Abstract
Aim:
To investigate the anti-cancer effects of p21WAF1/CIP1 transcriptional activation induced by dsRNAs in hepatocellular carcinoma (HCC) cell lines.
Methods:
HCC cell lines BEL7402, SMMC-7721, MHCC97L, MHCC97H, and MHCCLM3 were used. HCC cells were treated with dsP21-322 (50 nmol/L), dsControl (50 nmol/L), siP21 (50 nmol/L), or mock transfection. The expression of p21 was detected using quantitative PCR and Western blot. The effects of RNA activation on HCC cells were determined using cell viability assays, apoptosis analyses and clonogenic survival assays. Western blot was also conducted to detect the expression of Bcl-xL, survivin, cleaved caspase-3, cleaved caspase-9 and cleaved PARP.
Results:
At 72 to 120 h following the transfection, dsP21-322 markedly inhibited the viability of HCC cells and clone formation. At the same times, dsP21-322 caused a significant increase in HCC cell apoptosis, as demonstrated with cytometric analysis. The phenomena were correlated with decreased expression levels of the anti-apoptotic proteins Bcl-xL, surviving, and increased expression of cleaved caspase-3, cleaved caspase-9 and cleaved PARP.
Conclusion:
RNA-induced activation of p21 gene expression may have significant therapeutic potential for the treatment of hepatocellular carcinoma and other cancers.
Keywords: hepatocellular carcinoma, small activating RNA (saRNA), RNA-induced gene activation (RNAa), p21WAF1/CIP1, cell viability, apoptosis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common and aggressive malignancies worldwide, ranking among the top five leading causes of cancer death in the world, and is the second highest cause in China1, 2. HCC now accounts for over 626 000 new cases annually1, and the incidence has more than doubled over the last two decades3. Whereas the incidence of HCC has been steadily increasing, the emergence of new and effective therapeutic agents remains relatively stagnant. High mortality rates are due to the severity of underlying liver disease, late diagnosis resulting in advanced stage disease, lack of effective treatment options, and disease progression even after surgical therapies or locoregional procedures4. Even in well-selected patients with operable disease, the long-term prognosis is still unsatisfactory because of frequent recurrences after resection. Therefore, there is a great need for novel treatments and better interventions for HCC.
Noncoding RNA (ncRNA) molecules, such as small interfering RNA (siRNA), microRNAs (miRNAs), and short-hairpin RNAs (shRNAs), were recognized initially for their powerful ability to down-regulate specific genes at both the post-transcriptional5, 6, 7 and the transcriptional levels8, 9. Other reports have claimed the opposite effect10, 11, 12, 13, 14. Li et al reported that double stranded RNA (dsRNA) molecules could induce sequence-specific transcriptional gene activation, termed this phenomenon RNA-induced gene activation (RNAa) and termed the molecules small activating RNAs (saRNA)10. Although two mechanistic models related to RNA activation have been proposed10, 11, 12, 13, 14, 15, 16, very little is known about what makes one molecule a silencer and another an activator17. Nevertheless, what is becoming clear is that RNAa has the potential to be a powerful biological tool and could lead to new therapies for diseases such as cancer18, 19. Among those genes that can be modulated through RNAa10, 11, 12, 13, 14, the p21WAF1/CIP1 (p21) gene product is special because it is a potent cyclin-dependent kinase inhibitor that binds to and inhibits the activity of cyclin-CDK2 or cyclin-CDK4 complexes. It thus functions as a regulator of cell cycle progression at the G1 stage20. The p21 gene product may also play a regulatory role in S-phase DNA replication and DNA damage repair by interacting with proliferating cell nuclear antigen (PCNA), a DNA polymerase accessory factor21. Although the role of p21 in apoptosis is still controversial, with contradictory findings of both stimulation and inhibition of apoptosis22, there are studies indicating that p21 also possesses pro-apoptotic functions against cancer19, 23. Previous studies have also shown that decreased p21 expression may be involved in a variety of carcinomas, especially in cases of altered p53 expression24, 25. Therefore, p21 is a potential candidate for RNAa-mediated cancer therapy.
In this study, we sought to investigate the anticancer effects of RNAa-mediated p21 activation in HCC cells. Our study has shown that up-regulation of p21 triggered by an saRNA resulted in the significant inhibition of proliferation and survival and in the induction of apoptosis in HCC cells.
Materials and methods
dsRNAs
dsP21-322, 21 nucleotides long, corresponding to the promoter region of p21, was designed as described previously by Li et al10. siP21, which targets p21 mRNA, was designed with the BLOCK-iT™ RNAi Designer (Invitrogen, Carlsbad, CA, USA). A dsRNA lacking significant homology to all known human sequences (dsControl) was used as a non-specific control. The dsRNAs were chemically synthesized by Genepharma (Shanghai, China). All dsRNA sequences are listed in Table 1.
Table 1. Sequences for dsRNAs and PCR primers.
| dsRNA/Primer | Sequence (5′–3′) |
|---|---|
| dsP21-322 S | CCA ACU CAU UCU CCA AGU A[dT][dT] |
| dsP21-322 AS | UAC UUG GAG AAU GAG UUG G[dT][dT] |
| dsP21+271 S | GAA CUU CGA CUU UGU CAC CGA GAC A |
| dsP21+271 AS | UGU CUC GGU GAC AAA GUC GAA GUU C |
| dsControl S | ACU UAC GAG UGA CAG UAG A[dT][dT] |
| dsControl AS | UCU ACU GUC ACU CGU AAG U[dT][dT] |
| GAPDH-For | GCA CCG TCA AGG CTG AGA AC |
| GAPDH-Rev | TGG TGA AGA CGC CAG TGG A |
| P21-For | AAG ACC ATG TGG ACC TGT CAC TGT |
| P21-Rev | GAA GAT CAG CCG GCG TTT G |
S, sense; AS, antisense; For, forward; Rev, reverse.
Cell culture and transfection
The human hepatoma cell lines BEL7402, SMMC-7721, MHCC97L, MHCC97H, and MHCCLM3 used in this study were a generous gift from the Liver Cancer Institute and Zhongshan Hospital, Fudan University (Shanghai, China). MHCC97L, MHCC97H, and MHCCLM3 cell lines were established from the same parent human HCC cell line of MHCC97 in the Liver Cancer Institute and Zhongshan Hospital, Fudan University. Cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS, penicillin (100 units/mL), and streptomycin (0.1 mg/mL) and were incubated at 37 °C in a humidified incubator under an atmosphere of 5% CO2. The day before transfection, cells were plated in growth medium without antibiotics at a density of 50% to 70%. The dsRNA was transfected at a concentration of 50 nmol/L using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
The cultures were collected 48 or 72 h later, cells were counted to determine viability, and mRNA and protein were obtained.
Cell viability assay
Phase-contrast images of treated cells were taken with an Olympus X51 microscope (Olympus, Tokyo, Japan) at 40× magnification. Cell viability was measured by a colorimetric assay based on the cleavage of the tetrazolium salt WST-1 by mitochondrial dehydrogenases (cell proliferation reagent WST-1; Molecular Biochemicals, Mannheim, Germany). MHCCLM3 cells were seeded onto 96-well culture plates at approximately 6000–8000 cells in 200 μL DMEM medium with 10% FBS. Cells were cultured for 24 h under 5% CO2 at 37 °C. After overnight incubation, cells were treated with dsP21-322 (50 nmol/L), siP21 (50 nmol/L), dsControl (50 nmol/L), or mock transfection. Twelve hours later, the incubation medium was removed and replaced with 200 μL fresh medium with 10% FBS. At various time points following transfection (24, 48, 72, 96, or 120 h), 10 μL WTS-1 was added to each well. After incubation at 37 °C for 2 h, viable cells were detected by measuring absorbance at 440 nm using a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA, USA). Cell viability was expressed as a percentage of the absorbance of cells after transfection at various time points relative to the values before transfection. Cells present 24 h after seeding were considered 100% viable. The reduction in viability of dsP21-322 or dsControl-treated MHCCLM3 cells was compared to the mock transfection group. The dsP21-322 and dsControl treatment groups were compared to the mock transfection group to obtain P values. Each assay was repeated three times.
Apoptosis assay
Cells were plated in 6-well plates at a density of 0.5×106 cells/mL and incubated overnight. Transfections were performed and then cells were incubated for 12 h before changing the transfection medium to fresh medium containing 10% FBS. Cells were harvested at 72 h following transfection, washed twice with pre-chilled PBS, and resuspended in 100 μL 1× binding buffer at a concentration of 1×106 cells/mL. Annexin V and PI double-staining was performed using an Annexin V-FITC Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer's protocol. Cell apoptosis analysis was performed by an EPICS ALTRA Flow Cytometry System with CXP Software (Beckman Coulter, Fullerton, CA, USA) within 1 h.
Quantitative PCR
Total RNA was extracted from cells by TRIzol (Invitrogen) after 48 h of transfection (mock, 50 nmol/L dsControl, 50 nmol/L dsP21-322 or 50 nmol/L siP21) and reverse transcription was performed with a PrimeScript RT reagent Kit (Takara Biotechnology, Dalian, China). qRT-PCR was performed with SYBR Green PCR reagent kits (Toyobo Co, Osaka, Japan) at a constant annealing temperature (64 °C) according to the manufacturer's protocol. Specific primer sets used in the real-time PCR directed against human p21 and GAPDH were designed and generated by Takara Biotechnology Co (Dalian, China) (listed in Table 1). Data were recorded and analyzed using the real-time PCR analysis software Bio-Rad iQ5. Endogenous gene expression was normalized to GAPDH levels in the cells.
Western blot analysis
Cells were harvested at 72 h following dsRNAs treatment as described above and then washed and lysed with M-PER extraction buffer (Pierce Biotechnology) containing protease inhibitors. Protein lysates, quantified using a BCA assay (Sangon Biotech Co, Ltd, Shanghai, China), were separated on reducing SDS-polyacrylamide gels and transferred to polyvinyl difluoride membranes (PVDF, Millipore). The membranes were blocked with 5% nonfat milk TBS buffer for 2 h at room temperature and incubated with primary antibodies overnight at 4 °C. Beta-actin levels were used to normalize loading. Primary immunoblotting antibodies (anti-Bcl-xL rabbit monoclonal antibody, anti-p21WAF1/CIP1 rabbit monoclonal antibody, anti-survivin rabbit monoclonal antibody, anti-cleaved caspase-9 rabbit polyclonal antibody, anti-cleaved caspase-3 rabbit polyclonal antibody, anti-cleaved PARP rabbit polyclonal antibody, or anti-β-actin antibody) were obtained from Cell Signaling Technology (Cell Signaling, Beverly, MA, USA) and used at 1:1000 dilutions. The first antibody exposure was followed by incubation with an anti-rabbit IgG, HRP-linked secondary antibody (Cell Signaling, Beverly, MA, USA). Antigen-antibody complexes were visualized by an enhanced chemiluminescence detection method (ECL kit, Thermo, Waltham, MA, USA).
Clonogenic survival assay
The day before transfection, MHCCLM3 cells were plated as single-cell suspensions at a density of 5000 cells per well in a 6-well plate. Cells were transfected 2 times with 50 nmol/L dsRNA (dsControl and dsP21-322) or mock transfection using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) at 24 h intervals, and the transfection medium was replaced with normal complete medium after 6 h and changed every three days. At d 12, the medium was removed and colonies were stained with 0.5% w/v crystal violet solution for 30 min. Images were taken for analysis using a single-lens reflex digital camera (K100D Super Pentax, Japan).
Statistical analysis
All experiments were performed at least three times. Statistical significance of the differences between various treatment groups and controls was calculated by applying Student's two-tailed t-test. The level of statistical significance was set at P<0.05.
Results
dsP21-322 induced p21 gene expression in HCC cell lines
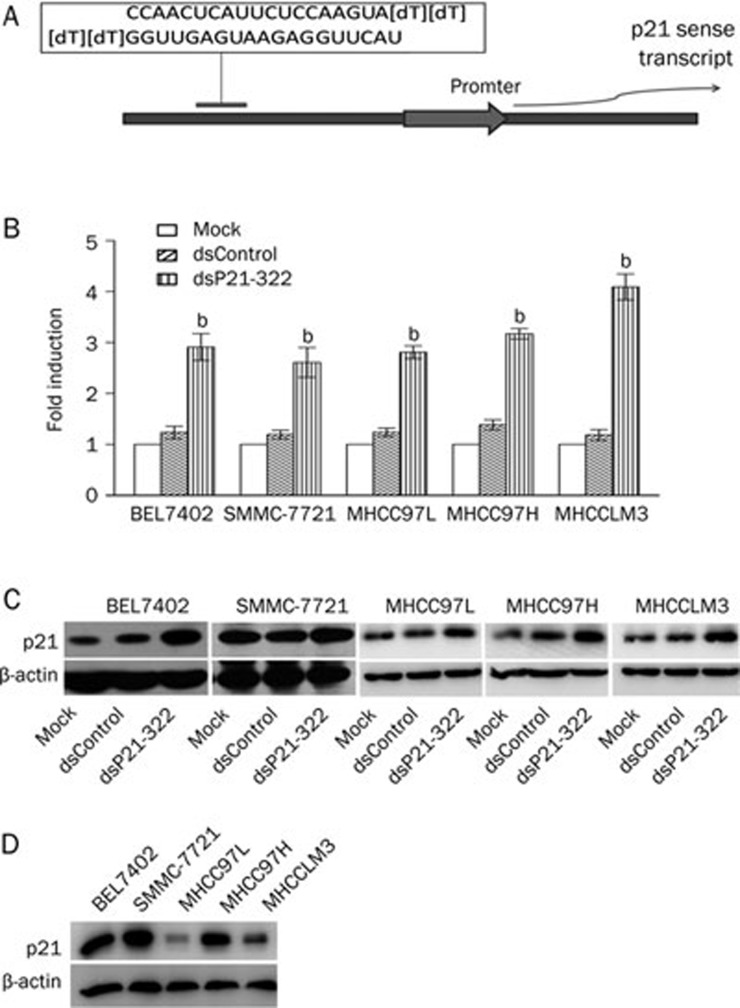
A dsRNA targeting the p21 gene promoter at position-322 relative to the transcription start site (dsP21-322)10 (Figure 1A) and a nonspecific control dsRNA (dsControl), which lacked significant homology to all known human sequences, were transfected into HCC cell lines of BEL7402, SMMC-7721, MHCC97L, MHCC97H, and MHCCLM3. After 48 and 72 h, mRNA and protein were obtained and analyzed by real-time quantitative PCR and Western blotting, respectively. As shown in Figure 1B, at 48 h, dsP21-322 caused a significant increase in p21 mRNA expression. Compared with mock transfections, the increases were 2.9-, 2.6-, 2.8-, 3.2-, and 4.0-fold higher in BEL7402, SMMC-7721, MHCC97L, MHCC97H, and MHCCLM3 cells, respectively (P<0.05, Figure 1B). Western blot analysis revealed that the levels of p21 protein elevated significantly when compared with mock and dsControl transfection, strongly correlating to the observed increase in p21 mRNA levels (Figure 1C). Although BEL7402, SMMC-7721 and MHCC97H cells had high baseline levels of p21 (Figure 1D), a moderate and consistent increase in p21 expression after dsP21-322 treatment was observed at both the mRNA and the protein levels. These results were reproducible in at least three independent experiments.
Figure 1.
Effects of dsRNAs targeting the p21 gene promoter on p21 mRNA and protein levels in HCC cell lines. (A) A schematic diagram showing the target sequence positions of dsP21-322 relative to the transcriptional start site. (B) Induction of p21 mRNA expression was detected by real-time quantitative RT-PCR. HCC cells were transfected with 50 nmol/L dsP21-322 and dsControl for 2 d. Mock transfections were also done in the absence of saRNA. Expression levels were measured as fold-induction relative to mock transfections. The results were normalized to GAPDH and presented as mean±SD of three independent experiments. bP<0.05 vs mock transfections. (C) The induction of p21 protein levels were detected by Western blot analysis at 72 h following dsRNA treatment, as described above. A representative blot is shown from three independent experiments with identical results. Beta-actin levels were also detected and served as endogenous controls. (D) Detection of p21 protein levels in the HCC cell lines of BEL7402, SMMC-7721, MHCC97L, MHCC97H, and MHCCLM3.
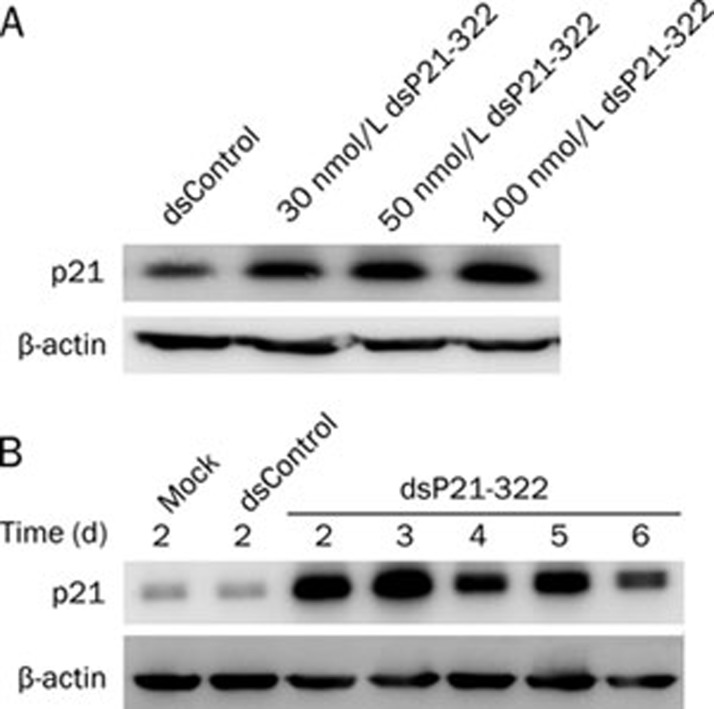
To determine whether activation of p21 by saRNA was dose- and time-dependent, concentration gradient and time-response studies were performed with MHCCLM3 cells. Figure 2A shows the up-regulation of p21 with various concentrations of dsP21-322 for 3 d. MHCCLM3 cells were transfected with 50 nmol/L dsP21-322 for the indicated lengths of time (Figure 2B). These data indicated that dsP21-322 exerted a significant activating effect upon MHCCLM3 cells in a dose- and time-dependent manner.
Figure 2.
Concentration dependence and the time course of activation by dsP21-322 in MHCCLM3 cells. (A) Western blot analysis showing the levels of p21 protein after transfection with various concentrations of dsRNAs into MHCCLM3 cells. A representative blot is shown from three independent experiments with identical results. (B) MHCCLM3 cells were treated with 50 nmol/L dsP21-322, dsControl or mock transfection at various time points (2–6 d). p21 protein and beta-actin expression were detected by Western blotting.
dsP21-322 inhibited HCC cell proliferation and survival
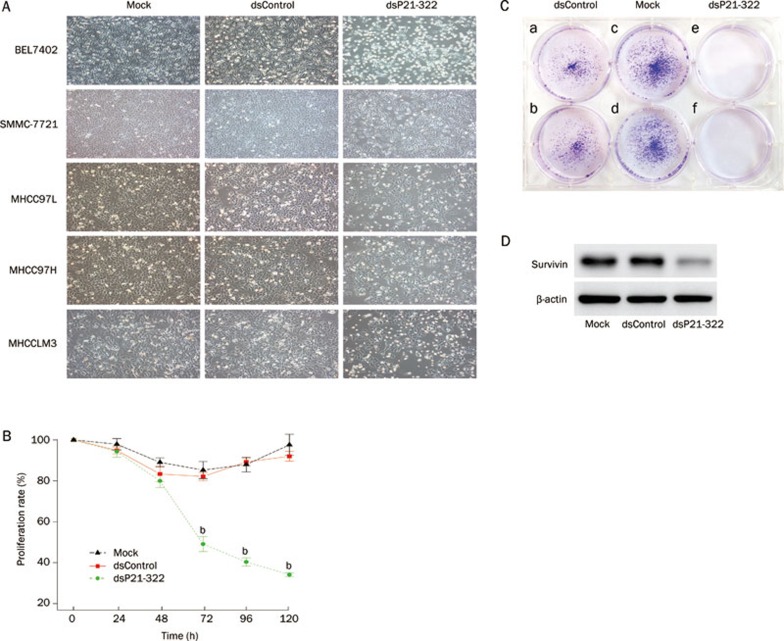
After transfection, mock-transfected and dsControl-transfected cells maintained healthy growth, whereas cells transfected with dsP21-322 gradually lost viability after d 2. Morphological changes were also observed (Figure 3A). There were more rounded floating cells in the supernatant of cells transfected with dsP21-322 while the adherent cells had become enlarged and flattened with decreased cell density. Typical apoptotic morphological changes were also seen, in which cells became smaller, rounded, and deformed with nuclear condensation.
Figure 3.
dsRNA targeting the p21 promoter inhibits tumor cell proliferation and survival. (A) BEL7402, SMMC-7721, MHCC97L, MHCC97H, and MHCCLM3 cells were seeded in 6-well plates and transfected with 50 nmol/L dsP21-322, 50 nmol/L dsControl, or mock transfection 24 h later. Cell images were taken 72 h after transfection at 40×magnification. (B) Cell proliferation was determined by colorimetric WST-1 assays and assessed at 24, 48, 72, 96, and 120 h following transfection. The data are presented as the mean±SD relative to mock transfections. bP<0.05 vs mock transfections. (C) MHCCLM3 clonogenic survival assays were performed in 6-well plates with 5000 cells per well. Transfections were done as previously described. On d 12, cell colonies were stained with crystal violet for 30 min and photographed. (a, b) dsControl transfection; (c, d) mock transfection; (e, f) dsP21-322 transfection. (D) The effect of dsP21-322 on the expression of survivin in MHCCLM3 cells. Cells were harvested after 72 h of transfection. Beta-actin levels served as an endogenous control. The data shown are from a representative experiment repeated three times with similar results.
Further quantitation performed in MHCCLM3 cells by WST-1 colorimetric assay demonstrated that the dsP21-322 transfection induced a significant loss in cell viability. As shown in Figure 3B, there were detectable reductions in MHCCLM3 cell viability after dsP21-322 transfection at a concentration of 50 nmol/L. After 48 h, proliferation of the dsP21-322 transfection group decreased dramatically from 65.9% to 20.1%. The time point of the strong growth-inhibitory effect substantially correlated with the kinetics of RNAa. By contrast, viability of the mock group and dsControl group decreased slightly during the 48 h after transfection, after which they recovered. These observations indicated that dsP21-322 was an effective inhibitor of HCC cell growth, and this inhibitory effect was time dependent.
The clonogenic survival assay or colony formation assay is a widely used method to determine the effectiveness of other cytotoxic agents. It tests every cell in the population for its ability to undergo “unlimited” division26. After 12 d of incubation, colonies were stained and photographed. As shown in Figure 3C, no colony formation was observed in cells transfected with dsP21-322, while numerous colonies were stained in both mock and dsControl-transfected cells. These results were consistent with a previous report in T24 bladder cancer cells18.
Previous studies have shown that survivin expression was strongly correlated with the proliferation index in HCC tissues, and overexpression of survivin resulted in both significant cell proliferation and survival promotion in carcinoma cells27. Hence, to determine whether the decrease in cell viability and survival in the induction of p21 by RNAa occurred through a survivin-dependent pathway, changes in the levels of survivin were investigated after treatment. As can be seen in Figure 3D, a dramatic decrease in survivin protein levels was found in the dsP21-322-treated group when compared with the control groups. The data showed that the up-regulation of p21 strongly correlated with decreased survivin levels in MHCCLM3 cells.
dsP21-322 significantly increased apoptosis and levels of apoptosis-related proteins in HCC cells
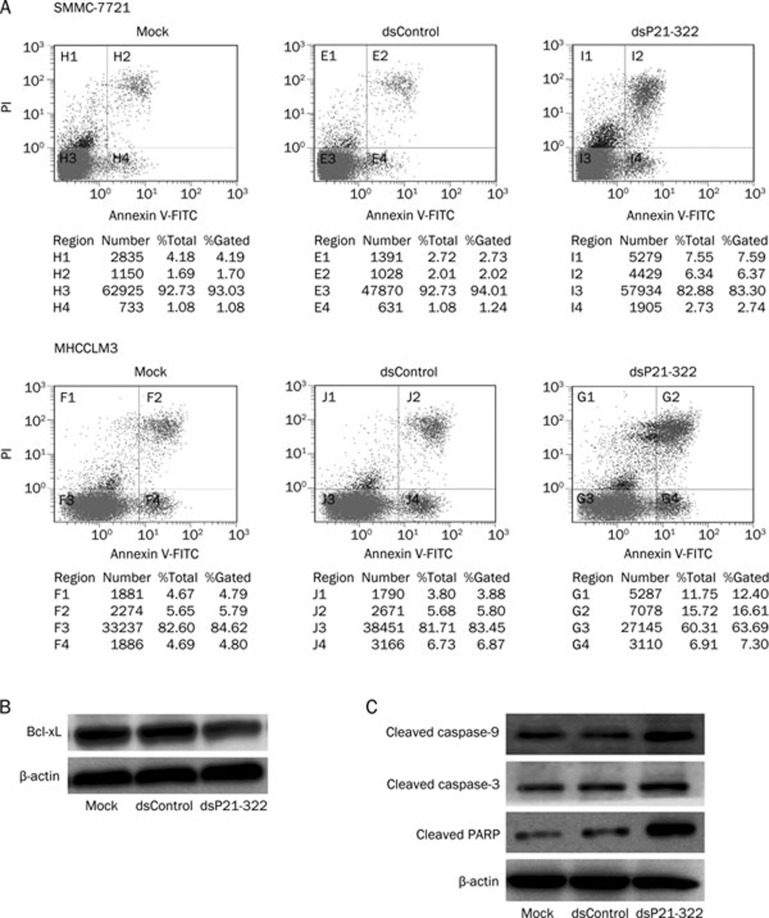
The extent of apoptosis was quantified by flow-cytometric analysis of saRNA-treated cells labeled with PI and annexin V. As shown in Figure 4A, treatment of SMMC-7721 and MHCCLM3 cells with 50 nmol/L dsP21-322 for 72 h resulted in 9.11% and 23.91% of total cells becoming apoptotic, respectively. In the mock and dsControl-transfected SMMC-7721 and MHCCLM3 cells, 2.78% and 3.26%, and 10.59% and 12.67%, were found to be apoptotic, respectively. These results revealed that treatment of SMMC-7721 and MHCCLM3 cells with dsP21-322 resulted in a significant increase in apoptosis.
Figure 4.
dsP21-322 transfection-induced apoptosis in HCC cells. (A) SMMC-7721 or MHCCLM3 cells were double-stained with annexin V and PI 72 h after treatment with dsP21-322 (50 nmol/L), dsControl (50 nmol/L), or mock transfection and analyzed by flow cytometry. Gate settings distinguished between live (bottom left), necrotic (top left), early apoptotic (bottom right), and late apoptotic (top right) cells. (B) Changes in the levels of Bcl-xL anti-apoptotic gene products in MHCCLM3 cells 72 h after treatment with 50 nmol/L dsP21-322. (C) Effects of dsP21-322 transfection-activated p21 levels on cleavage of apoptosis-related proteins caspase-3, caspase-9, and poly (ADP-ribose) polymerase (PARP). Beta-actin levels served to normalize loading. The data shown are from a representative experiment repeated three times with similar results.
Cells undergoing apoptosis follow a death program by inhibiting the anti-apoptosis protein Bcl-xL and activating a hierarchy of caspases. Caspase-9 and caspase-3 are at least partially responsible for the proteolytic cleavage of many key proteins, such as the nuclear enzyme poly (ADP-ribose) polymerase (PARP), which plays a central role in the execution phase of cell apoptosis. In our study, besides a remarkable decrease in Bcl-xL, significant increases in cleaved caspase-9 and caspase-3 as well as cleaved PARP were observed in the dsP21-322-transfected MHCCLM3 cells (Figure 4B). Such significant changes in apoptosis execution further confirm the observed features of the apoptotic phenotype and the results from the flow cytometry assay.
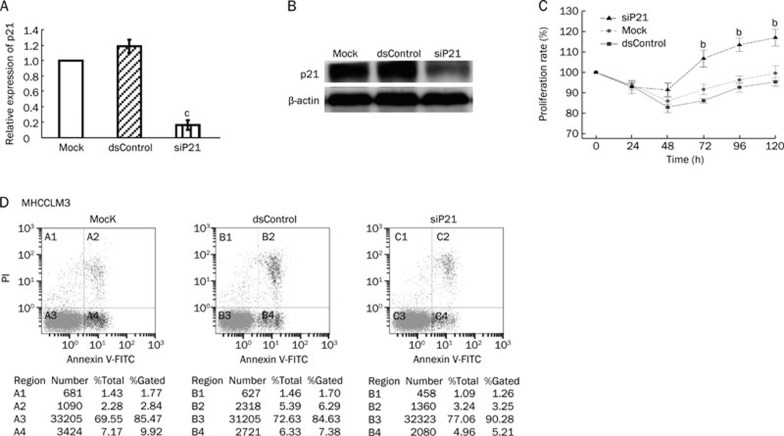
p21 knockdown by siP21 resulted in the induction of proliferation and inhibition of apoptosis in MHCCLM3 cells
To confirm the role of p21 in the inhibition of proliferation and the induction of apoptosis, a siRNA strategy was used to knockdown expression of p21 in MHCCLM3 cells. The siRNA knockdown efficiency of p21 mRNA expression was 83% after 48 h of transfection (Figure 5A). Figure 5B showed that p21 levels decreased significantly in the siP21 transfection group. Next, cell viability and apoptosis assays were used to investigate the effect of p21 knockdown on the growth of MHCCLM3 cells. As shown in Figure 5C, inhibition of p21 expression is sufficient to promote survival and cell proliferation in 50 nmol/L siP21-transfected cells. Similar results were obtained with a flow cytometry assay, where 8.46% of the total cells became apoptotic 72 h after treatment of MHCCLM3 cells with 50 nmol/L siP21, which was substantially lower than the mock and dsControl groups, whose apoptotic rates were 12.76% and 13.67%, respectively (Figure 5D). These findings further confirm that the role of the p21 gene in growth control of MHCCLM3 cells is mediated by an apoptotic mechanism.
Figure 5.
Effects of p21 knockdown on cell proliferation and apoptosis of MHCCLM3 cells. Either 48 or 72 h after cells were treated with siP21 (50 nmol/L), dsControl (50 nmol/L), or mock transfection, messenger RNA levels (A) were determined by qPCR analysis, and Western blot (B) was used to analyze p21 knockdown efficiency. Cell proliferation (C) and apoptosis (D) were assessed as described previously. bP<0.05 vs mock transfections.
Discussion
RNA activation in human cells was first observed with dsRNAs targeted to the promoters of E-cadherin, vascular endothelial growth factor (VEGF), and p21 and was confirmed in studies on the progesterone receptor (PR)11. Subsequently, microRNA-373 was found to induce expression of E-cadherin and cold-shock domain-containing protein C2 (CSDC2) with complementary promoter sequences12. When it comes to the exact mechanism underlying dsRNA induced RNAa, several models have been reported or proposed, including transcriptional activation by directly targeting the transcriptional start site of the desired gene10, 12 or the activating siRNAs-mediated loss of an endogenous repressor antisense non-coding RNA, resulting in a loss of epigenetic regulation for the sense/mRNA partner, which ultimately resulted in a marked increase in transcription15, 28. However, due to the lack of a deeper understanding of the detailed mechanism of RNAa, more research will certainly be required. Regardless, what is becoming clear is that small RNAs can be used to mediate the activation of a wide array of genes. RNAa offers a promising new therapeutic strategy for diseases that can be corrected by stimulating gene expression18, 19.
In our study, we found that dsP21-322 can cause a 2.6- to 4-fold increase in p21 mRNA levels in BEL7402, SMMC-7721, MHCC97L, MHCC97H, and MHCCLM3 cells. We also noticed that, although the BEL7402, SMMC-7721, and MHCC97H cells normally expressed high levels of p21, a moderate and consistent increase in p21 levels occurred after dsP21-322 treatment. Data from both time- and concentration-dependence experiments in MHCCLM3 cells showed that the stimulatory effect of dsP21-322 was quite potent. Those results revealed that RNAa can be manipulated to up-regulate gene expression in hepatocellular carcinoma. To our knowledge, this is the first report that demonstrates dsRNA-mediated gene activation in HCC, which suggests that RNAa may be used to manipulate genes, especially tumor suppressor genes (TSG), in the treatment of HCC.
Clonogenic assays permit the study of the effectiveness of drugs or radiation on the survival and proliferation of cells26. The results from our study in the clonogenic assay indicate that colony formation in MHCCLM3 cells was completely inhibited by the up-regulation of p21, which is in agreement with the findings of the study by Zhong et al in bladder cancer cells18. However, the proliferation assay showed that dsP21-322-transfected cells decreased proliferation by approximately 70%. This discrepancy may be because, in the clonogenic assay, MHCCLM3 cells were transfected twice with 50 nmol/L dsRNA (dsControl and dsP21-322) or without dsRNA (mock transfection) using Lipofectamine 2000 (Invitrogen) at 24 h intervals. In addition, a density of 5000 cells per well also limited the survival of single cells in the clonogenic assay.
Survivin, a unique member of the inhibitors-of-apoptosis gene family, is characterized by its abundant expression in transformed cells and a variety of human cancers. However, it is barely detectable in terminally differentiated normal cells or tissues29. Besides inhibiting apoptosis in cells exposed to diverse apoptotic stimuli29, survivin was found to promote cell proliferation in human HCC cells by interacting with cyclin-dependent kinase4 (CDK4)30. In the current study, a significant decrease in survivin levels was found following dsP21-322 transfection, which is consistent with the inhibition of proliferation and survival. This suggests that the dsRNA-induced stimulation of p21 may lead to decreased survival of HCC cells through a survivin-dependent mechanism. However, the causality and association between p21 and survivin need to be explored in future studies.
Through the proliferation assay, as well as the clonogenic assay, our study has shown that dsP21-322 can significantly inhibit the growth of HCC cells. These effects may be due mainly to the action of p21 as a potent cyclin-dependent kinase inhibitor (CKI) that binds to and inhibits the activity of cyclin-CDK2 or -CDK4 complexes. Thus, it functions as a mediator of cell-cycle G1-phase arrest in response to a variety of stress stimuli31, 32.
There have been contradictory results claiming both the promotion and the inhibition of apoptosis by p2122, 23. Our results showed that apoptosis was increased both early and late after transfection, which was also consistent with a previous study on bladder cancer19. The decrease in Bcl-xL as well as the activation of caspase proteins contributed to the effect of p21 up-regulation in HCC cell lines. Then knockdown of p21 by siRNA resulted in the reverse of proliferation and the reduction of apoptosis, which supports the role of p21 in the induction of apoptosis and the inhibition of proliferation.
In summary, transfection with dsP21-322 decreased cell viability, inhibited colony formation, and induced apoptosis of HCC cells. Evidence supports the hypothesis that a loss of survivin and activation of the caspase pathway ultimately results in the observed anti-cancer effects in the HCC cell lines.
Taken together, the induction of p21 by small RNA molecules targeted to the promoter regions of p21 can cause anti-cancer effects in HCC cell lines. These results show that RNAa is also activated in HCC cells, pointing to a potential therapeutic use for dsRNA-induced gene activation in HCC.
Author contribution
Gang CHEN designed research; Zhi-ming WU, Chun DAI, and Cui-fang ZHENG performed research; Ying HUANG, Qiong-zhu DONG, Guan WANG, Xiao-wen LI, Xiao-fei ZHANG, and Bin LI contributed new analytical tools and data analysis; Zhi-ming WU wrote the paper.
Acknowledgments
This work was supported by grants from both the Shanghai Natural Science Fund (Grant No 08ZR1402400) and the Fudan University Institute of Biomedical Science Opening Fund (Grant No IBS0837). We wish to express our deepest gratitude and indebtedness to Prof Lun-xiu QIN, Zhongshan Hospital, the Liver Cancer Institute, Fudan University, Shanghai, for his generous help throughout the entire course of this project, without which it would not have been possible for us to complete this work.
References
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- He J, Gu DF, Wu XG, Reynolds K, Duan XF, Yao CH, et al. Major causes of death among men and women in China. N Engl J Med. 2005;353:1124–34. doi: 10.1056/NEJMsa050467. [DOI] [PubMed] [Google Scholar]
- Hashem BE, Andrew CM. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745–50. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–36. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–58. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Chan SWL, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–92. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- Khraiwesh B, Asif MA, Seumel GI, Ossowski S, Weigel D, Reski R. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–22. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006;103:17337–42. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–73. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen MP, Lehtola T, Heinonen SE, Assefa GS, Korpisalo P, Gienary R, et al. Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: A novel example of epigenetherapy. Circ Res. 2009;105:604–9. doi: 10.1161/CIRCRESAHA.109.200774. [DOI] [PubMed] [Google Scholar]
- Huang V, Qin Y, Wang J, Wang XL, Place RF, Lin GT, et al. RNAa is conserved in mammalian cells. PLoS One. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional Transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–8. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber K. Small RNAs reveal an activating side. Science. 2006;341:741–2. doi: 10.1126/science.314.5800.741a. [DOI] [PubMed] [Google Scholar]
- Zhong C, Place RF, Jia ZJ, Pookot D, Dahiya R, Li LC. Antitumor effect of dsRNA-induced p21WAF1/CIP1 gene activation in human bladder cancer cells. Mol Cancer Ther. 2008;7:698–703. doi: 10.1158/1535-7163.MCT-07-2312. [DOI] [PubMed] [Google Scholar]
- Yang K, Zheng XY, Qin J, Wang YB, Bai Y, Mao QQ, et al. Up-regulation of p21WAF1/CIP1 by saRNA induces G1-phase arrest and apoptosis in T24 human bladder cancer cells. Cancer Lett. 2008;265:206–14. doi: 10.1016/j.canlet.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Sekiguchi T, Hunter T. Induction of growth arrest and cell death by overexpression of the cyclin-Cdk inhibitor p21 in hamster BHK21 cells. Oncogene. 1998;16:369–80. doi: 10.1038/sj.onc.1201539. [DOI] [PubMed] [Google Scholar]
- Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–8. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- Gorospe M, Wang X, Guyton KZ, Holbrook NJ. Protective role of p21(WAF1/CIP1)against prost agland in A2-mediated apoptosis of Human colorectal carcinoma cells. Mol Cell Biol. 1996;16:6654–60. doi: 10.1128/mcb.16.12.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther. 2002;1:639–49. [PubMed] [Google Scholar]
- Shi YZ, Hui AM, Takayama T, Li X, Makuuchi M. Reduced p21WAF1/CIP1 protein expression is predominantly related to altered p53 in hepatocellular carcinomas. Br J Cancer. 2000;83:50–5. doi: 10.1054/bjoc.2000.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui AM, Kanai Y, Sakamoto M, Tsuda H, Hirohashi S. Reduced p21WAF1/CIP1 expression and p53 mutation in hepatocellular carcinomas. Hepatology. 1997;25:575–9. doi: 10.1002/hep.510250314. [DOI] [PubMed] [Google Scholar]
- Franken NAP, Rodermond HM, Stap J, Haveman J, Bree CV. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–9. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- Ito T, Shiraki K, Sugimoto K, Yamanaka T, Fujikawa K, Ito M, et al. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology. 2000;31:1080–5. doi: 10.1053/he.2000.6496. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, et al. Antisence transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–8. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah NK, Khan Z, Khan GJ, Bisen PS. Structural, functional and therapeutic biology of survivin. Cancer Lett. 2006;244:164–71. doi: 10.1016/j.canlet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–21. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–7. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–16. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]