Abstract
Interferon regulatory factor 3 (IRF3), one member of the IRF family, plays a central role in induction of type I interferon (IFN) and regulation of apoptosis. Controlled activity of IRF3 is essential for its functions. During reverse transcription (RT)-PCR to clone the full-length open reading frame (ORF) of IRF3, we cloned a full-length ORF encoding an isoform of IRF3, termed as IRF3-CL, and has a unique carboxyl-terminus of 125 amino acids. IRF3-CL is ubiquitously expressed in distinct cell lines. Overexpression of IRF3-CL inhibits Sendai virus (SeV)-triggered induction of IFN-β and SeV-induced and inhibitor of NF-κB kinase-ε (IKKε)-mediated nuclear translocation of IRF3. When IKKε is overexpressed, IRF3-CL is associated with IRF3. These results suggest that IRF3-CL, the alternative splicing isoform of IRF-3, may function as a negative regulator of IRF3.
Keywords: interferon regulatory factor 3, negative regulation, splicing variant
Introduction
The interferon regulatory factor (IRF) family of transcriptional factors plays versatile roles in many biological processes, including innate and adaptive immune responses, cell growth control, apoptosis and hematopoietic development.1, 2 To date, nine members of the mammalian IRF family have been identified and characterized, each containing a highly conserved DNA-binding domain of ∼120 amino acids (aa) at the amino terminus, and a variable carboxyl-terminal IRF association domain (IAD).2 IRF3, one member of the IRF family, possesses a transactivation domain (aa 134–394) and two autoinhibition domains.3 After viral infection, the latent IRF3, which resides primarily in the cytoplasm, is phosphorylated by the inhibitor of NF-κB kinase-ε (IKKε) and TANK-binding kinase 1 (TBK1) and forms a homo- or heterodimer with other transcriptional factors which then translocates into the nucleus.4, 5, 6 The activated IRF3 associates with the transcriptional cofactors cyclic AMP-responsive element-binding protein and P300 to bind to the specific DNA targets, thus leading to transcriptional initiation of target genes with other cofactors simultaneously recruited.7
Accumulating evidence demonstrates that IRF3 plays an essential role in the virus- or bacterium-mediated induction of interferon (IFN)-β and a subset of IFN-stimulated genes through the Toll-like receptors or the cytosolic receptors pathway.8, 9, 10 Furthermore, IRF3 has been shown to function as a vital signaling regulator in the innate immune response, development of immune cells and apoptosis.11 As rapid and controlled cellular responses are pivotal to host defense, the activity of IRF3 should be regulated at multiple levels.
A single gene is capable of generating multiple transcripts from a common mRNA precursor through alternative splicing, which may produce distinct protein isoforms with diverse and even antagonistic functions. Several genome-wide analyses indicate that over 50% of human genes present alternatively spliced isoforms.12 In the past decades, various isoforms of many important signaling molecules including different IRFs have been found and functionally characterized.13, 14 These studies demonstrate that alternative splicing plays a crucial role in regulating the functions of proteins.
In an attempt to characterize IRF3, we isolated a full-length open reading frame (ORF) from HEK293 cells encoding an isoform of IRF3, which was denoted as IRF3-CL and has a unique carboxyl-terminus of 125 aa. Our research shows that IRF3-CL is ubiquitously expressed in all cell lines tested, and functions in a dominant-negative manner to regulate virus-triggered IFN-β signaling pathway.
Materials and methods
Cell lines and virus
HEK293 and HeLa cells were maintained in HEPES-buffered DMEM (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA), 100 U/ml penicillin and 100 mg/ml streptomycin. THP-1, K562 and HL60 cells were maintained in HEPES-buffered RPMI-1640 (Invitrogen) with 10% fetal bovine serum, 100 U/ml penicillin and 100 mg/ml streptomycin. Sendai virus (SeV) was kindly provided by Dr Hanzhong Wang (Wuhan Institute of Virology, Chinese Academy of Sciences).
Plasmid construction
The cDNA fragments of IRF3 and IRF3-CL were amplified from HEK293 cells by reverse transcription (RT)-PCR using the primer pair F1 and R1 (Table 1) and confirmed by sequencing. The IRF3 and IRF3-CL ORF were individually subcloned into pEGFP-C1, pDsRed-Express-C1 (BD Sciences Clontech, Palo Alto, CA, USA), pcDNA3.1-Myc and pcDNA3.1-3×Flag vectors, derived from pcDNA3.1-HisA (Invitrogen), respectively, and fused to the 5′ terminal tag by standard molecular cloning procedure. The sequences of PCR primers were shown in Table 1. The pRK-HA-IKKε and the reporter plasmid pISRE-Luc were gifts from Professor Hongbing Shu (Wuhan University). The reporter plasmid pIFN-Luc was kindly provided by Professor Reich (Stony Brook University).
Table 1. Sequences of PCR primers.
| Name | Sequence (5′ to 3′)a | |
|---|---|---|
| F1 | ATGGGAACCCCAAAGCCACG | For cloning of IRF 3 and IRF3-CL ORF |
| R1 | TTATTGGTTGAGG TGGTGGGGAAC | |
| LCH-17F | GCTCAAGCTTCGAATTCTGGAACCC CAA AGCCACGGATC | Fusion of IRF3-CL with EGFP |
| LCH-18R | GCCCGCGGTACCGTCGACTTATTGGTTGAGGTGGTGGGGAAC | |
| LCH-17F | GCTCAAGCTTCGAATTCTGGAACCCCA AAGCCACGGATC | Fusion of IRF3 with DsRed |
| LCH-20R | GCCCGCGGTACCGTCGACTCAGGTCTC CCCAGGGCCCT | |
| LCH-25F | TTACCGGAACCCCAAAGCCACGGATC | Fusion of IRF3-CL with Myc tag |
| LCH-26R | TGACTTATTGGTTGAGGTGGTGGGGAAC | |
| LCH-25F | TTACCGGAACCCCAAAGCCACGGATC | Fusion of IRF3 with Myc tag |
| LCH-28R | TGACTCAGGTCTCCCCAGGGCCCT | |
| LCH-29F | CGCGAAGGAACCCCAAAGCCACGGATC | Fusion of IRF3-CL with Flag tag |
| LCH-30R | CGATTATTGGTTGAGGTGGTGGGGAAC | |
| LCH-29F | CGCGAAGGAACCCCAAAGCCACGGATC | Fusion of IRF3-CL with Flag tag |
| LCH-32R | CGATCAGGTCTCCCCAGGGCCCT |
Abbreviations: EGFP, enhanced green fluorescent protein; IRF3, interferon regulatory factor-3; ORF, open reading frame.
The sequence underlined at the 5′ end of each primer is the additional non-specific sequence used for cloning.
Preparation of anti-IRF3-CL antiserum
The carboxyl-terminal fragment of IRF3-CL ORF, encoding the unique amino acids (327-452 aa), was amplified by PCR and subcloned into the pET-28a vector (Novagen, Madison, WI, USA) in-frame to the 5′ terminal His tag. The expressed His-tagged polypeptide was purified with Ni-column (Pharmacia, Piscataway, NJ, USA) from Escherichia coli BL21 strain, and then used as an antigen to produce antiserum in rabbit. The titer and specificity of the antiserum were determined by western blotting.
Infection
HEK293 cells were infected with SeV (100 hemagglutinating units (HAU) per ml) in serum-free medium. After incubation for 2 h, cells were added with the same volume of medium containing 20% fetal bovine serum. After infection, cells were collected and used for various experiments.
Transfection and reporter gene assay
HEK293 cells were seeded on 24-well plates and transfected the following day using FuGENE HD Transfection Reagent (Roche Diagnostics, Mannheim, Germany). In the same experiment, where necessary, the respective empty vector was used to adjust the total amount of DNA. To normalize the transfection efficiency, 0.02 µg of pRL-TK Renilla luciferase reporter plasmid was added to each transfection. Approximately 24 h after transfection, cultures were mock infected or infected with SeV (100 HAU per ml) for an additional period of 8 h before luciferase activities were measured. Luciferase assays were performed using a Dual-Specific Luciferase Assay Kit (Promega, Madison, WI, USA). All reporter assays were repeated at least three times.
Reverse transcription and quantitative real-time PCR (Q-PCR)
Total RNA was extracted from cultured cells with TRIzol (Invitrogen). Q-PCR analysis was performed using the ABI StepOne Real-Time PCR system. The primers for human Ifnb (QT00203763) and the kit used for Q-PCR were purchased from Qiagen (Hamburg, Germany). The human β-actin primers (sense: 5′-GAAATCGTGCGTGACATTAA-3′ antisense: 5′-AAGGAAGGCTGGAAGAGTG-3′) were designed as control in Q-PCR. Data were normalized according to the level of β-actin expression in each sample.
Immunoprecipitation and western blot analysis
HEK293 cells (1×106) were transiently transfected with the corresponding plasmids. After 24 h, cells were lysed in lysis buffer (20 mM Tris-HCl (pH7.5), 150 mM NaCl, 1 mM EGTA, 1 mM Na2EDTA, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1% Triton X-100, and a protease inhibitors cocktail (Sigma, St Louis, MO, USA)). Following the removal of insoluble materials by centrifugation, cell lysates were precleared with protein G-agarose slurry (Amersham, Piscataway, NJ, USA) and incubated with specific antibodies at 4 °C overnight. The immunoprecipitates were washed three times with lysis buffer, and proteins were removed from the protein G beads by boiling for 10 min in SDS sample buffer. The recovered proteins were separated on 10% SDS–polyacrylamide gel electrophoresis (PAGE) and electrotransferred onto a nitrocellulose membrane (Millipore, Billerica, MA, USA). Western blotting was performed using specific antibodies, and blots were developed by enhanced chemiluminescence using SuperSignal West Femo Maximum Sensitivity Substrate (Pierce, Rockford, IL, USA).
Native PAGE
Native PAGE (7.5%) for detecting protein dimers was performed as described by Iwamura et al.15 Anti-Myc monoclonal antibody (Sigma) and rabbit anti-IRF3-CL antiserum were used to detect exogenous expressed or endogenous IRF3-CL, respectively.
Fluorescence imaging
HEK293 cells (1×105) plated on coverslips were fixed with 4% paraformaldehyde for 20 min, permeabilized in 0.25% Triton X-100 for 10 min, blocked in 10% goat serum, and incubated with anti-HA monoclonal antibody (1∶200; Sigma) for 1 h. Secondary antibody conjugated to rhodamine (Jackson ImmunoResearch Laboratories West Grove, PA, USA) was applied at 1∶100 dilution for 1 h. Nuclei were stained with Bisbenzimid H 33342 (Fluka, Buchs, Switzerland). Cells were examined with a laser scanning confocal microscope (Zeiss, LSM S10 META).
Results
IRF3-CL is ubiquitously expressed
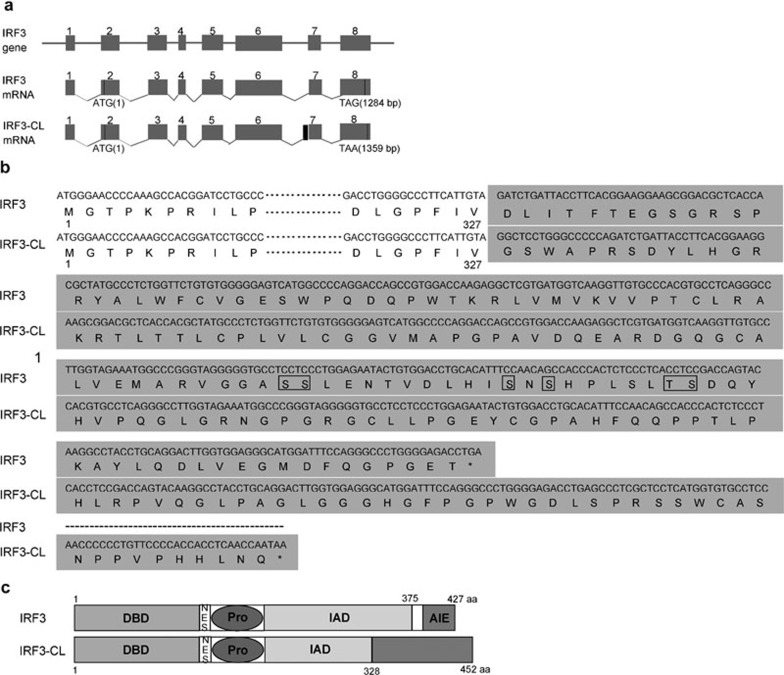
During RT-PCR to clone the full-length ORF of IRF3 from HEK293 cells, sequencing results revealed a different ORF. Sequence analysis indicated that this transcript used the AG, 16 nucleotides upstream of the 3′ splice site for IRF3 ORF, as the 3′ splice site for exon 7, leading to an addition of 16 nucleotides for exon 7 (Figure 1a). The 16 nucleotides further resulted in a frame shift and produced a spliced isoform of 1359 nucleotides ending in a stop codon TAA (nucleotides 1357–1359) (Figure 1a and b), which encodes a 452 aa polypeptide-termed IRF3-CL (Figure 1b). IRF3-CL shares a common amino terminus (1–327 aa) with IRF3, but has a unique carboxyl-terminus (328–452 aa) with no homology to IRF3 carboxyl-terminus and other proteins (Figure 1b and c). Thus, IRF3-CL does not contain the autoinhibition element and the consensus motif SxSxxxS targeted for phosphorylation.
Figure 1.
Schematic diagram of IRF3 and IRF3-CL. (a) Intron-exon borders for IRF3 gene and its alternative splicing. Boxes represent exons and lines represent introns. The black box designates the addition of 16 bp upstream exon 7 resulting from alternative splicing. (b) Schematic representation of the C-terminal regions of IRF3 and IRF3-CL. The mRNA and the corresponding amino acids of IRF3-CL were aligned against the C-terminus of IRF3. IRF3-CL encodes 452 aa residues, of which the first 327 aa residues are identical to the N-terminus of IRF3 and the remaining 125 aa residues do not resemble any proteins in the database. The shaded region represents the different C-termini of two proteins. The phosphorylation sites within the carboxyl-terminus of IRF3 are boxed. (c) Schematic alignment of IRF3 and IRF3-CL. IRF3 contains a conserved DBD, a NES, a Pro, an IAD and a C-terminal AIE (Mamane et al., 1999). IRF3-CL shares a common N-terminus with IRF3 and contains a partial IAD, but has a unique C-terminus (328–452 aa). aa, amino acids; AIE, autoinhibition element; DBD, DNA-binding domain; IAD, IRF association domain; IRF3, interferon regulatory factor-3; NES, nuclear export signal; Pro, proline-rich domain.
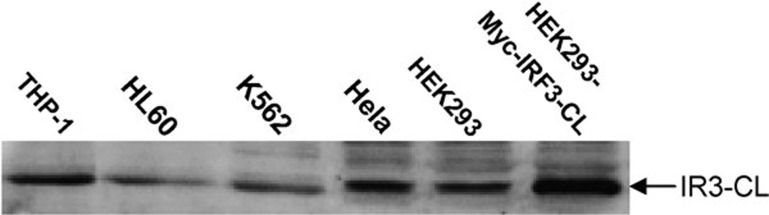
IRF3 is expressed ubiquitously.1 To examine the distribution pattern of IRF3-CL, we first examined its expression at the mRNA level by RT-PCR in various cell lines and by PCR in a cDNA library. DNA sequencing results indicated that the IRF3-CL transcript could be detected both in normal Chang liver cell line and tumor cell lines such as HeLa, Huh7, HT29, and even from the adult human liver cDNA library (Invitrogen) (data not shown), suggesting its ubiquitous expression at the mRNA level. Furthermore, two full-length mRNA sequences (BC009395 and BC000660) corresponding to IRF3-CL from human brain and kidney tumor tissues, respectively, are found in GenBank. To confirm the expression of IRF3-CL at the protein level, anti-IRF3-CL antiserum was prepared in rabbit and used for western blotting. As shown (Figure 2), a protein of ∼55 kDa was detected in all cell lines tested suggesting the ubiquitous expression of IRF3-CL at the protein level.
Figure 2.
Western blot analysis of expression of IRF3-CL. Various cell lines as indicated were used. Lysate of HEK293 cells expressing Myc-IRF3-CL was designed as a positive control. Western blotting was performed using anti-IRF3-CL antiserum. IRF3, interferon regulatory factor-3.
IRF3-CL inhibits virus-triggered induction of IFN-β
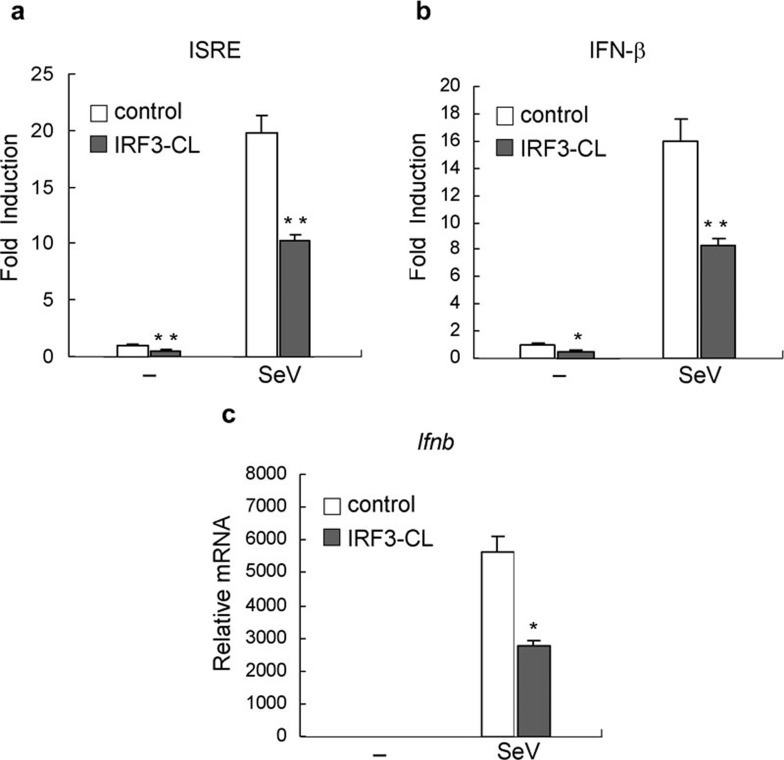
Studies to date indicate that IRF3 plays an essential role in virus-induced transcription of Ifnb gene.8, 11 Because IRF3-CL is an alternative splicing isoform of IRF3, which has a unique C-terminus, we investigated whether IRF3-CL functions in the regulation of virus-triggered activation of IRF3. In reporter gene assays, overexpression of IRF3-CL inhibited SeV-induced activation of interferon-stimulated response element (ISRE) and the IFN-β promoter (Figure 3a and b). Furthermore, we measured the effect of IRF3-CL on expression of Ifnb gene by Q-PCR. As shown in Figure 3c, IRF3-CL inhibited SeV-induced expression of Ifnb. These data suggest that IRF3-CL inhibits SeV-induced expression of IFN-β.
Figure 3.
IRF3-CL suppresses SeV-triggered induction of IFN-β. HEK293 cells (1×105) were transiently transfected with an expression plasmid for IRF3-CL (filled bars) or an empty expression vector (empty bars). After 24 h, cells were infected with SeV or left uninfected for 8 h. (a, b) Cells were cotransfected with the indicated luciferase reporter plasmid. After infection, cells were subjected to the dual-luciferase assay. (c) After infection, total RNA was extracted and subjected to quantitative real-time PCR using the primers for human Ifnb gene. In panels a and b, data are expressed as relative folds activation to that of non-stimulated sets and are means±SEM of three independent experiments. In panel c, data shown are averages and deviations of relative mRNA from three independent experiments. *P<0.05; **P<0.01. IFN, interferon; IRF3, interferon regulatory factor-3; SeV, Sendai virus.
IRF3-CL inhibits IKKε-mediated activation of ISRE and the IFN-β promoter
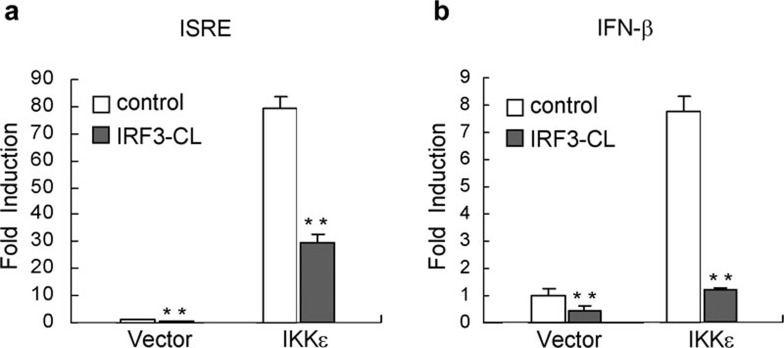
Previous studies indicated that kinase IKKε plays key roles in phosphorylation, activation and degradation of IRF3.16 Coexpression of IKKε phosphorylates and activates IRF3 leading to gene expression regulated by ISRE and the IFN-β promoter.17 To elucidate the mechanism of the negative effect of IRF3-CL on SeV-induced activation of IRF3, we examined whether IRF3-CL plays an inhibitory role in IKKε-mediated IFN-β signaling. The effect of IRF3-CL on the activity of the inducible elements was evaluated using the dual luciferase reporter system. As shown in Figure 4, overexpression of IRF3-CL significantly reduced IKKε-mediated activation of ISRE and the IFN-β promoter, indicating that IRF3-CL targeted IKKε-mediated activation of IRF3.
Figure 4.
IRF3-CL suppresses IKKε-mediated activation of ISRE and the IFN-β promoter. HEK293 cells (1×105) were transiently transfected with the indicated luciferase reporter plasmid, an expression plasmid for IRF3-CL (filled bars) or an empty expression vector (empty bars), and an expression plasmid for IKKε or an empty expression vector. Reporter assays were performed 24 h after transfection. Data are expressed as relative folds activation to that of non-stimulated sets and are means±SEM of three independent experiments. *P<0.05; **P<0.01. IFN, interferon; IKKε, inhibitor of NF-κB kinase-ε IRF3, interferon regulatory factor-3; ISRE, interferon-stimulated response element.
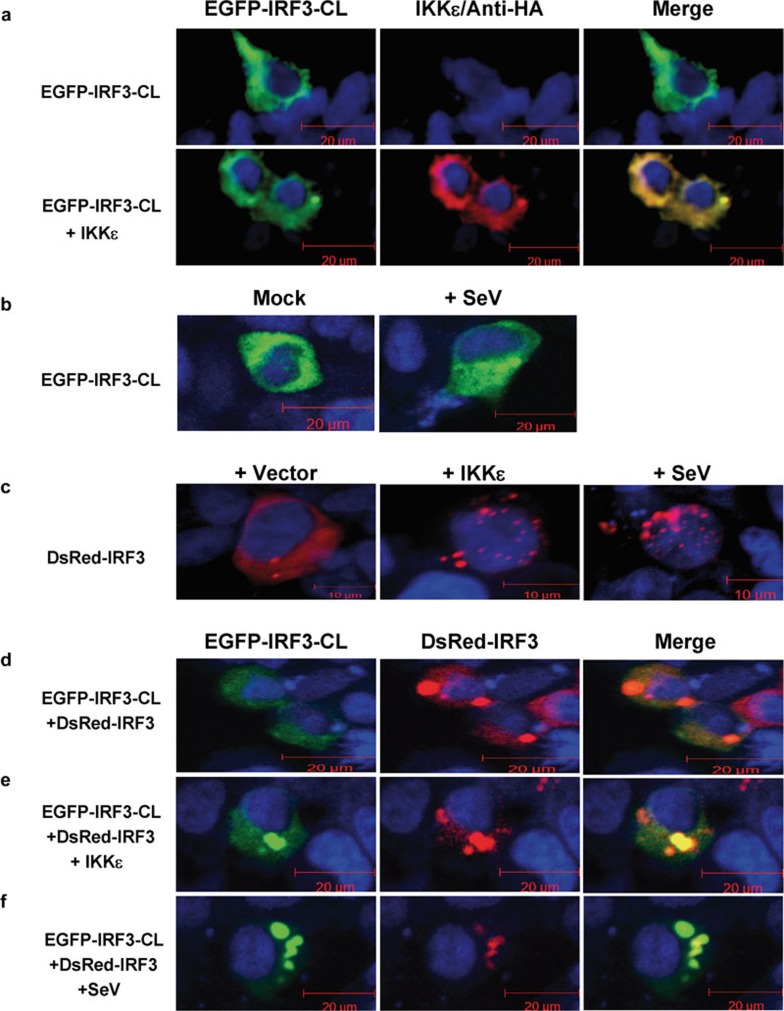
IRF3-CL inhibits IKKε-mediated and SeV-triggered nuclear translocation of IRF3
The shuttling of IRF3 between the cytoplasm and nucleus is regulated by a nuclear localization sequence and a nuclear export sequence at the amino terminus of IRF3, thus regulating expression of the inducible genes.18 As IRF3-CL is unique in the carboxyl-terminus compared to IRF3 and inhibits SeV-triggered and IKKε-mediated activation of IRF3 (Figures 3 and 4), we examined its localization and the effect on IKKε- and SeV-induced nuclear translocation of IRF3 by visualizing the behavior of EGFP-IRF3-CL or DsRed-IRF3 with fluorescent microscopy. IRF3-CL was clearly detected in the cytoplasm, and overexpression of IKKε and infection of SeV did not induce its translocation into nucleus from cytoplasm (Figure 5a and b), which occurs in the case of IRF3 (Figure 5c). When IRF3-CL was overexpressed, IRF3 did not translocate from the cytoplasm into the nucleus (Figure 5d), even at the induction by overexpression of IKKε (Figure 5e) and infection of SeV (Figure 5f). These observations show that overexpression of IRF3-CL inhibits IKKε- and SeV-triggered nuclear translocation of IRF3.
Figure 5.
IRF3-CL inhibits IKKε-mediated and SeV-triggered nuclear translocation of IRF3. (a) HEK293 cells were transfected with pEGFP-IRF3-CL with or without pRK-HA-IKKε. The expression of IKKε was confirmed by immunofluorescence of the same cells with anti-HA antibody. (b) HEK293 cells were transfected with pEGFP-IRF3-CL. After 24 h, cells were infected with SeV or left uninfected for 8 h. (c) HEK293 cells were transfected with pDsRed-IRF3 with or without pRK-HA-IKKε. After 24 h, cells were infected with SeV or left uninfected for 8 h. (d) HEK293 cells were cotransfected with pDsRed-IRF3 and pEGFP-IRF3-CL. (e) HEK293 cells were transfected with pDsRed-IRF3, pEGFP-IRF3-CL and pRK-HA-IKKε. (f) HEK293 cells were cotransfected with pDsRed-IRF3 and pEGFP-IRF3-CL. After 24 h, cells were infected with SeV or left uninfected for 8 h. Images represent 70–100% of the cells in the cultures. HA, hemagglutinin; IKKε, inhibitor of NF-κB kinase-ε IRF3, interferon regulatory factor-3; Sev, Sendai virus.
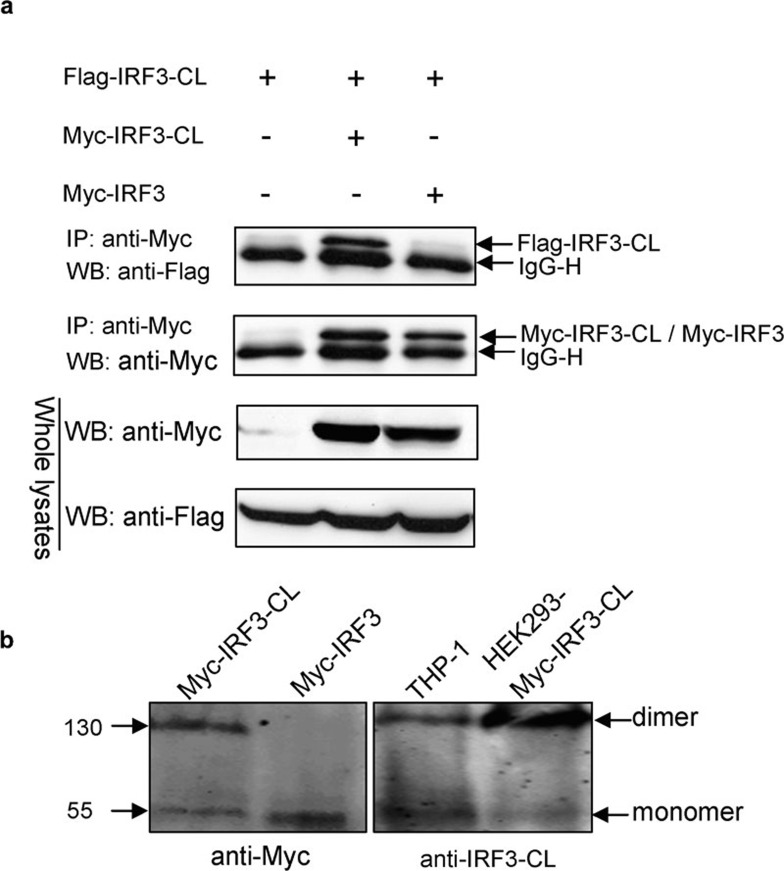
IRF3-CL forms a homodimer but not a heterodimer with IRF3 constitutively
Structure and mutation studies indicated that the amino terminus of IRF3 plays a critical role in homodimer formation.19, 20 Since IRF3-CL and IRF3 share a common amino terminus, we investigated whether IRF3-CL could form a homo- or a heterodimer with IRF3 constitutively. It was clear that Flag-tagged IRF3-CL was co-immunoprecipitated with Myc-tagged IRF3-CL but not with Myc-tagged IRF3 (Figure 6a), thus indicating the association between IRF3-CL molecules, but not between IRF3-CL and IRF3. In addition, after native PAGE, two bands with sizes of 110 and 55 kDa, corresponding to the dimer and monomer, respectively, were detected from lysates of HEK293 expressing Myc-tagged IRF3-CL using an anti-Myc antibody, and from lysates of THP-1 and HEK293 expressing Myc-tagged IRF3-CL using anti-IRF3-CL antiserum, respectively (Figure 6b). These results indicate that IRF3-CL can form a homodimer rather than a heterodimer with IRF3 constitutively.
Figure 6.
IRF3-CL forms a homodimer but not a heterodimer with IRF3 constitutively. (a) HEK293 cells were transfected with plasmids as indicated. After 24 h of transfection, co-immunoprecipitation was performed with anti-Myc antibody followed by WB with antibody as indicated. (b) Lysates from HEK293 cells transfected with plasmids as indicated and from THP-1 cells were resolved on native PAGE. Western blot was performed with anti-Myc or anti-IRF3-CL antiserum as indicated. IP, immunoprecipitation; IRF3, interferon regulatory factor-3; PAGE, polyacrylamide gel electrophoresis; WB, western blotting.
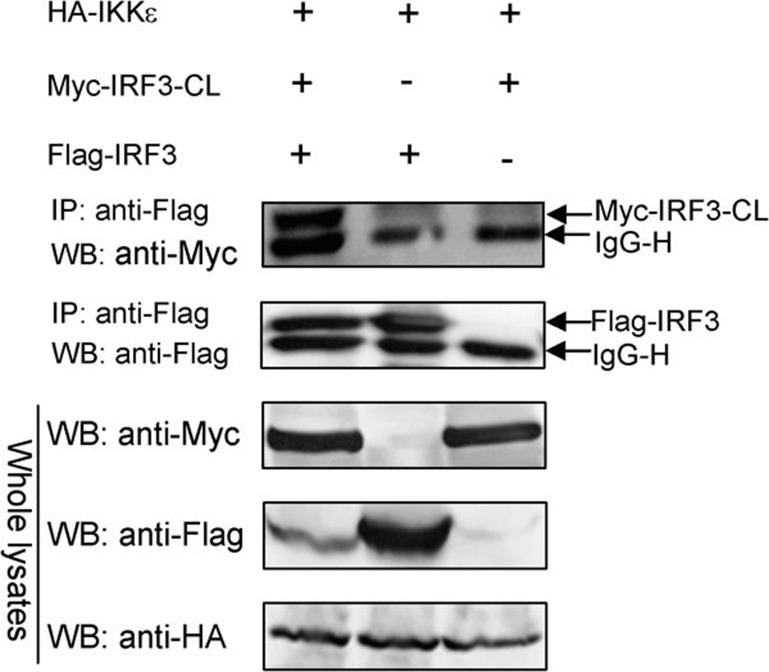
IRF3-CL is associated with IRF3 when IKKε is overexpressed
Upon activation, IRF3 is phosphorylated leading to relief of its intramolecular autoinhibition and formation of a homo- or heterodimer.3, 8 As IRF3-CL did not interact with IRF3 constitutively (Figure 6a) and overexpression of IRF3-CL inhibited IKKε- and SeV-induced nuclear translocation of IRF3 (Figure 5e and f), we further investigated whether IRF3-CL is associated with the activated IRF3. As shown (Figure 7), while IKKε was overexpressed, Myc-tagged IRF3-CL was co-immunoprecipitated with Flag-tagged IRF3. The result demonstrates that IRF3-CL interacts with IRF3 when IKKε is overexpressed.
Figure 7.
IRF3-CL forms a heterodimer with IRF3 when IKKε is overexpressed. HEK293 cells were transfected with plasmids as indicated. After 24 h, co-immunoprecipitation was performed with anti-Flag antibody (Sigma) followed by WB with antibody as indicated. IKKε, inhibitor of NF-κB kinase-ε IP, immunoprecipitation; IRF3, interferon regulatory factor-3; WB, western blotting.
Discussion
IRF3 has been functionally characterized as a vital transcriptional factor and a key regulator in the immune response to pathogen infection and DNA damage. Different mechanisms of regulation of activity of IRF3 have been reported, including positive-feedback regulation by IRF7,21 regulation of phosphorylation and dimerization of IRF3 by Cyclophilin B,22 negative regulation by PIN1 to facilitate the degradation of IRF3,23 and by SIKE to disrupt interactions of IKKε or TBK1 with TRIF, RIG-I and IRF3.24 In addition, several splicing isoforms of IRF3, such as IRF-3a (Ref. 13) and IRF3-nirs3,25 have been identified and functioned in the negative modulation of the activity of IRF3. In this study, we describe another splicing variant of IRF3 and suggest that the alternative splicing of the IRF3 primary transcript plays a key role in the fine-tuning of the activity of IRF3.
In the splicing of pre-mRNA to produce IRF3-CL, a different 3′ splice site was used, which is common in aberrant splicing but different with production of IRF-3a ORF.26 Like IRF3 and IRF-3a,IRF3-CL is also ubiquitously expressed in distinct cell lines. However, the transcript level of IRF3-CL is lower than that of IFR3, as we found that averagely one IRF3-CL cDNA clone out of nine IRF3 cDNA clones from sequencing results (data not shown), which is similar to the low ratio of IRF-3a to IRF3.26 Several Ser/Thr residues in the unique C-terminal region of IRF3-CL were predicted to be potential phosphorylation sites, which might also account for the slightly higher weight of 55 kDa than calculated 49 kDa.
In reporter gene and Q-PCR assays, overexpression of IRF3-CL inhibits SeV-triggered and IKKε-mediated activation of ISRE and the IFN-β promoter, as well as expression of IFN-β induced by SeV infection (Figures 3 and 4), indicating that IRF3-CL negatively regulates the activity of IRF3. Previous studies indicated that the formation of a homodimer of IRF3 is mediated by the IAD and only a portion of IAD is required.1, 3 Sharma et al. demonstrated that expression of IKKε or TBK1 induced phosphorylation of IRF3, resulting in IRF3 dimerization and translocation into the nucleus to induce transcription of target genes.5 In our results, overexpression of IRF3-CL inhibited IKKε- and SeV-induced nuclear translocation of IRF3 and overexpression of IKKε induced the interaction of IRF3 with IRF3-CL. Thus, the activated IRF3 might be sequestered in the cytoplasm by IRF3-CL resulting in the inhibition of its nuclear translocation, then reducing its transcriptional activity. Likewise, IRF-3a inhibited activity of the IFN-β promoter through forming a heterodimer with IRF3 following virus infection,13 suggesting the association of an isoform with IRF3 may be a common way for isoforms to negatively affect IRF3.
Unlike the variant IRF3-nirs3, which translocated into the nucleus upon activation, IRF3-CL is localized in the cytoplasm even with coexpression of IKKε and infection of SeV (Figure 5a and b). Thus, it is impossible to speculate that IRF3-CL negatively regulates the activity of IRF3 through competing with IRF3 for binding to DNA, by which the other variants of IRF3 were postulated to downregulate the activity of IRF3. Since IRF3-CL was detected to be nearly colocalized with IKKε in the cytoplasm (Figure 5a) and target IKKε-mediated IFN-β signaling, we speculated that IRF3-CL might compete with IRF3 for association with IKKε within the cytoplasm to reduce the activity of IRF3. Further studies will be necessary to clarify the precise mechanism.
Acknowledgments
We thank Rongjuan Pei for technical help. This work was supported by grants from the China National Human Liver Proteomics Project (2004BA711A19), the China National High-Tech 863 Program (2006AA 02A310), the Natural Science Foundation of Hubei Province of China (2009CDB012) and the Educational Commission of Hubei Province of China (D20091004).
References
- Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant MJ, LePage C, et al. Interferon regulatory factors: the next generation. Gene. 1999;237:1–14. doi: 10.1016/s0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- Lin R, Mamane Y, Hiscott J. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol Cell Biol. 1999;19:2465–2474. doi: 10.1128/mcb.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci USA. 2004;101:233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Hiscott J, Pitha P, Genin P, Nguyen H, Heylbroeck C, Mamane Y, et al. Triggering the interferon response: the role of IRF-3 transcription factor. J Interferon Cytokine Res. 1999;19:1–13. doi: 10.1089/107999099314360. [DOI] [PubMed] [Google Scholar]
- Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Mak TW, Sen G, Li X. Toll-like receptor 3-mediated activation of NF-kappaB and IRF3 diverges at Toll-IL-1 receptor domain-containing adapter inducing IFN-beta. Proc Natl Acad Sci USA. 2004;101:3533–3538. doi: 10.1073/pnas.0308496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, Materna T, et al. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J Immunol. 2004;173:7416–7425. doi: 10.4049/jimmunol.173.12.7416. [DOI] [PubMed] [Google Scholar]
- Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Karpova AY, Ronco LV, Howley PM. Functional characterization of interferon regulatory factor 3a (IRF-3a), an alternative splice isoform of IRF-3. Mol Cell Biol. 2001;21:4169–4176. doi: 10.1128/MCB.21.13.4169-4176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Jo M, Park J, Zhang W, Lee JH. Alternative splicing variants of IRF-1 lacking exons 7, 8, and 9 in cervical cancer. Biochem Biophys Res Commun. 2006;347:882–888. doi: 10.1016/j.bbrc.2006.06.145. [DOI] [PubMed] [Google Scholar]
- Iwamura T, Yoneyama M, Yamaguchi K, Suhara W, Mori W, Shiota K, et al. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells. 2001;6:375–388. doi: 10.1046/j.1365-2443.2001.00426.x. [DOI] [PubMed] [Google Scholar]
- Bibeau-Poirier A, Gravel SP, Clement JF, Rolland S, Rodier G, Coulombe P, et al. Involvement of the IkappaB kinase (IKK)-related kinases TANK-binding kinase 1/IKKi and cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J Immunol. 2006;177:5059–5067. doi: 10.4049/jimmunol.177.8.5059. [DOI] [PubMed] [Google Scholar]
- Cheng TF, Brzostek S, Ando O, van Scoy S, Kumar KP, Reich NC. Differential activation of IFN regulatory factor (IRF)-3 and IRF-5 transcription factors during viral infection. J Immunol. 2006;176:7462–7470. doi: 10.4049/jimmunol.176.12.7462. [DOI] [PubMed] [Google Scholar]
- Kumar KP, McBride KM, Weaver BK, Dingwall C, Reich NC. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol Cell Biol. 2000;20:4159–4168. doi: 10.1128/mcb.20.11.4159-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K, Suzuki NN, Horiuchi M, Mori M, Suhara W, Okabe Y, et al. X-ray crystal structure of IRF-3 and its functional implications. Nat Struct Biol. 2003;10:922–927. doi: 10.1038/nsb1001. [DOI] [PubMed] [Google Scholar]
- Yang H, Lin CH, Ma G, Orr M, Baffi MO, Wathelet MG. Transcriptional activity of interferon regulatory factor (IRF)-3 depends on multiple protein-protein interactions. Eur J Biochem. 2002;269:6142–6151. doi: 10.1046/j.1432-1033.2002.03330.x. [DOI] [PubMed] [Google Scholar]
- Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Obata Y, Yamamoto K, Miyazaki M, Shimotohno K, Kohno S, Matsuyama T. Role of cyclophilin B in activation of interferon regulatory factor-3. J Biol Chem. 2005;280:18355–18360. doi: 10.1074/jbc.M501684200. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T, et al. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- Huang J, Liu T, Xu LG, Chen D, Zhai Z, Shu HB. SIKE is an IKK epsilon/TBK1-associated suppressor of TLR3- and virus-triggered IRF-3 activation pathways. Embo J. 2005;24:4018–4028. doi: 10.1038/sj.emboj.7600863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marozin S, Altomonte J, Stadler F, Thasler WE, Schmid RM, Ebert O. Inhibition of the IFN-beta response in hepatocellular carcinoma by alternative spliced isoform of IFN regulatory factor-3. Mol Ther. 2008;16:1789–1797. doi: 10.1038/mt.2008.201. [DOI] [PubMed] [Google Scholar]
- Karpova AY, Howley PM, Ronco LV. Dual utilization of an acceptor/donor splice site governs the alternative splicing of the IRF-3 gene. Genes Dev. 2000;14:2813–2818. doi: 10.1101/gad.813800. [DOI] [PMC free article] [PubMed] [Google Scholar]