Abstract
AIM
To solidify the involvement of Saa-related pathway in corneal neovascularization (CorNV). The pathogenesis of inflammatory CorNV is not fully understood yet, and our previous study implicated that serum amyloid A (Saa) 1 (Saa1) and Saa3 were among the genes up-regulated upon CorNV induction in mice.
METHODS
Microarray data obtained during our profiling project on CorNV were analyzed for the genes encoding the four SAA family members (Saa1-4), six reported SAA receptors (formyl peptide receptor 2, Tlr2, Tlr4, Cd36, Scarb1, P2rx7) and seven matrix metallopeptidases (Mmp) 1a, 1b, 2, 3, 9, 10, 13 reportedly to be expressed upon SAA pathway activation. The baseline expression or changes of interested genes were further confirmed in animals with CorNV using molecular or histological methods. CorNV was induced in Balb/c and C57BL/6 mice by placing either three interrupted 10-0 sutures or a 2 mm filter paper soaked with sodium hydroxide in the central area of the cornea. At desired time points, the corneas were harvested for histology examination or for extraction of mRNA and protein. The mRNA levels of Saa1, Saa3, Fpr2, Mmp2 and Mmp3 in corneas were detected using quantitative reverse transcription-PCR, and SAA3 protein in tissues detected using immunohistochemistry or western blotting.
RESULTS
Microarray data analysis revealed that Saa1, Saa3, Fpr2, Mmp2, Mmp3 messengers were readily detected in normal corneas and significantly up-regulated upon CorNV induction. The changes of these five genes were confirmed with real-time PCR assay. On the contrary, other SAA members (Saa2, Saa4), other SAA receptors (Tlr2, Tlr4, Cd36, P2rx7, etc), or other Mmps (Mmp1a, Mmp1b, Mmp9, Mmp10, Mmp13) did not show consistent changes. Immunohistochemistry study and western blotting further confirmed the expression of SAA3 products in normal corneas as well as their up-regulation in corneas with CorNV.
CONCLUSION
SAA-FPR2 pathway composing genes were expressed in normal murine corneas and, upon inflammatory stimuli challenge to the corneas, their expressions were up-regulated, suggesting their roles in pathogenesis of CorNV. The potential usefulness of SAA-FPR2 targets in future management of CorNV-related diseases deserves investigation.
Keywords: corneal neovascularization, serum amyloid A, formyl peptide receptor, matrix metallopeptidase, inflammation
INTRODUCTION
Neovascularization occurs in response to injury of tissues, supposedly to favor reconstruction of the structure of the affected tissues. When neovascularization develops in the naturally vessel-deficient tissues or organs like cornea and cartilage, however, it may destroy the structures or functions of the tissues instead, either temporarily or permanently. Specifically, growth of vessels from limbal vascular plexus into the cornea blurs the light path needed for a good vision or biologically alters the refractory characteristics of the corneas. Often encountered insults that induce corneal neovascularization (CorNV) include hypoxia, burn, ischemia, infection, trauma, or even therapeutic operation[1]. Transient or mild CorNV might reverse when initial insults are removed, but lasting or serious CorNV causes heavy vision loss or blindness[2]. Among all pathological processes started by above etiology factors, inflammation is dominant[3]. Hence many studies are carried out using experimental CorNV in animals to dissect the interactions between inflammation and neovascularization. In a serial study addressing the molecular pathogenesis of experimental murine CorNV initiated by inflammatory stimuli, we used microarray strategy to monitor the transcriptome changes during the development of CorNV[4]. Through that project, we made some novel findings like that nonenzymatic crystallins are expressed at high level in normal corneas and might contribute to the maintenance or restoration of transparency of corneas[5],[6]. Here we report that, after mining of the microarray data, another family that is closely related with inflammation, namely serum amyloid A (SAA), might be of potential interest in the pathogenesis of neovascularization. Specifically, two members of SAA family, namely Saa1 and Saa3, and one of their receptor formyl peptide receptor 2 (Fpr2, also known as Fprl1), were up-regulated in the CorNV context[4]. In fact, as one of the main acute phase reactant families, SAA family members are involved in many pathological process and diseases like tumor[7]-[9], infection[10],[11], autoimmunity [12],[13], cardiovascular diseases[14],[15], etc. So far, reported receptors for SAA in various cells or tissues include FPR2 or its like[16],[17], CD36 or its like[18]-[20], TLR2[21] or TLR4[22], and purinergic receptor P2X, ligand-gated ion channel, 7 (P2RX7, also known as P2X7 receptor)[23]. In one of downstream pathways following coupling of SAA will their receptor(s), production of matrix metalloproteinases (MMPs) is among the main outcomes[13],[16]. While MMPs have been often associated with various corneal pathological processes, the precedent SAA-FPR signal pathway has not been addressed before in any cornea-related processes[1],[24],[25]. Thus this study was performed to confirm the involvement of SAA in cornea physiology and CorNV.
MATERIALS AND METHODS
Animal Model
The general design and procedure of the experiments were described elsewhere[4]. In brief, inbred Balb/c and C57BL/6 mice, female, 6-8wk old were purchased from Chinese Academy of Medical Sciences (Beijing, China) and used following the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and the institutional guideline. Mice were anesthetized with intraperitoneal chlorpromazine and ketamine plus topical application of Benoxil (Santen, Osaka, Japan). For suture-induced CorNV (S-CorNV), three interrupted stitches of 10-0 polypropylene suture (MANI Inc., Togichi, Japan) were placed at about 1 mm from the corneal apex. For induction of chemical burn-induced CorNV (CB-CorNV), a 2 mm paper filter soaked with 1.5 µL 1 mol/L NaOH was laid on central corneas for 40s, follow which the eye and the conjunctival sacs were rinsed with saline buffer. The sacrificed eyes were checked daily using a slit lamp to monitor the growth of blood vessels into the cornea. For all experiments, only one eye of each mouse was used for CorNV induction and the other eye was reserved as control. Pilot studies showed that new vessels grew most fast at day 5 (D5) in S-CorNV model and D6 in CB-CorNV, and reached maximum length around D10 in S-CorNV and D14 in CB-CorNV.
Microarray Data Retrieval and Analysis
The previous project concerning microarray profiling of experimental CorNV has been described and the data are deposited in the public Gene Expression Omnibus (GEO) of National Center for Biotechnology Information with an accession number GSE23347[4],[26]. In that study, totally twelve microarrays were used to track the gene expression change at different time points in different CorNV models, namely D5 or D10 after S-CorNV induction (in Balb/c mice), and D6 or D14 after CB-CorNV induction (in Balb/c and C57BL/6 mice). In current study, the normalized data of the genes related with SAA pathways were retrieved from the dataset and their expression levels or change folds in CorNV models were analyzed. These include genes encoding for another acute phase protein (C-reactive protein, CRP), four members of SAA family, six reported receptors for SAA, and seven MMPs (Table 1). Promising genes were subjected to further investigation by experimental studies.
Table 1. Summary of the genes associated with Saa and possibly involved in CorNV.
| Gene symbol | Gene ID | Gene description | Brief notes of interest | References |
| Crp | NM_007768 | C-reactive protein, pentraxin-related | Another acute-phase protein often accompanying thus compared with Saa | 27-29 |
| Saa1 | NM_009117 | Serum amyloid A1 | Main isoforms of inducible Saa, expressed in liver upon inflammation, stress, neoplasia, etc | 30-33 |
| Saa2 | NM_011314 | Serum amyloid A2 | ||
| Saa3 | NM_011315 | Serum amyloid A3 | Induced in multiple non-liver tissues in non-human mammals. Human Saa3 is a pseudogene | 34-37 |
| Saa4 | NM_011316 | Serum amyloid A4 | Constitutively expressed in liver as minor apolipoproteins | 38, 39 |
| Fpr2 | NM_008039 | Formyl peptide receptor 2 | Most studied receptor of Saa, orthologue of human FprL1 | 40, 41 |
| Cd36 | NM_007643 | Cd36 antigen, transcript variant 2 | Synonym=platelet glycoprotein IV, Scarb3 | 18-20 |
| Scarb1 | NM_016741 | Scavenger receptor class B, member 1, transcript variant 1 | Synonym=Cd36-like 1, SR-BI, Cla-1. Binding by Saa blocks functions of other Scarb1 ligands | 18, 20, 42, 43 |
| Tlr2 | NM_011905 | Toll-like receptor 2 | TLR-bound Saa acts like adjuvant and activates mainly monocytes | 21, 44, 45 |
| Tlr4 | NM_021297 | Toll-like receptor 4 | 22, 46-48 | |
| P2rx7 | NM_011027 | Purinergic receptor P2X, ligand-gated ion channel, 7, transcript variant 1 | By coupling P2X7R, SAA activates NLRP3 inflammasome pathway | 23, 49 |
| Mmp1a | NM_032006 | Matrix metallopeptidase 1a | Saa reportedly induces secretion of different Mmps from various cells or tissues, which in turn degrade Saa or other matrix components | 13, 48, 50, 51 |
| Mmp1b | NM_032007 | Matrix metallopeptidase 1b | ||
| Mmp2 | NM_008610 | Matrix metallopeptidase 2 | 51, 52 | |
| Mmp3 | NM_010809 | Matrix metallopeptidase 3 | 13, 48, 51, 52 | |
| Mmp9 | NM_013599 | Matrix metallopeptidase 9 | 16, 51 | |
| Mmp10 | NM_019471 | Matrix metallopeptidase 10 | 53 | |
| Mmp13 | NM_008607 | Matrix metallopeptidase 10 | 48, 51 |
Real Time-PCR Assay
At chosen time points, the corneas were excised using a 2 mm trephine and placed in ice-cold TRIzol reagent (Invitrogen, Gaithersburg, MD, USA) and total RNA was extracted using isopropanol precipitation, followed by purification with NucleoSpin RNA clean-up columns (MACHEREY-NAGEL, Düren, Germany). RNA from 3 corneas was pooled to yield one RNA sample, and three samples were included per group. One microgram total RNA from each pool was reverse transcribed into cDNA using a PrimeScript RT Reagent Kit [TaKaRa Biotechnology (Dalian) Co., Ltd, Dalian, China] following the instruction of the manufacturer. The expression levels of interested genes were detected using real time (RT)-PCR with the TaqMan probes and primers (Table 2). In brief, amplification for each sample was performed in triplicate in an ABI 7500 Detection System (Applied Biosystems, Foster City, CA, USA) and the amplification protocol comprised an initial 10min incubation at 95°C followed by 50 cycles of 15s at 95°C and 1min at 60°C. The data were analyzed using accompanying software and threshold cycle (Ct) values were obtained. The average of three duplicates was used to calculate the relative Ct against reference gene Rpl5 (dCt=CtGene-CtRpl5) for each sample. Then the average dCt for the three samples in control groups were used to calculate the ddCt of each CorNV samples (ddCt=dCtCorNV-dCtcontrol). The relative expression folds of genes in the CorNV samples were calculated as 1/2ddCt.
Table 2. Primers and probes for the genes detected in this study.
| Gene | Oligo | Sequences | Amplicon (bp) |
| Saa1 | F | GAGTCTGGGCTGCTGAGAA | 78 |
| R | TGGTGTCCTCATGTCCTCTG | ||
| P | FAM-TTCCTGAAAGGCCTCTCTTCCATCA-TAMRA | ||
| Saa3 | F | CTGGGCTGCTAAAGTCATCA | 73 |
| R | TGAGTCCTCTGCTCCATGTC | ||
| P | FAM-TGAACAGCCTCTCTGGCATCGC-TAMRA | ||
| Fpr2 | F | TTACAGCAGTTGTGGCTTCC | 77 |
| R | TAAACCAGACTGTGCCCAAA | ||
| P | FAM-TTTCCCTTTCAGCTTGTGGCCC-TAMRA | ||
| Mmp2 | F | CTGGGAGCATGGAGATGGATA | 96 |
| R | AAGTGAGAATCTCCCCCAACA | ||
| P | FAM-ACATGCCTTTGCCCCGGGCA-TAMRA | ||
| Mmp3 | F | GGAGATGCTCACTTTGACGA | 78 |
| R | TGAGCAGCAACCAGGAATAG | ||
| P | FAM-TGACATCCTCTGTCCATCGTATCGTTCATCA-TAMRA | ||
| Rpl5 | F | GGAAGCACATCATGGGTCAGA | 70 |
| R | TACGCATCTTCATCTTCCTCCATT | ||
| P | FAM-TGTGGCAGACTACATGCGCTACC-TAMRA |
Western Blotting
At desired time points after CorNV induction, corneas were harvested as above and three corneas were pooled as one sample. Total proteins were extracted using RIPA lysis buffer (50 mmol/L Tris pH 7.4, 150 mmol/L NaCl, 1%Triton X-100, 1% sodium deoxycholate, 1% sodium dodecyl sulfate, sodium orthovanadate, and sodium fluoride; Beyotime, Shanghai, China) as suggested by the manufacturer. Samples were quantified using bicinchoninic acid (BCA) method and 50 µg of proteins was resolved on 12% SDS-PAGE gels for 1.5h at 120 V and then transferred to nitrocellulose membranes (Millipore, Billerica, MA, USA). The blots were soaked for blocking in 5% nonfat dry milk in Tris-Buffered Sabline with Tween 20 (TBST buffer) for 1h, incubated with polyclonal rabbit anti-SAA (sc-20651, recognizing SAA1 and SAA2 of human but only SAA3 of mouse. Santa Cruz Biotechnology, Santa Cruz, CA, USA) or anti-GAPDH (KC-5G5; KangChen Biotech, Shanghai, China) antibodies in TBST for 1h, followed by incubation with horse-radish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibody (MAXIM BIO, Fuzhou, China) for 1h. All incubations were done at room temperature, and three washes with 10 mL TBST buffer were applied between each step. The membranes were then developed with SuperSignal West Femto Maximum Sensitivity substrate (Pierce Biotechnology, Rockford, IL, USA) and exposed to X-ray film (Kodak, Rochester, NY, USA). The bands were analyzed using NIH Image 1.62 software (NIH, Bethesda, MD, USA). For each sample, the levels of SAA3 were normalized to that of GAPDH.
Immunohistochemistry
As desired times, enucleated eyeballs were embedded in a paraffin block and subjected to routine immunohistochemistry. Three animals were used in each group and serial sections were prepared to ensure high quality of staining results. Polyclonal anti-SAA in combination with HRP-conjugated goat anti-rabbit IgG antibody as mentioned above was used. After developing with 3,3′-diaminobenzidine, the sections were counterstained with hematoxylin. All sections were observed using an E800 microscope (Nikon, Tokyo, Japan) with appropriate digital camera.
Statistical Analysis
Wherever statistical analysis was appropriate, Student's t-test was performed, and P<0.05 was considered significant for difference.
RESULTS
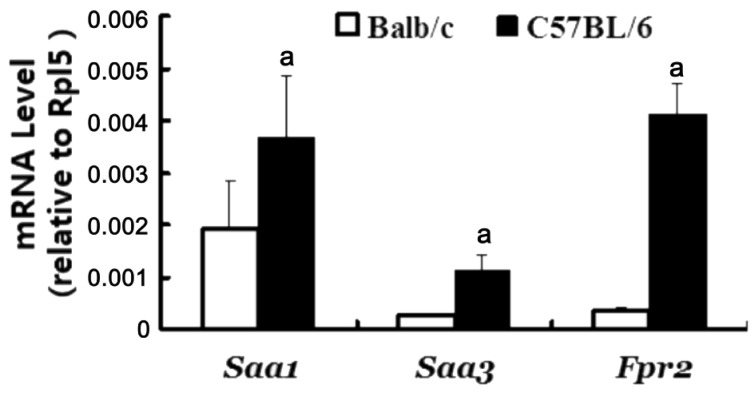
In a previous paper based on our microarray profiling projects performed on S- and CB-CorNV models in mice, Saa1, Saa3 and Fpr2 were listed among the up-regulated genes at D5 in CorNV and D6 in CB-CorNV, but not any discussion on these three genes was attempted[4]. Now, extending of vision to other time points and to other related genes demonstrated that these three genes as well as Mmp2, Mmp3, Mmp13 were significantly up-regulated in the inflammatory CorNV models in both mice strains at one or more time points (Table 3). On the contrary, Tlr2 and Mmp10 were detectable only in C57BL/6 mice, while Crp, Saa2, Saa4, Tlr4, Cd36, Scarb1, P2rx7, Mmp1a, Mmp1b, Mmp9 were not consistently detectable in any conditions studied here. Following this data mining step, the five genes with highest detection rates, namely the two SAA members (Saa1, Saa3), one receptor (Fpr2) and two respondents (Mmp2, Mmp3), were further detected with RT-PCR methodology. Except for Fpr2 data at D6 in CB-CorNV in C57BL/6 mice, all other changes recorded in microarray were confirmed by RT-PCR (Table 4). When the animal strain was taken into consideration, it is noteworthy that beside the change folds, the baseline expression level of Fpr2 was also significantly different between Balb/c and C57BL/6 mice (Figure 1), again alerting us of the genetic dependence of any pathological processes like CorNV. Furthermore, expression of SAA3 in either normal or vascularized corneas was also detectable at protein level, as illustrated in the immunohistochemistry and western blot assays (Figure 2). Confirmation of other gene products in cornea and their change were not attempted.
Table 3. Expression of Saa-Fpr2-Mmps in murine CorNV measured by microarraya.
| Gene symbol | S-CorNV (Balb/c) |
CB-CorNV (Balb/c) |
CB-CorNV (C57BL/6) |
||
| D5 (n=3) | D10 (n=2) | D6 (n=3) | D14 (n=2) | D6 (n=2) | |
| Saa1 | 27.71±12.37 | 12.19±5.18 | 9.47±1.08 | 13.34±1.13 | |
| Saa3 | 85.59±9.77 | 91.93±101.65 | 35.14±3.70 | 13.73±14.00 | 28.43±5.48 |
| Fpr2 | 26.28±15.33 | 26.33±2.52 | 16.59±6.13 | 23.57±0.66 | |
| Tlr2 | 5.81±0.14 | ||||
| Mmp2 | 5.20±3.98 | 7.26±2.02 | 3.86±0.73 | 4.21±0.36 | |
| Mmp3 | 25.20±5.84 | 24.78±0.97 | 50.79±21.63 | 17.39±4.74 | 33.73±8.59 |
| Mmp10 | 22.08±4.35 | ||||
| Mmp13 | 40.86±38.30 | 25.39±6.81 | 59.37±41.41 | ||
aThe results presented in this table were summarized from our data that had been deposited in GEO[26]. Data of Saa1, Saa3 and Fpr2 on D5 of S-CorNV and D6 of CB-CorNV were previously reported elsewhere, but listed here for better comparison[4]. Numbers in brackets denote the number of arrays for that group in original experiments.
Table 4. Relative expression levels of genes in murine CorNV as detected by RT-PCR.
| Gene symbol | S-CorNV (Balb/c) |
CB-CorNV(Balb/c) |
CB-CorNV(C57BL/6) |
|||
| D5 | D10 | D6 | D14 | D6 | D14 | |
| Saa1 | 23.2±2.2a | 9.0±1.7 | 8.2±1.2 | 5.6±0.7 | 7.9±1.4 | 13.6±2.1 |
| Saa3 | 1643.7±209.8 | 176.8±45.3 | 365.9±91.5 | 68.2±12.4 | 26.7±2.9 | 358.5±31.0 |
| Fpr2 | 65.5±2.8 | 77.0±3.0 | 24.6±2.2 | 18.2±0.2 | 1.3±0.3b | 5.4±0.5 |
| Mmp2 | 59.3±3.1 | 106.5±1.6 | 23.5±0.9 | 58.2±6.5 | 4.2±0.7 | 4.5±0.2 |
| Mmp3 | 1255.1±73.0 | 2139.9±58.3 | 316.3±73.0 | 490.3±50.3 | 685.8±66.1 | 1821.1±161.6 |
aFolds, average±standard error of three samples in each experimental group. In the rationale described in the methods, the expression level of each gene normalized against Rpl5 in untreated corneas was set at 1. Experiments were performed twice with similar conclusions. bThis number, and only this number, is much below the fold change (23.57±0.66, refer to Table 3) observed in microarray analysis under same condition.
Figure 1. Expression levels of Saa1, Saa3 and Fpr2 in murine corneas.
The relative mRNA intensities of genes were obtained by real-time PCR assay, via 1/2dCt, where dCt=CtGene-CtRpl5. Each sample was run in triplicate in PCR reaction, and the error bars represented standard errors for three samples in each group. Shown was one of two experiments with similar results. aP<0.05 between Balb/c and C57BL/6 strains.
Figure 2. Clinical manifestation of inflammatory corneal neovascularization and SAA3 expression in the corneas.
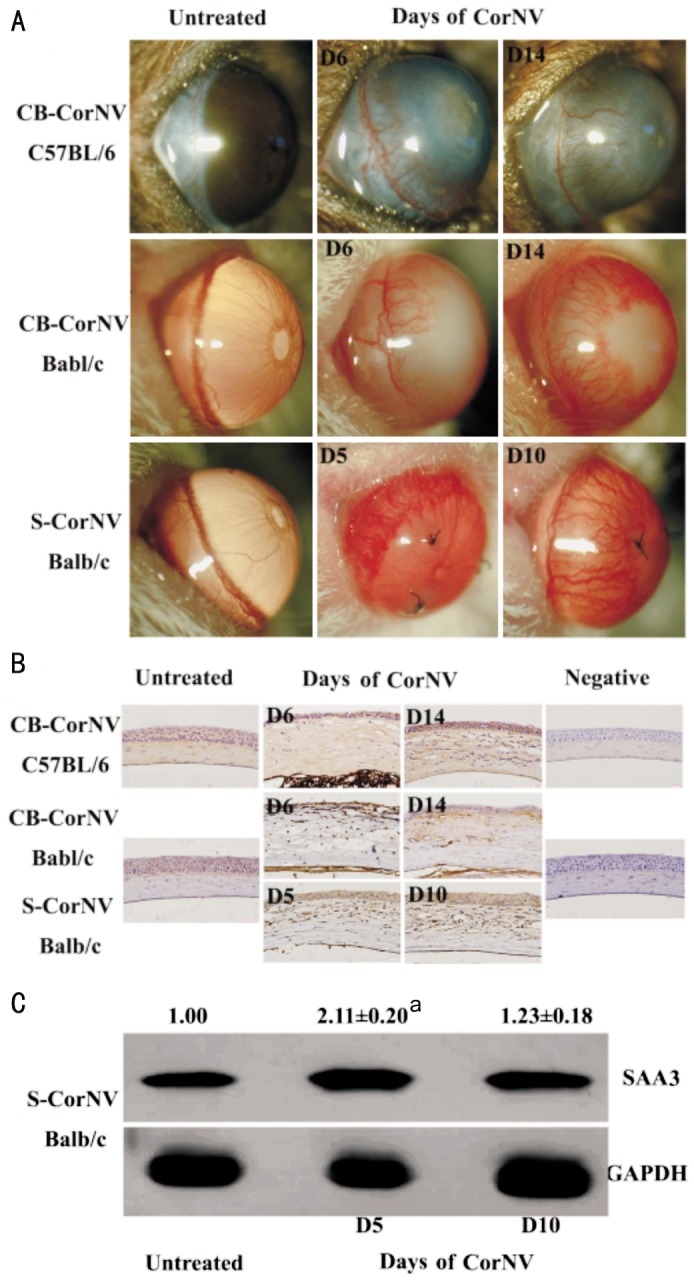
A: To compare the potential effect of animal strain or inflammatory stimuli on the expression pattern of SAA3, Balb/c and C57BL/6 mice were subjected to S-CorNV or CB-CorNV respectively; B: At desired time points, eyeballs were harvested for immunohistochemistry. Three corneas were included in each group, and shown were images from representative tissues; C: Corneas were harvested for protein extraction and western blotting. Intensity of GAPDH in untreated corneas was arbitrarily set at 1.00, and that of CorNV corneas was compared to untreated corneas. Experiments were performed three times with similar results. Numbers above the SAA3 bands (mean±standard error) were obtained from three independent experiments, respectively, and aP<0.05 vs untreated control.
DISCUSSION
Though simple and straightforward, the findings reported here are not of less significance. First, like other acute-phase proteins, SAA is mainly produced by hepatocytes though many other tissues reportedly express SAA to various abundance[33],[54]. To the best of our knowledge, this is the first study to show that Saa genes and their receptor Fpr2 are expressed in corneas, thus expanding our knowledge about the distribution and functions of this pair of players. In specific, cornea is avascular and immune-privileged hence deserves a protective system that would respond quickly and efficiently to either acute or chronic inflammation caused by exogenous insults like trauma or infective. Existence of Saa mRNA and protein product makes SAA perfect candidates of such protective component. Contrary to Saa, however, the other main acute phase protein, namely Crp, was undetectable by microarray assay in our study system, implying differential involvement of these two classes of acute phase proteins in corneal physiopathology, just like noted in other conditions[29].
As with the possible SAA receptors that cooperate with SAA1 or SAA3 in CorNV, FPR2 was the only one that manifested significant mRNA changes at all detected time points of CorNV in both strains, and Tlr2 only in C57BL/6 mice (Tables 3, 4), while Cd36, Scarb1, Tlr4 and P2rx7 were not detectable in any conditions. Actually we also looked at several other receptors (e.g. Fpr1, Fpr-rs1, Fpr-rs3, Fpr-rs4, Fpr3) that shown sequence homology and functional similarity with FPR2, and found that none of them manifested detectable expression in the detected samples, leaving FPR2 as the only candidate receptor for SAA1/SAA3 in the corneas. Taking a step further, once FPR2 are to be activated, MMP2 and MMP3 are likely among the effector molecules produced in the studied CorNV context. In another word, though MMP2/MMP3 were also possibly induced by other inflammatory mediators via other pathways, the SAA-FPR2-MMP pathway reportedly to function in other environments might as well function in cornea inflammation[13],[52]. Considering that CorNV is one of the faithful angiogenesis or neovascularization models, and that evidence are coming up to show that SAA stimulate angiogenesis via direct action on vascular endothelial cells, we propose that therapies or protocols targeting SAA, FPR2 or MMP should be tested for their potency in managing inflammation or resultant neovascularization-related diseases[55],[56]. This strategy is in line with, thus supported by, a dozen of patents that target SAA for novel treatments of inflammation-derived diseases in either human or animals[57]. To address the contribution of SAA-FPR2-MMP pathway to the overall CorNV pathogenesis, more experimental studies, such as using Fpr2-deficient mice, are required to check whether interfering SAA-FPR2-MMP pathway helps to prevent or cure inflammatory CorNV or other related diseases.
Acknowledgments
Foundation: Supported by National Natural Science Foundation of China (No.81200664; 81271050)
Conflicts of Interest: Ren SW, None; Qi X, None; Jia CK, None; Wang YQ, None.
REFERENCES
- 1.Ellenberg D, Azar DT, Hallak JA, Tobaigy F, Han KY, Jain S, Zhou Z, Chang JH. Novel aspects of corneal angiogenic and lymphangiogenic privilege. Prog Retin Eye Res. 2010;29(3):208–248. doi: 10.1016/j.preteyeres.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qazi Y, Wong G, Monson B, Stringham J, Ambati BK. Corneal transparency: genesis, maintenance and dysfunction. Brain Res Bull. 2010;81(2–3):198–210. doi: 10.1016/j.brainresbull.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clements JL, Dana R. Inflammatory corneal neovascularization: etiopathogenesis. Semin Ophthalmol. 2011;26(4–5):235–245. doi: 10.3109/08820538.2011.588652. [DOI] [PubMed] [Google Scholar]
- 4.Jia C, Zhu W, Ren S, Xi H, Li S, Wang Y. Comparison of genome-wide gene expression in suture- and alkali burn-induced murine corneal neovascularization. Mol Vis. 2011;17:2386–2399. [PMC free article] [PubMed] [Google Scholar]
- 5.Ren S, Liu T, Jia C, Qi X, Wang Y. Physiological expression of lens alpha-, beta-, and gamma-crystallins in murine and human corneas. Mol Vis. 2010;16:2745–2752. [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu W, Qi X, Ren S, Jia C, Song Z, Wang Y. alphaA-crystallin in the pathogenesis and intervention of experimental murine corneal neovascularization. Exp Eye Res. 2010;98:44–51. doi: 10.1016/j.exer.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Moshkovskii SA. Why do cancer cells produce serum amyloid A acute-phase protein? Biochemistry (Mosc) 2012;77(4):339–341. doi: 10.1134/S0006297912040037. [DOI] [PubMed] [Google Scholar]
- 8.Malle E, Sodin-Semrl S, Kovacevic A. Serum amyloid A: an acute-phase protein involved in tumour pathogenesis. Cell Mol Life Sci. 2009;66(1):9–26. doi: 10.1007/s00018-008-8321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramankulov A, Lein M, Johannsen M, Schrader M, Miller K, Loening SA, Jung K. Serum amyloid A as indicator of distant metastases but not as early tumor marker in patients with renal cell carcinoma. Cancer Lett. 2008;269(1):85–92. doi: 10.1016/j.canlet.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 10.Harr KE, Rember R, Ginn PE, Lightsey J, Keller M, Reid J, Bonde RK. Serum amyloid A (SAA) as a biomarker of chronic infection due to boat strike trauma in a free-ranging Florida manatee (Trichechus manatus latirostris) with incidental polycystic kidneys. J Wildl Dis. 2011;47(4):1026–1031. doi: 10.7589/0090-3558-47.4.1026. [DOI] [PubMed] [Google Scholar]
- 11.Falsey AR, Walsh EE, Francis CW, Looney RJ, Kolassa JE, Hall WJ, Abraham GN. Response of C-reactive protein and serum amyloid A to influenza A infection in older adults. J Infect Dis. 2001;183(7):995–999. doi: 10.1086/319275. [DOI] [PubMed] [Google Scholar]
- 12.Pertovaara M, Jylhava J, Uusitalo H, Pukander J, Helin H, Hurme M. Serum amyloid A and C-reactive protein concentrations are differently associated with markers of autoimmunity in patients with primary Sjogren's syndrome. J Rheumatol. 2009;36(11):2487–2490. doi: 10.3899/jrheum.090300. [DOI] [PubMed] [Google Scholar]
- 13.O'Hara R, Murphy EP, Whitehead AS, FitzGerald O, Bresnihan B. Local expression of the serum amyloid A and formyl peptide receptor-like 1 genes in synovial tissue is associated with matrix metalloproteinase production in patients with inflammatory arthritis. Arthritis Rheum. 2004;50(6):1788–1799. doi: 10.1002/art.20301. [DOI] [PubMed] [Google Scholar]
- 14.King VL, Thompson J, Tannock LR. Serum amyloid A in atherosclerosis. Curr Opin Lipidol. 2011;22(4):302–307. doi: 10.1097/MOL.0b013e3283488c39. [DOI] [PubMed] [Google Scholar]
- 15.Kisilevsky R, Tam SP. Acute phase serum amyloid A, cholesterol metabolism, and cardiovascular disease. Pediatr Pathol Mol Med. 2002;21(3):291–305. doi: 10.1080/02770930290056523. [DOI] [PubMed] [Google Scholar]
- 16.Lee HY, Kim MK, Park KS, Bae YH, Yun J, Park JI, Kwak JY, Bae YS. Serum amyloid A stimulates matrix-metalloproteinase-9 upregulation via formyl peptide receptor like-1-mediated signaling in human monocytic cells. Biochem Biophys Res Commun. 2005;330(3):989–998. doi: 10.1016/j.bbrc.2005.03.069. [DOI] [PubMed] [Google Scholar]
- 17.Bjorkman L, Karlsson J, Karlsson A, Rabier MJ, Boulary F, Fu H, Bylund J, Dahlgren C. Serum amyloid A mediates human neutrophil production of reactive oxygen species through a receptor independent of formyl peptide receptor like-1. J Leukoc Biol. 2008;83(2):245–253. doi: 10.1189/jlb.0607-408. [DOI] [PubMed] [Google Scholar]
- 18.Mullan RH, McCormick J, Connolly M, Bresnihan B, Veale DJ, Fearon U. A role for the high-density lipoprotein receptor SR-B1 in synovial inflammation via serum amyloid-A. Am J Pathol. 2010;176(4):1999–2008. doi: 10.2353/ajpath.2010.090014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baranova IN, Bocharov AV, Vishnyakova TG, Kurlander R, Chen Z, Fu D, Arias IM, Csako G, Patterson A, Eggerman TL. CD36 is a novel serum amyloid A (SAA) receptor mediating SAA binding and SAA-induced signaling in human and rodent cells. J Biol Chem. 2010;285(11):8492–8506. doi: 10.1074/jbc.M109.007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baranova IN, Vishnyakova TG, Bocharov AV, Kurlander R, Chen Z, Kimelman ML, Remaley AT, Csako G, Thomas F, Eggerman TL, Patterson A. Serum amyloid A binding to CLA-1 (CD36 and LIMPII analogous-1) mediates serum amyloid A protein-induced activation of ERK1/2 and p38 mitogen-activated protein kinases. J Biol Chem. 2005;280(9):8031–8040. doi: 10.1074/jbc.M405009200. [DOI] [PubMed] [Google Scholar]
- 21.Cheng N, He R, Tian J, Ye PP, Ye RD. Cutting edge: TLR2 is a functional receptor for acute-phase serum amyloid A. J Immunol. 2008;181(1):22–26. doi: 10.4049/jimmunol.181.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandri S, Rodriguez D, Gomes E, Monteiro HP, Russo M, Campa A. Is serum amyloid A an endogenous TLR4 agonist? J Leukoc Biol. 2008;83(5):1174–1180. doi: 10.1189/jlb.0407203. [DOI] [PubMed] [Google Scholar]
- 23.Niemi K, Teirila L, Lappalainen J, Rajamaki K, Baumann MH, Oorni K, Wolff H, Kovanen PT, Matikainen S, Eklund KK. Serum amyloid A activates the NLRP3 inflammasome via P2X7 receptor and a cathepsin B-sensitive pathway. J Immunol. 2011;186(11):6119–6128. doi: 10.4049/jimmunol.1002843. [DOI] [PubMed] [Google Scholar]
- 24.Zou Y, Zhang H, Li H, Chen H, Song W, Wang Y. Strain-dependent production of interleukin-17/interferon-gamma and matrix remodeling-associated genes in experimental Candida albicans keratitis. Mol Vis. 2012;18:1215–1225. [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon GM, Austin JS, Sklar AL, Feuer WJ, LaGier AJ, Fini ME. Comprehensive gene expression profiling and functional analysis of matrix metalloproteinases and TIMPs, and identification of ADAM-10 gene expression, in a corneal model of epithelial resurfacing. J Cell Physiol. 2011;226(6):1461–1470. doi: 10.1002/jcp.22306. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Wang C, Jia C, Ren S, Yang L. Gene Expression profiling in murine corneal neovascularization models. GSE23347. 2011 http://www.ncbi.nlm.gov/geo. [Google Scholar]
- 27.Steel DM, Whitehead AS. The major acute phase reactants: C-reactive protein, serum amyloid P component and serum amyloid A protein. Immunol Today. 1994;15(2):81–88. doi: 10.1016/0167-5699(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 28.Pizzini C, Mussap M, Plebani M, Fanos V. C-reactive protein and serum amyloid A protein in neonatal infections. Scand J Infect Dis. 2000;32(3):229–235. doi: 10.1080/00365540050165848. [DOI] [PubMed] [Google Scholar]
- 29.Wilson PG, Thompson JC, Webb NR, de Beer FC, King VL, Tannock LR. Serum amyloid A, but not C-reactive protein, stimulates vascular proteoglycan synthesis in a pro-atherogenic manner. Am J Pathol. 2008;173(6):1902–1910. doi: 10.2353/ajpath.2008.080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knebel FH, Albuquerque RC, Massaro RR, Maria-Engler SS, Campa A. Dual effect of serum amyloid A on the invasiveness of glioma cells. Mediators Inflamm. 2013;2013:509089. doi: 10.1155/2013/509089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Campos JL, Calero C, Rojano B, Lopez-Porras M, Saenz-Coronilla J, Blanco AI, Sanchez-Lopez V, Tobar D, Montes-Worboys A, Arellano E. C-reactive protein and serum amyloid a overexpression in lung tissues of chronic obstructive pulmonary disease patients: a case-control study. Int J Med Sci. 2013;10(8):938–947. doi: 10.7150/ijms.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsui S, Yamane T, Kobayashi-Hattori K, Oishi Y. Calcitonin Gene-Related Peptide Upregulates Serum Amyloid A Synthesis through Activation of Interleukin-6. Biosci Biotechnol Biochem. 2013;77(10):2151–2153. doi: 10.1271/bbb.130450. [DOI] [PubMed] [Google Scholar]
- 33.Urieli-Shoval S, Linke RP, Matzner Y. Expression and function of serum amyloid A, a major acute-phase protein, in normal and disease states. Curr Opin Hematol. 2000;7(1):64–69. doi: 10.1097/00062752-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 34.Reigstad CS, Backhed F. Microbial regulation of SAA3 expression in mouse colon and adipose tissue. Gut Microbes. 2010;1(1):55–57. doi: 10.4161/gmic.1.1.10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kluve-Beckerman B, Drumm ML, Benson MD. Nonexpression of the human serum amyloid A three (SAA3) gene. DNA Cell Biol. 1991;10(9):651–661. doi: 10.1089/dna.1991.10.651. [DOI] [PubMed] [Google Scholar]
- 36.Soler L, Luyten T, Stinckens A, Buys N, Ceron JJ, Niewold TA. Serum amyloid A3 (SAA3), not SAA1 appears to be the major acute phase SAA isoform in the pig. Vet Immunol Immunopathol. 2011;141(1–2):109–115. doi: 10.1016/j.vetimm.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 37.Geurts J, Vermeij EA, Pohlers D, Arntz OJ, Kinne RW, van den Berg WB, van de Loo FA. A novel Saa3-promoter reporter distinguishes inflammatory subtypes in experimental arthritis and human synovial fibroblasts. Ann Rheum Dis. 2011;70(7):1311–1319. doi: 10.1136/ard.2010.135665. [DOI] [PubMed] [Google Scholar]
- 38.de Beer MC, de Beer FC, Gerardot CJ, Cecil DR, Webb NR, Goodson ML, Kindy MS. Structure of the mouse Saa4 gene and its linkage to the serum amyloid A gene family. Genomics. 1996;34(1):139–142. doi: 10.1006/geno.1996.0253. [DOI] [PubMed] [Google Scholar]
- 39.de Beer MC, Yuan T, Kindy MS, Asztalos BF, Roheim PS, de Beer FC. Characterization of constitutive human serum amyloid A protein (SAA4) as an apolipoprotein. J Lipid Res. 1995;36(3):526–534. [PubMed] [Google Scholar]
- 40.He R, Sang H, Ye RD. Serum amyloid A induces IL-8 secretion through a G protein-coupled receptor, FPRL1/LXA4R. Blood. 2003;101(4):1572–1581. doi: 10.1182/blood-2002-05-1431. [DOI] [PubMed] [Google Scholar]
- 41.Su SB, Gong W, Gao JL, Shen W, Murphy PM, Oppenheim JJ, Wang JM. A seven-transmembrane, G protein-coupled receptor, FPRL1, mediates the chemotactic activity of serum amyloid A for human phagocytic cells. J Exp Med. 1999;189(2):395–402. doi: 10.1084/jem.189.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai L, de Beer MC, de Beer FC, van der Westhuyzen DR. Serum amyloid A is a ligand for scavenger receptor class B type I and inhibits high density lipoprotein binding and selective lipid uptake. J Biol Chem. 2005;280(4):2954–2961. doi: 10.1074/jbc.M411555200. [DOI] [PubMed] [Google Scholar]
- 43.Lavie M, Voisset C, Vu-Dac N, Zurawski V, Duverlie G, Wychowski C, Dubuisson J. Serum amyloid A has antiviral activity against hepatitis C virus by inhibiting virus entry in a cell culture system. Hepatology. 2006;44(6):1626–1634. doi: 10.1002/hep.21406. [DOI] [PubMed] [Google Scholar]
- 44.He RL, Zhou J, Hanson CZ, Chen J, Cheng N, Ye RD. Serum amyloid A induces G-CSF expression and neutrophilia via Toll-like receptor 2. Blood. 2009;113(2):429–437. doi: 10.1182/blood-2008-03-139923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ather JL, Ckless K, Martin R, Foley KL, Suratt BT, Boyson JE, Fitzgerald KA, Flavell RA, Eisenbarth SC, Poynter ME. Serum amyloid A activates the NLRP3 inflammasome and promotes Th17 allergic asthma in mice. J Immunol. 2011;187(1):64–73. doi: 10.4049/jimmunol.1100500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho K, Pham TN, Crivello SD, Jeong J, Green TL, Greenhalgh DG. Involvement of CD14 and toll-like receptor 4 in the acute phase response of serum amyloid A proteins and serum amyloid P component in the liver after burn injury. Shock. 2004;21(2):144–150. doi: 10.1097/01.shk.0000108398.56565.ae. [DOI] [PubMed] [Google Scholar]
- 47.Tamamoto T, Ohno K, Goto-Koshino Y, Tsujimoto H. Feline serum amyloid A protein as an endogenous Toll-like receptor 4 agonist. Vet Immunol Immunopathol. 2013;155(3):190–196. doi: 10.1016/j.vetimm.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 48.de Seny D, Cobraiville G, Charlier E, Neuville S, Esser N, Malaise D, Malaise O, Calvo FQ, Relic B, Malaise MG. Acute-phase serum amyloid a in osteoarthritis: regulatory mechanism and proinflammatory properties. PLoS One. 2013;8(6):e66769. doi: 10.1371/journal.pone.0066769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christenson K, Bjorkman L, Tangemo C, Bylund J. Serum amyloid A inhibits apoptosis of human neutrophils via a P2X7-sensitive pathway independent of formyl peptide receptor-like 1. J Leukoc Biol. 2008;83(1):139–148. doi: 10.1189/jlb.0507276. [DOI] [PubMed] [Google Scholar]
- 50.Mullan RH, Bresnihan B, Golden-Mason L, Markham T, O'Hara R, FitzGerald O, Veale DJ, Fearon U. Acute-phase serum amyloid A stimulation of angiogenesis, leukocyte recruitment, and matrix degradation in rheumatoid arthritis through an NF-kappaB-dependent signal transduction pathway. Arthritis Rheum. 2006;54(1):105–114. doi: 10.1002/art.21518. [DOI] [PubMed] [Google Scholar]
- 51.Connolly M, Mullan RH, McCormick J, Matthews C, Sullivan O, Kennedy A, FitzGerald O, Poole AR, Bresnihan B, Veale DJ, Fearon U. Acute-phase serum amyloid A regulates tumor necrosis factor alpha and matrix turnover and predicts disease progression in patients with inflammatory arthritis before and after biologic therapy. Arthritis Rheum. 2012;64(4):1035–1045. doi: 10.1002/art.33455. [DOI] [PubMed] [Google Scholar]
- 52.Migita K, Kawabe Y, Tominaga M, Origuchi T, Aoyagi T, Eguchi K. Serum amyloid A protein induces production of matrix metalloproteinases by human synovial fibroblasts. Lab Invest. 1998;78(5):535–539. [PubMed] [Google Scholar]
- 53.Zhao Y, Zhou S, Heng CK. Celecoxib inhibits serum amyloid a-induced matrix metalloproteinase-10 expression in human endothelial cells. J Vasc Res. 2009;46(1):64–72. doi: 10.1159/000139134. [DOI] [PubMed] [Google Scholar]
- 54.Upragarin N, Landman WJ, Gaastra W, Gruys E. Extrahepatic production of acute phase serum amyloid A. Histol Histopathol. 2005;20(4):1295–1307. doi: 10.14670/HH-20.1295. [DOI] [PubMed] [Google Scholar]
- 55.Connolly M, Marrelli A, Blades M, McCormick J, Maderna P, Godson C, Mullan R, FitzGerald O, Bresnihan B, Pitzalis C, Veale DJ, Fearon U. Acute serum amyloid A induces migration, angiogenesis, and inflammation in synovial cells in vitro and in a human rheumatoid arthritis/SCID mouse chimera model. J Immunol. 2010;184(11):6427–6437. doi: 10.4049/jimmunol.0902941. [DOI] [PubMed] [Google Scholar]
- 56.Cai X, Freedman SB, Witting PK. Serum amyloid A stimulates cultured endothelial cells to migrate and proliferate: inhibition by the multikinase inhibitor BIBF1120. Clin Exp Pharmacol Physiol. 2013;40(9):662–670. doi: 10.1111/1440-1681.12148. [DOI] [PubMed] [Google Scholar]
- 57.Lakota K, Mrak-Poljsak K, Rozman B, Sodin-Semrl S. Serum amyloid A and its potential physiological/pathological functions-an overview of patents. Recent Pat Endoocr Metab & Immune Drug Discov. 2010;4:89–99. [Google Scholar]