Abstract
microRNAs are a novel group of small, conserved, non-coding RNA molecules that are present in all species. These molecules post-transcriptionally regulate gene expression by targeting mRNAs for degradation or by repressing the translation of the mRNAs. A good understanding of miRNA-mediated gene regulation is critical to gain a comprehensive view of many physiological processes and disease states. Emerging evidence demonstrates that miRNAs play an important role in the differentiation and function of the adaptive immune system. This review provides an overview of the diverse functions of miRNAs in modulating immune responses and in immune cell development, particularly the development of Th17 cells, and explores the involvement of miRNAs in several autoimmune diseases including multiple sclerosis (MS), rheumatoid arthritis (RA), inflammatory bowel disease (IBD) and diabetes.
Keywords: autoimmune diseases, microRNAs, Th17 cells
Introduction
CD4+ T cells play an important role in the initiation of adaptive immunity by providing help to other cells and by taking on a variety of effector functions over the course of an immune response. Upon antigenic stimulation, naive CD4+ T cells activate, expand and differentiate into different effector cell subsets referred to as T helper (Th) 1, Th2 and Th17. These T-cell subsets are characterized by their distinct cytokine production profiles and effector functions. Interferon-γ is the signature cytokine produced by Th1 cells and is responsible for immunity against intracellular pathogens. IL-4, IL-5 and IL-13 are secreted by Th2 cells, which play important roles in clearing extracellular pathogens and mediating allergic responses.1 Recently, a new subset of effector T cells that preferentially produce IL-17, but not interferon-γ or IL-4, was classified and is referred to as the Th17 cell subset.2 Th17 differentiation is directed by lineage-specific transcription factors, including RORγt and RORα, and is controlled by the coordinated activity of a series of positive and negative regulators. Th17 cells are essential for the clearance of foreign pathogens and for inducing tissue inflammation in autoimmune diseases. However, what factors link the generation of Th17 cells to pathological conditions remains largely unknown.2
microRNAs (miRNAs) are endogenously encoded single-stranded RNAs of about 22 nucleotides (nt) in length and have been implicated in the regulation of gene expression that is essential for organ development, cellular differentiation, homeostasis, immune regulation and function. miRNAs regulate genes by targeting mRNAs for either degradation or translational repression.3, 4, 5 To date, over 800 miRNAs have been predicted to exist in the human genome and as many as one-third of all mRNAs are bioinformatically predicted to be regulated by miRNAs.6 The analysis of miRNA expression and function in specific physiological and pathological contexts can broaden our knowledge of the regulatory mechanisms by which miRNAs fine-tune cell differentiation and cellular functions. miRNAs have been recently recognized as important new players in the normal development and physiology of the immune system.7 This review will focus on the roles of miRNAs in Th17 cell development and differentiation, as well as in the pathogenesis of human diseases. We are especially interested in the reported involvement of miRNAs in autoimmune diseases.
miRNAs biogenesis and the mechanisms of miRNA-mediated gene silencing
Although many miRNAs are derived from independent transcriptional units, miRNA genes can also be located in the introns of protein-coding genes.8 A large percent of miRNAs genes are located in the genome as individual cluster that is first transcribed by RNA polymerase II as primary miRNA transcripts.9, 10 The majority of miRNA maturation occurs by a two-step process involving two ribonuclease III enzymes, Drosha and Dicer (Figure 1). In the first step, long primary miRNA transcripts are processed into corresponding pre-miRNA stem-loop structures that are approximately 60 nt in length by the nuclear-specific ‘microprocessor' complex, which is comprised of Drosha and its binding partner, DiGeorge syndrome critical region 8. In the second step, the pre-miRNA is exported from the nucleus into the cytoplasm by exportin-5 in a ras-related nuclear protein-guanosine triphosphate-dependent manner and is then further processed into 22-nt duplexes by the cytoplasmic Dicer. In addition, a relatively small percent of miRNA maturation is independent of Drosha. These intronic miRNA precursors, termed ‘mirtrons', are processed in the nucleus by the usual RNA splicing machinery without Drosha-mediated cleavage.11, 12, 13, 14 The functional miRNA strand is then selectively loaded into the RNA-induced silencing complex (RISC). Once loaded into RISC, a miRNA will bind to its target mRNA, usually at the three prime untranslated region (3′UTR), resulting in degradation or translational repression of the mRNA.15
Figure 1.
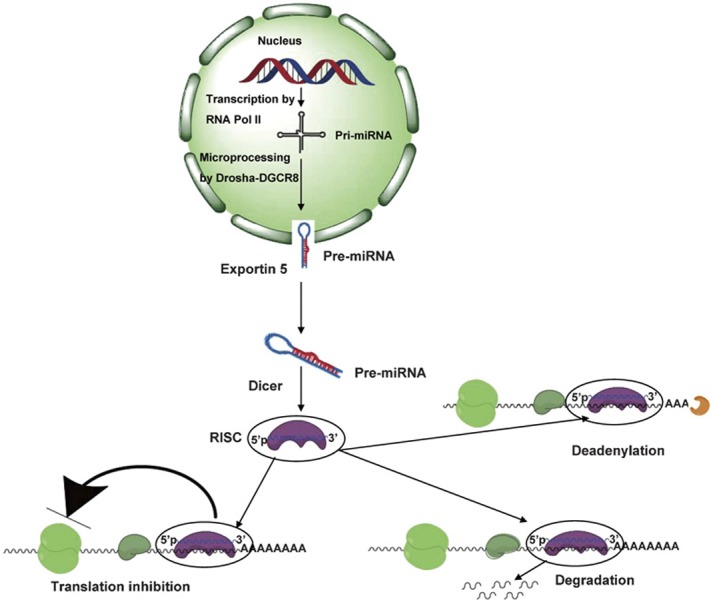
MicroRNA biogenesis and the mechanism of microRNA-mediated gene silencing. MicroRNAs (miRNAs) are short, double-stranded RNA molecules of approximately 19–23 nt in length. The primary transcripts of miRNAs (pri-miRNAs) are transcribed by RNA polymerase II from varied genomic loci. Primary miRNA transcripts are processed into precursor miRNA (pre-miRNA) stem-loops of approximately 60 nt in length by the nuclear RNase III enzyme, Drosha, which is assisted by DiGeorge syndrome critical region gene 8 (DGCR8). These pre-miRNAs are then exported into the cytoplasm by a Ran-GTP-dependent nuclear transport receptor, exportin 5, where they are further processed into approximately 22-nt miRNA duplexes by the type III RNase, Dicer. The final step of miRNA maturation is the selective loading of the functional strand of the small RNA duplex into the RNA-induced silencing complex (RISC). Mature miRNAs then guide the complex to their complementary mRNAs for regulation of gene expression. Regulation is be mediated by various mechanisms including repression of translation, mRNA cleavage and deadenylation. Ran-GTP, ras-related nuclear protein-guanosine triphosphate.
Several mechanisms have been proposed for miRNA-mediated gene regulation that would result in a reduction in the total amount of target protein. In one mechanism, the miRNA represses target gene expression at the translational level; mRNA levels remain constant, but the level of the encoded protein declines. This effect is achieved by the miRNA-mediated inhibition of either initiation, post-initiation (elongation) or the miRNA-mediated enhancement of ribosomal drop-off from translating polysomes during protein translation.7 In another mechanism, miRNAs repress target gene expression by triggering the degradation of target mRNAs. Individual miRNAs regulate the expression of target genes by forming base-pair interactions with target sites located in the UTRs of their target mRNAs. Multiple miRNA binding sites in the target UTR are required for efficient regulation, and these target sites are evolutionarily conserved. Several groups have postulated that miRNA target sites in mRNAs include a core sequence that forms a perfect or near-perfect base-pair match with seven or eight bases near the 5′ end of an miRNA (known as the ‘seed nucleotides').16, 17 However, it has been suggested that the RNA secondary structure and RNA sequences outside of mature miRNA binding regions have potent effects on target recognition,18 indicating a previously unappreciated complexity with regard to miRNA interactions with target mRNAs.
miRNAs in the development and function of immune cells
Given the detailed conceptual and experimental framework of our present understanding of the immune response, it is not surprising that there is another unrecognized layer of regulation by miRNAs in the development and physiology of the immune system. Genetic ablation of miRNA machinery and deregulation of individual miRNAs have provided direct evidence to support that miRNAs play a fundamental role in regulating the immune system. Deletion of Dicer, the essential protein component of the miRNA processing machine, in immature double negative thymocytes (DN3 stage), leads to a 10-fold reduction in total thymocyte numbers. Very few peripheral T cells are detectable when Dicer is deleted at the double-positive stage of thymocyte differentiation, and the peripheral CD4+ T-cell population is reduced by twofold.19 These peripheral CD4+ T cells exhibit reduced proliferation and increased cell death upon activation and demonstrate a bias toward Th1 differentiation, reflecting the failure of these cells to repress interferon-γ expression.20 The cell subset that is most affected by Dicer deletion at the double-positive stage is regulatory T cells (Tregs). The sixfold reduction of Tregs in both the thymus and the periphery causes severe immunopathology in mutant mice and is characterized by splenomegaly, enlarged intestinal lymph nodes and colitis.21 When Dicer is deleted in the B-cell lineage from the earliest stage of B-cell development, the pro-B to pre-B transition is almost completely blocked. This is, at least in part, due to the apoptosis of Dicer-deficient pro-B cells.22 Furthermore, the conditional deletion of Ago2, a component of RISC, in hematopoietic cells, results in a partial deficiency in miRNAs and compromised development of B cells and erythroid cells.23
The precise deletion of individual miRNA genes in mice has been performed to study the role of miRNAs in the development and function of the immune system (Figure 2, and Table 1). miRNA (miR)-181a is highly expressed in the thymus, but is expressed at a lower level in the heart, lymph nodes and bone marrow. Furthermore, miR-181a expression decreases during the development of B cells from the pro-B to the pre-B cell stages. The expression of miR-181a in hematopoietic stem and progenitor cells results in an increase in CD19+ B cells and a decrease in CD8+ T cells, which demonstrates that lineage-specific miRNAs might play a role in the regulation of lymphocyte development.24 Additionally, miR-181a alone, or in combination with other miRNAs, does not completely block the differentiation of lymphoid and myeloid cell types, indicating that this miRNA acts as a lineage modulator rather than a developmental switch. This might differ from the effects of lineage-specific transcription factors or oncogenes, which can completely shut down the differentiation of a cell lineage.
Figure 2.
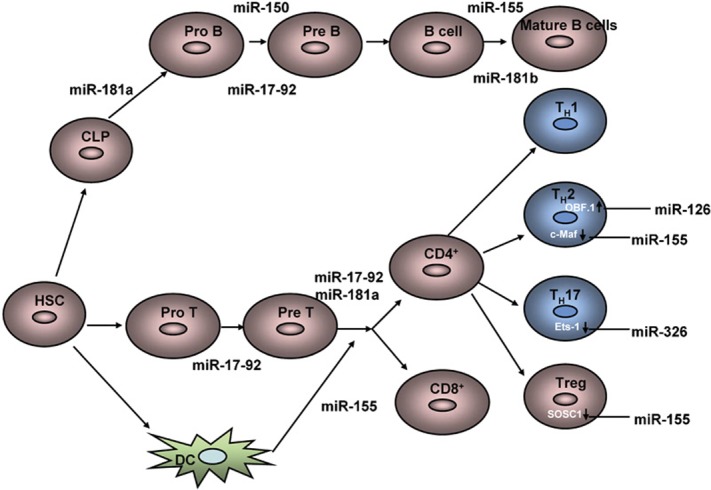
Roles of miRNAs in the development and function of the adaptive immune system. See text for detailed explanations. CLP, common lymphocyte progenitor; DC, dendritic cells; Ets-1, E26 transformation-specific-1; HSC, hematopoietic stem cells; Treg, regulatory T cell.
Table 1. Summary of miRNAs involvement in adaptive immune response.
| miRNA | Function | Targets | References |
|---|---|---|---|
| miR-181a | Positive regulator of B-cell development and CD4+ T-cell selection, activation and sensitivity | SHP-2, PTPN22, DUSP5, DUSP6 | 19, 20 |
| miR-181b | Class switch recombination in activated B cells | AID | 21 |
| miR-155 | Required for T-cell differentiation, germinal center B-cell responses and responses to bacterial and viral infection | Maf, PU.1 | 25, 26, 27, 28 |
| miR-150 | Increased expression leads to suppression of B-cell formation by blocking in pro-B to pre-B cell transition. Decreased expression in chronic lymphocytic leukemia (CLL) | c-Myb | 22 |
| miR-146 | Negative regulator of TLR-NF-κB signaling in response to bacterial infections | IRAK1, TRAF6 | 35 |
| miR-326 | Involved in the regulation of Th17 differentiation and pathogenesis of autoimmune disease | Ets-1 | 31 |
| miR-126 | Involved in suppression of the effector function of Th2 cells and the development of allergic airways disease | POU domain class 2-associated factor 1 | 28 |
Abbreviations: AID, activation-induced cytidine deaminase; Ets-1, E26 transformation-specific-1; miRNA, microRNA; Th, T helper; TLR, Toll-like receptors.
Interestingly, recent work has demonstrated that miR-181a critically modulates the strength and threshold of T-cell receptor signaling by downregulating multiple phosphatases that negative regulate distinct steps in the T-cell receptor signaling pathway.25 The protein tyrosine phosphatase PTPN22 dephosphorylates and inactivates Lck and ZAP70, two crucial components of proximal T-cell receptor signaling. The dual specificity phosphatases DUSP5 and DUSP6 dephosphorylate and inactivate the kinase extracellular signal-regulated kinase 1/2, which are essential regulators of positive selection. Overexpression of miR-181a in mature T cells similarly increases extracellular signal-regulated kinase activity, while the treatment of thymocytes with an antagomir against miR-181a (antagomir-181a), which degrades endogenous miR-181a, reduces extracellular signal-regulated kinase activity and impairs positive selection of thymocytes.25 Furthermore, miR-181a expression is high in the immature T-cell population but low in more differentiated T-cell populations, such as in Th1 and Th2 effector cells. The inhibition of this miRNA by antagomir-181a significantly impairs double-positive cell sensitivity and disrupts positive and negative selection, suggesting that properly tuning T-cell sensitivity at various developmental stages might be critical for regulating the development of tolerance and effector cells.
In contrast to miR-181a, miR-181b regulates class switching in activated B cells. The expression of miR-181b in activated B cells impairs class switching and downregulates the activation-induced cytidine deaminase, both at the mRNA and protein levels. This suggests that restricted activation-induced cytidine deaminase activity might be important to prevent malignant B-cell transformation.26 It was recently reported that the expression of miR-150 increases during both B-cell maturation in the bone marrow and T-cell maturation in the thymus, but decreases rapidly when naive T cells differentiate into Th1 or Th2 cells. The ectopic expression of miR-150 in lymphocyte progenitors blocks B-cell development at the transition from the pro-B to pre-B cell stage, leading to severe defects in the production of mature B cells. miR-150-deficient mice have an approximately twofold increase in the number of splenic B1 cells but have no apparent defects in the development of other lymphoid-derived T- and B-cell types.27
Interestingly, miR-155 is largely restricted to hematopoietic cells and B-cell malignancies in humans, including Hodgkin's disease and Burkitt lymphomas, indicating that this miRNA is related to hematopoietic cancers.28 As a corollary to these clinical observations, B cell-restricted expression in miR-155 transgenic mice leads to pre-leukemic proliferation of B lineage cells that progresses to a severe B-cell malignancy.29 The deletion of miR-155 results in a diminished germinal center response and impaired B-cell memory formation.30 Notably, Tregs express a high level of miR-155, and a loss of miR-155 profoundly diminishes Treg numbers but apparently does not affect their suppressive functions.31 miR-155-deficient dendritic cells fail to induce efficient T-cell activation in response to antigens, while activated miR-155-deficient T cells have impaired IL-2 production and display skewed differentiation towards the Th2 lineage. In addition to these phenotypic findings, there is a coordinated increase in the level of the Th2 transcription factor c-Maf, a miR-155 target.32 miR-126 has a similar role to miR-155 in influencing Th2 cell responses. Blocking miR-126 results in the diminishment of Th2 responses, including inflammation, airway hyper-responsiveness, eosinophil recruitment and mucus hypersecretion33 This study demonstrates that miR-126 blockade augments the expression of the POU domain class 2-associated factor 1, which activates the transcription factor PU.1. PU.1 alters Th2 cell function by negatively regulating GATA3 expression. The authors suggest that targeting miRNAs in the airway might be a feasible anti-inflammatory treatment for allergic asthma.
The miR-17-92 cluster is composed of six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b1 and miR-92-1) and is highly expressed in B and T precursor cells. While miR-17-92 and Dicer knockout mice have indicated that this cluster is required for the pro-B to pre-B cell transition during B-cell development,34 mice with ectopic expression of the miR-17-19 cluster in the lymphocyte compartment develop severe lymphoproliferative disease and autoimmunity. The major cell expansion in these mice is comprised of activated CD4+ T cells and, to a lesser extent, CD8+ T cells. Bim, a proapoptotic member of the antiapoptotic Bcl-2 family, has been identified as a target of the miR-17-92 cluster.35
miRNAs regulate Th17-cell differentiation
In contrast to the relatively low levels of miR-326 that is expressed in Th1, Th2 and inducible Tregs, miR-326 is more highly expressed in Th17 cells, suggesting that miR-326 might be relevant to Th17 lineage commitment. In central nervous system infiltrates that are isolated from mice with experimental autoimmune encephalomyelitis (EAE), a mouse model that pathologically mimics the human disease multiple sclerosis (MS), both the frequency and absolute number of Th17 cells are higher than in mice with less severe forms of the disease. Notably, EAE mice that overexpress miR-326 show an increased number of Th17 cells in central nervous system infiltrates. The expression levels of the Th17 lineage marker genes (Rorc and Il17a) and the presence of IL-17 in the culture supernatants are dramatically increased by miR-326 overexpression. Likewise, human CD4+CCR6+ cells, which are reported to be enriched in IL-17 producing cells and home to sites of inflammation, have an approximately fivefold greater expression of miR-326 than their CCR6− counterparts.36 These data indicate that miR-326 might regulate Th17-cell development and migration and thus influence the development of EAE pathology. In contrast, Th1-cell numbers are not affected by miR-326 overexpression in EAE mice.36 Consistent with these observations, gene expression profiles demonstrate that genes related to the Th17 lineage, Il17a, Il17f, I122, Il21 and Il23r, are significantly upregulated in T cells that overexpress miR-326, while other immune genes, such as Ifng, Il4 and Tgfb, are not affected by miR-326.
E26 transformation-specific-1 (Ets-1) is a functional target of miR-326
Ets-1, the prototype member of the Ets family of transcription factors, has been shown to play a role in hematopoietic development, angiogenesis and tumor progression, and is a negative regulator of Th17 differentiation.37 Ets-1-deficient T cells increase IL-17 production after differentiation in the presence of transforming growth factor-β1 and IL-6, but produce less IL-2 and are resistant to IL-2 due to a defect in the phosphorylation of the downstream effector STAT5. Furthermore, Ets-1 knockout mice spontaneously express an abnormally high level of IL-17 in their lungs and their airway epithelial cells overproduce mucus.37 Although the latest miRNA-binding site prediction programs, Targetscan and Miranda, indicate that both Ets-1 and Foxp3, another negative regulator of Th17 differentiation, have putative miR-326-binding elements in their 3′UTR regions, our previous findings demonstrate that miR-326 inhibits the activity of a reporter containing the Ets-1 3′UTR but does not inhibit the activity of a Foxp3 3′UTR-containing reporter. Furthermore, the lentiviral overexpression of miR-326 in naive CD4+ T cells does not affect the transforming growth factor-β1-driven induction of Foxp3, but rather miR-326 suppresses Ets-1 expression in a dose-dependent manner.36 These observations indicate that Ets-1, not Foxp3, is the target of miR-326. Additional studies suggest that miR-326 inhibits the translation of Ets rather than destabilizing its transcripts. Interestingly, Ets-1 expression has been found to be substantially downregulated by miR-326 in human patients with relapsing MS.36 This implies that miR-326 is associated with the differentiation and function of both mouse and human Th17 cells and that Ets-1 acts as a key target of miR-326 in the development of mouse models of EAE as well as in human MS.
miRNAs in autoimmune diseases
It is becoming increasingly clear from cell culture and animal studies that proper miRNA regulation is critical for the prevention of autoimmunity. Several studies have investigated whether miRNA dysregulation is linked to autoimmune disease pathogenesis in human patients, specifically in MS,18 rheumatoid arthritis (RA), inflammatory bowel disease (IBD) and diabetes (Table 2).
Table 2. Summary of miRNAs involvement in autoimmune diseases.
| miRNA | Cell subsets influenced | Targets | Disease | References |
|---|---|---|---|---|
| miR-155 | Treg cells, activated B cells and monocytes, macrophages | SOCS1, PU.1, c-Maf, IKKε, FADD | RA | 35, 36 |
| miR-146 | Activated monocytes | IRAK1, TRAF6 | RA | 36 |
| miR-192 | Epithelial cell in colon tissues | MIP-2α | UC | 38 |
| miR-21 | Epithelial cell in colon tissues | ND | UC | 38 |
| miR-375 | β cells in pancreatic endocrine cells | Myotrophin | Diabetes | 41, 42 |
| miR-9 | ND | Pancreas granuphilin/Slp4 | Diabetes | 43 |
| miR-124a | β cells in pancreas | Foxa2 | Diabetes | 44 |
| miR-326 | Th17 cells | Ets-1 | MS | 31 |
| miR-18b | Human blood | ND | MS | 33 |
| miR-599 | Human blood | ND | MS | 33 |
| miR-96 | Human blood | ND | MS | 33 |
Abbreviations: Ets-1, E26 transformation-specific-1; MIP, macrophage-inflammatory peptide; miRNA, microRNA; MS, multiple sclerosis; ND, not determined; RA, rheumatoid arthritis; UC, ulcerative colitis; Th, T helper.
miRNAs in MS
As discussed above, miR-326 expression is significantly higher in relapsing–remitting MS patients during acute relapsing.36 Enhanced miR-326 expression has been specifically found in CD4+ T cells but not CD8+ T cells or non-T-cell populations from MS patients. Notably, the transcription levels of Il17a, but not Ifng, Il4 or Tgfb, are increased in CD4+ T cells that overexpress miR-326. The upregulation of Il17a is observed in CD4+ T cells from patients with relapsing MS. During the acute phase of the mouse model of EAE, higher expression of miR-326 is also found in CD4+ T cells. These observations indicate that miR-326 expression in Th17 cells significantly correlates with MS in both humans and the mouse disease model. Further analysis illustrates that EAE severity in the mice efficiently overexpresing miR-326 was greatly increased compared with that in the control mice received miR-326 inhibitors, which demonstrates that miR-326 expression and function correlate with the development of EAE. Another study compared the expression pattern of miRNAs in peripheral blood leukocytes obtained from MS patients in either relapse status or remission status to that obtained from healthy controls. These data revealed that two miRNAs, hsa-miR-18b and hsa-miR-599, might be important at the time of relapse and that hsa-miR-96 might be involved in MS remission.38
miRNAs in RA
RA is a systemic autoimmune disorder characterized by chronic inflammation of synovial tissue that results in irreversible joint damage. Inflammatory cytokines, especially tumor-necrosis factor-α, IL-1β, and IL-6, are known to play important roles in RA pathogenesis. The inhibition of these cytokines can ameliorate disease in some patients.39 Recently, several studies have examined miRNA expression in RA synovial tissue and fibroblasts. Stanczyk et al. reported that miR-155 and miR-146 expression is increased in synovial fibroblasts from RA patients compared to those from osteoarthritis patients.40 The expression level of miR-155 is also higher in synovial monocytes compared to peripheral blood monocytes from RA patients. The expression of miR-155 in RA synovial fibroblasts revealed that matrix metalloproteinase 3 is a potential target of miR-155, suggesting that miR-155 might modulate downstream tissue damage. Another group reported that the expression levels of miR-146a, miR-155, miR-132 and miR-16 are increased in peripheral blood leukocytes from RA patients. There is no correlation between the observed increase in miRNA expression and patient age, race or medications in this study.12 Further investigation is needed to elucidate the mechanisms of these miRNAs in the regulation of RA pathogenesis.
miRNAs in IBD
Crohn's disease and ulcerative colitis (UC) are the two main subtypes of IBD and are chronic, relapsing inflammatory disorders of the gastrointestinal tract.41 Patients typically suffer from frequent and chronically relapsing flares, resulting in diarrhea, abdominal pain, rectal bleeding and malnutrition. Multiple global gene expression profiles demonstrate that IBD is associated with an increase in the expression of genes involved in inflammation and fibrosis, including cytokines, growth factors, inflammatory mediators, extracellular matrix proteins, matrix metalloproteinases, antimicrobial factors and cell cycle regulators. Recently, miRNA expression was assessed in patients with active UC, inactive UC, Crohn's disease, irritable bowel syndrome and infectious colitis, and in healthy subjects.42 Compared to healthy control tissues, three miRNAs (miR-192, miR-375 and miR-422b) are significantly decreased and eight miRNAs (miR-16, miR-21, miR-23a, miR-24, miR-29a, miR-126, miR-195 and let-7f) are significantly increased in active UC tissues. miRNA microarray analysis and qRT-PCR verification identified miR-21 as the most highly expressed UC-associated miRNA in human colon tissues. In situ hybridization studies revealed that miR-192, the miRNA with decreased expression in active UC, is predominantly localized to colonic epithelial cells and that macrophage-inflammatory peptide 2α is a target of miR-192. These data suggest that miRNAs regulate colonic epithelial cell-derived chemokine expression.42 A deeper understanding of the regulatory roles of miRNAs in acute and chronic inflammatory diseases might lead to the development of miRNA-based diagnoses or treatments for chronic inflammatory diseases.
miRNAs in diabetes
Diabetes is a deadly global health problem and is expected to affect 366 million patients by the year 2030.43 A balance between normal insulin secretion and function maintains appropriate levels of circulatory glucose. Dysregulation at any step of this finely tuned system can result in the initiation of type 1 diabetes, while insulin resistance culminates in type 2 diabetes.44 Although miRNAs are notably absent from the list of genes that are associated with type 2 diabetes risk, emerging evidence suggests that miRNAs play significant roles in insulin production, action and secretion, and are involved in diverse aspects of both glucose and lipid metabolism.44
Serious developmental defects are observed in all pancreatic cell types when Dicer is conditionally deleted at an early stage of pancreatic development. Additional studies have been performed in an effort to identify the precise roles that individual miRNAs play in the development and function of the pancreas and in diabetes. miR-375 is selectively expressed in pancreatic endocrine cell lines. Overexpression of miR-375 suppresses glucose-stimulated insulin secretion, while inhibition of miR-375 enhances insulin secretion.45 Phosphoinositide-dependent kinase-1, a protein serine/threonine kinase that can phosphorylate protein kinase B (also known as Akt) and glycogen synthase kinase-3, is a potential target of miR-375.46 Overexpression of miR-375 in β-cell lines leads to reduced protein kinase B and glycogen synthase kinase-3 phosphorylation. Conversely, inhibition of miR-375 increases phosphoinositide-dependent kinase-1 levels and elevates glucose-dependent insulin mRNA levels and β-cell proliferation. Another target of miR-375 is myotrophin, a protein implicated in exocytosis. miR-9 is another miRNA that has been shown to regulate insulin exocytosis from the pancreas. Elevated levels of miR-9 correlate with glucose-stimulated insulin release. Granuphilin/Slp4, the Rab GTPase effector, is one of the targets of miR-9 that is inhibited by Onecut2. Silencing Onecut2 replaces the effect of miR-9 on insulin exocytosis.47 Furthermore, Baroukh et al. found that miR-124a is also strongly correlated with mouse pancreatic development, suggesting that it has a role in β-cell differentiation.48
Conclusion
Recent studies have demonstrated that miRNAs function as ‘fine-tuners' for immune responses and play crucial roles in the development and function of different effector T-cell subsets, including Th17 cells and Tregs. Future investigations will uncover the detailed mechanisms for how miRNAs regulate the development and functions of effector T cell subsets. Given the current understanding of miRNAs in the pathogenesis and progression of autoimmune diseases, continued exploration of specific mutations or deletions of individual miRNAs in autoimmune diseases might shed light on their genetic mechanisms. A more detailed understanding of specific miRNAs in a particular type of autoimmune diseases might allow us to develop new diagnostic approaches. The appropriate therapeutic interventions targeting dysregulated levels of specific miRNAs might offer valuable novel tools for the treatment of autoimmune diseases.
Acknowledgments
We thank Miss C. Liu for critically reading this manuscript. This work was supported by the Shanghai Natural Science Foundation (No. 10ZR1435300).
References
- Dong C. Differentiation and function of pro-inflammatory Th17 cells. Microbes Infect. 2009;11:584–588. doi: 10.1016/j.micinf.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miossec P, Korn T, Kuchroo VK. Interleukin-17 and type 17 helper T cells. N Engl J Med. 2009;361:888–898. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–194. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- Cobb BS, Nesterova TB, Thompson E, Hertweck A, O'Connor E, Godwin J, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- de Yébenes VG, Belver L, Pisano DG, González S, Villasante A, Croce C, et al. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci USA. 2009;106:18704–18709. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho IC. Ets-1 is a negative regulator of Th17 differentiation. J Exp Med. 2007;204:2825–2835. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaegui D, Baranzini SE, Armañanzas R, Calvo B, Muñoz-Culla M, Khankhanian P, et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS One. 2009;4:e6309. doi: 10.1371/journal.pone.0006309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst DE, Keystone EC, Kirkham B, Kavanaugh A, Fleischmann R, Mease P, et al. Updated consensus statement on biological agents for the treatment of rheumatic diseases, 2008. Ann Rheum Dis. 2008;67:iii2–iii25. doi: 10.1136/ard.2008.100834. [DOI] [PubMed] [Google Scholar]
- Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Agarwal P, Kaur K, Datta M. MicroRNAs in diabetes: tiny players in big disease. Cell Physiol Biochem. 2009;23:221–232. doi: 10.1159/000218169. [DOI] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E. miR-375 targets 3′-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes. 2008;57:2708–2717. doi: 10.2337/db07-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, Regazzi R. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J Biol Chem. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R, et al. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem. 2007;282:19575–19588. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]


