enhancer-binding protein can significantly enhance the activity of the mda-7/IL-24 promoter in melanoma cells.12 In addition, sequence analysis by the GCG program has identified binding sites for several additional transcription factors, including Sp1, CREB, CBP, Elk1 and p53.5 In this study, we found that HDAC4 participated in the regulation of mda-7/IL-24 expression (Figure 2a), and there is another report showing that HDAC4 inhibited the transcriptional activation of the p21 gene induced by Sp1.11 Moreover, we also demonstrated that the transcription factor Sp1 bound to the mda-7/IL-24 promoter (Figure 3b) and upregulated mda-7/IL-24 expression (Figure 2b), whereas HDAC4 treatment inhibited the binding of Sp1 (Figure 3c) and downregulated mda-7/IL-24 expression (Figure 2c). One of the possible mechanistic explanations for this observation may be that histone acetylation plays an important role in the expression of mda-7/IL-24 because histone hyperacetylation may result in an ‘open' chromatin conformation that facilitates the binding of transcription factors to the gene's promoter, thus stimulating transcription. Histone deacetylation tends to form a ‘closed' chromatin structure that prevents transcription factors from binding to the promoter. Based on these data, we conclude that histone acetylation modification affects the transcription regulation of mda-7/IL-24, and Sp1 participates in this process.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 30671184) and the Program for Changjiang Scholars and Innovative Research Team in University (No. IRT0519).
References
- Caudell EG, Mumm JB, Poindexter N, Ekmekcioglu S, Mhashilkar AM, Yang XH, et al. The protein product of the tumor suppressor gene, melanoma differentiation-associated gene 7, exhibits immunostimulatory activity and is designated IL-24. J Immunol. 2002;168:6041–6046. doi: 10.4049/jimmunol.168.12.6041. [DOI] [PubMed] [Google Scholar]
- Chada S, Sutton RB, Ekmekcioglu S, Ellerhorst J, Mumm JB, Leitner WW, et al. MDA-7/IL-24 is a unique cytokine – tumor suppressor in the IL-10 family. Int Immunopharmacol. 2004;4:649–667. doi: 10.1016/j.intimp.2004.01.017. [DOI] [PubMed] [Google Scholar]
- Kotenko SV. The family of IL-10-related cytokines and their receptors: related, but to what extent. Cytokine Growth Factor Rev. 2002;13:223–240. doi: 10.1016/s1359-6101(02)00012-6. [DOI] [PubMed] [Google Scholar]
- Ekmekcioglu S, Ellerhorst J, Mhashilkar AM, Sahin AA, Read CM, Prieto VG, et al. Down-regulated melanoma differentiation associated gene (mda-7/IL-24) expression in human melanomas. Int J Cancer. 2001;94:54–59. doi: 10.1002/ijc.1437. [DOI] [PubMed] [Google Scholar]
- Madireddi MT, Dent P, Fisher PB. Regulation of mda-7/IL-24 gene expression during human melanoma differentiation. Oncogene. 2000;19:1362–1368. doi: 10.1038/sj.onc.1203424. [DOI] [PubMed] [Google Scholar]
- Lebedeva IV, Su ZZ, Chang Y, Kitada S, Reed JC, Fisher PB. The cancer growth suppressing gene mda-7/IL-24 induces apoptosis selectively in human melanoma cells. Oncogene. 2002;21:708–718. doi: 10.1038/sj.onc.1205116. [DOI] [PubMed] [Google Scholar]
- Sauane M, Lebedeva IV, Su ZZ, Choo HT, Randolph A, Valerie K, et al. Melanoma differentiation associated gene-7/interleukin-24 promotes tumor cell-specific apoptosis through both secretory and nonsecretory pathways. Cancer Res. 2004;64:2988–2993. doi: 10.1158/0008-5472.can-04-0200. [DOI] [PubMed] [Google Scholar]
- Mhashilkar AM, Schrock RD, Hindi M, Liao J, Sieger K, Kourouma F, et al. Melanoma differentiation associated gene-7 (mda-7/IL-24): a novel anti-tumor gene for cancer gene therapy. Mol Med. 2001;7:271–282. [PMC free article] [PubMed] [Google Scholar]
- Sauane M, Gopalkrishnan RV, Sarkar D, Su ZZ, Lebedeva IV, Dent P, et al. mda-7/IL-24: novel cancer growth suppressing and apoptosis inducing cytokine. Cytokine Growth Factor Rev. 2003;14:35–51. doi: 10.1016/s1359-6101(02)00074-6. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mottet D, Pirotte S, Lamour V, Hagedorn M, Javerzat S, Bikfalvi A, et al. HDAC4 represses p21(WAF1/Cip1) expression in human cancer cells through a Sp1-dependent, p53-independent mechanism. Oncogene. 2009;28:243–256. doi: 10.1038/onc.2008.371. [DOI] [PubMed] [Google Scholar]
- Madireddi MT, Dent P, Fisher PB. AP-1 and C/EBP transcription factors contribute to mda-7/IL-24 gene promoter activity during human melanoma differentiation. J Cell Physiol. 2000;185:36–46. doi: 10.1002/1097-4652(200010)185:1<36::AID-JCP3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]


