Abstract
Aim:
To identify the key proteins involved in the nephrotoxicity induced by andrographolide sodium bisulfite (ASB).
Methods:
Male ICR mice were intravenously administrated with ASB (1000 or 150 mg·kg−1·d−1) for 7 d. The level of malondialdehyde (MDA) and the specific activity of superoxide dismutase (SOD) in kidneys were measured. The renal homogenates were separated by two-dimensional electrophoresis, and the differential protein spots were identified using a matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF)/TOF mass spectrometry.
Results:
The high dose (1000 mg/kg) of ASB significantly increased the MDA content, but decreased the SOD activity as compared to the control mice. The proteomic analysis revealed that 6 proteins were differentially expressed in the high-dose group. Two stress-responsive proteins, ie heat shock cognate 71 kDa protein (HSC70) and peroxiredoxin-6 (PRDX6), were regulated at the expression level. The remaining 4 proteins involving in cellular energy metabolism, including isoforms of methylmalonyl-coenzyme A mutase (MUT), nucleoside diphosphate-linked moiety X motif 19 (Nudix motif19), mitochondrial NADH dehydrogenase 1 alpha subcomplex subunit 10 (NDUFA10) and nucleoside diphosphate kinase B (NDK B), were modified at the post-translational levels.
Conclusion:
Our findings suggest that the mitochondrion is the primary target of ASB and that ASB-induced nephrotoxicity results from oxidative stress mediated by superoxide produced by complex I.
Keywords: andrographolide sodium bisulfite, Lianbizhi injection, nephrotoxicity, proteomics, reactive oxygen species, peroxiredoxin-6
Introduction
Andrographis paniculata (Burm f) Nees (Acanthaceae) (A paniculata, Chuanxinlian) is a widely used Chinese medicinal herb that is believed to be effective for relieving fever, inflammation and pain according to traditional Chinese medicine. Compared with other Chinese medicinal herbs, A paniculata has been well studied. The major bioactive compounds are diterpenoids, flavonoids and polyphenols1, 2. A paniculata has a variety of health benefits, including anti-inflammation, anti–cancer and immunity enhancement. Andrographolide (C20H30O5), the major diterpenoid of A paniculata, has multiple pharmacological properties and is a potential chemotherapeutic agent3.
Andrographolide sodium bisulfite (ASB, C20H29O7SNa, 436.23) is a water-soluble sulfonate of andrographolide that is synthesized by an addition reaction with sodium bisulfite. Lianbizhi (LBZ) injection, which contains ASB as its sole component, has been used clinically in mainland China to treat infectious diseases such as bacillary dysentery, mumps, laryngitis, tonsillitis and upper respiratory tract infections4. However, reports of adverse drug reactions (ADRs) have increased in the past decade, leading to the issue of a drug use warning from the State Food and Drug Administration of China5. The clinical ADRs to LBZ injection are diverse, the most serious of which is acute renal failure4, 6, 7, 8. There are various explanations for the cause of ADRs to LBZ, such as allergic reactions6, cross-reactions with aminoglycosides in combination therapy4 and nephrotoxicity7, but extensive studies of the toxicology of LBZ injection are still scarce.
Injection agents for Chinese medicinal herbs generally include bioactive ingredients and additives. Bioactive ingredients are usually synthesized, semi-synthesized or extracted from herbs; additives are added to increase the hydrophilicity of the active ingredient. The toxicities of the injection agents could therefore be due to the toxicities of the bioactive ingredients, additives, and/or impurities resulting from the manufacturing process. The potential nephrotoxicity of LBZ injections has been suggested by several recent toxicological studies. Renal damage triggered by a single intravenous injection of two kinds of LBZ with different purities has been reported9. We previously used purified ASB (>99% pure), the raw material for LBZ injection, to investigate its toxicity in kidneys10. Administration of ASB at a high dose (1000 mg/kg) for 7 d resulted in renal dysfunction and an increase in both the serum creatinine and blood urea nitrogen levels. In addition, microscopic examination revealed the presence of tubular interstitial injury and cloudy swelling in the proximal tubule in the high-dose group. However, the mechanism of renal toxicity of ASB remains unclear.
A global proteomics approach typically links separation and identification technologies to create a protein profile or differential protein display. Proteomic techniques have been used in kidney toxicity studies of chemicals such as the antibiotic gentamicin and the chemotherapeutic agent cisplatin and have provided insight into the mechanisms governing key proteins in critical biological pathways that create adverse drug effects11.
In the present study, a differential proteomic analysis of the mouse kidney was performed to identify the key proteins involved in kidney dysfunction induced by ASB administration and to investigate the mechanism of ADRs to LBZ. A two-dimensional (2-D) electrophoresis proteomic approach revealed that six proteins were significantly differentially expressed in the murine kidney upon exposure to ASB.
Materials and methods
Animals and treatment
Male ICR mice weighing 18–22 g were purchased from the Zhejiang Experimental Animal Center. All animals were housed with a 12-h light/12-h dark cycle at 22 °C and 55%±5% relative humidity. Food (Zhejiang Experimental Animal Center, China) and tap water were provided ad libitum. All experiments were carried out according to the guidelines of China for the care and use of laboratory animals. Each of the mice was randomly assigned to one of three experimental groups (each n=10): high-dose ASB (1000 mg/kg), low-dose ASB (150 mg/kg) and a control group. The ASB groups were induced with daily intravenous (iv) injections of 20 mL/kg body weight of ASB (99%, Zhejiang Jiuxu Pharmaceutical Co, Ltd, China) in a 0.9% sodium chloride injection solution (Huadong Pharmaceutical Co, Ltd, Hangzhou, China) for 7 d. The control group received an equal volume of 0.9% sodium chloride injection solution. Mice were sacrificed 1 h after the last injection, and the kidneys were surgically removed for further analysis.
Measurement of malondialdehyde content and superoxide dismutase activity
One removed kidney from each animal was homogenized with saline 1:10 (w/v) and centrifuged at 3000×g for 10 min. The supernatant was used to measure the content of malondialdehyde (MDA), one of the end products of lipid peroxidation and an indicator of reactive oxygen species (ROS) production, and the specific activity of superoxide dismutase (SOD), using commercially available kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) in accordance with the manufacturer's protocols. The data were statistically analyzed using Student's t-test to compare the means of two different groups.
Protein extraction for proteomic analysis
Renal proteins were extracted as previously described12. After removing the renal capsule, the kidney was excised into several thin slices and washed in ice-cold PBS to remove the contaminating blood. The tissue was frozen in liquid N2, ground to powder, and resuspended in lysis buffer containing 7 mol/L urea, 2 mol/L thiourea, 4% (w/v) CHAPS, 2% (v/v) ampholyte (pH 3–10), 120 mmol/L DTT, 40 mmol/L Tris-base and protease inhibitor cocktail (50 μL for 1 mL lysis buffer, Sigma), followed by a 30-min incubation at RT. After centrifugation (10 000×g for 5 min at 4 °C), the supernatant was stored at -80 °C as a 500-μL aliquot, and the protein concentration was determined using the Bradford method with a Bio-Rad Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA, USA).
2-D electrophoresis and staining
Immobiline™ DryStrips, pH 3–10 NL (non-linear), 13 cm long (Amersham Biosciences, Uppsala, Sweden) were rehydrated overnight with rehydration buffer (250 μL per strip) containing 7 mol/L urea, 2 mol/L thiourea, 2% (w/v) CHAPS, 0.5% (v/v) ampholytes (pH 3–10), 40 mmol/L DTT, 40 mmol/L Tris-base, and 0.002% (w/v) bromophenol blue. The first-dimension separation, isoelectric focusing (IEF), was performed in an Ettan IPGphor III IEF System (Amersham Biosciences, Uppsala, Sweden) at 20 °C using the stepwise mode to reach 23 100 V·h with cup loading. For each sample, 400 μg total protein was premixed with the rehydration buffer, followed by a short spin-down before loading a sample cup with 100 μL. After IEF, the proteins on the strip were equilibrated for 15 min in buffer containing 6 mol/L urea, 130 mmol/L DTT, 30% (v/v) glycerol, 75 mmol/L Tris-base, 2% (w/v) SDS, and 0.002% (w/v) bromophenol blue, and then in buffer containing 6 mol/L urea, 135 mmol/L iodoacetamide, 30% (v/v) glycerol, 75 mmol/L Tris-base, 2% (w/v) SDS, and 0.002% (w/v) bromophenol blue for 15 min. The IPG strip was then transferred onto a 12.5% acrylamide slab gel for the second-dimension separation, which was performed in an Ettan DALTsix Electrophoresis Unit (Amersham Biosciences, Uppsala, Sweden) with a current of 12 mA/gel for 4 h at 25 °C. The separated protein spots were visualized with CBB G-250 stain using the Colloidal Coomassie staining protocol in the GE Healthcare handbook (80-6429-60).
Spot analysis, matching and statistical analysis
Image Master 2D Platinum 7.0 (Amersham Biosciences) software was used to match and analyze protein spots on 2-D gels and for the statistical analysis. The parameters used for spot detection were (i) mini-mal area=5 pixels; (ii) smooth factor=3.0; and (iii) saliency=2000.
Protein identification
The protein spots of interest were analyzed by matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF)/TOF mass spectrometry (MS) by a commercial company (Beijing Proteome Research Center, Beijing, China). The proteins were subsequently identified with the MASCOT search algorithm (http://www.matrixscience.com) using a combination of peptide mass fingerprint profiles and MS/MS sequencing data. The UniProt Knowledgebase and NCBI databases were used.
Results
Index of renal oxidation
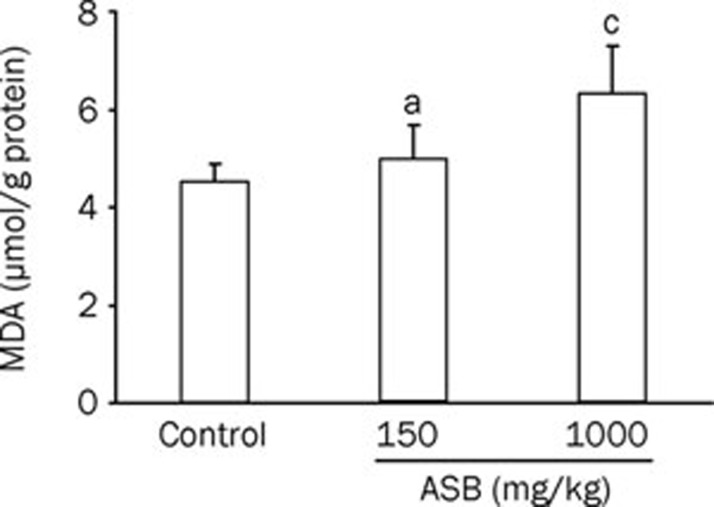
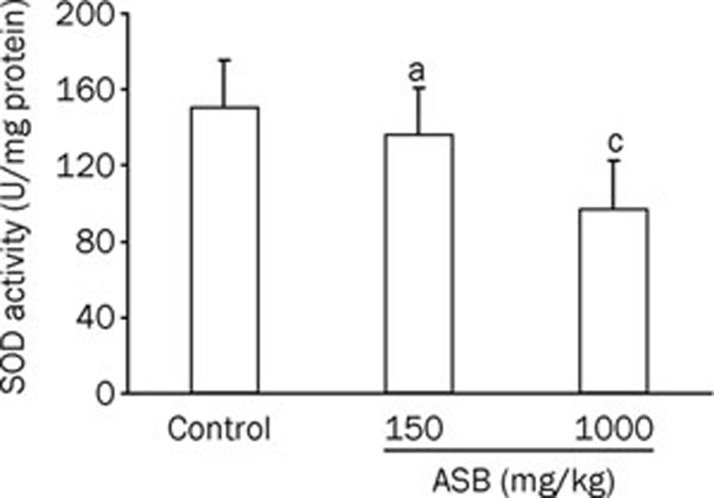
Oxidative stress is a pathway for renal injury13. To investigate whether the renal damage induced by ASB is caused by ROS, the levels of SOD and MDA in the renal tissue were analyzed (Figure 1, 2). Upon ASB treatment, SOD activity decreased, whereas the MDA content increased. The downregulation of SOD activity and the upregulation of MDA were both dose-dependent. However, only the group treated with the high dose of ASB was significantly different from the control group, suggesting that the toxicity is only trigged by a high concentration of ASB, consistent with our previous results10.
Figure 1.
Concentration changes in MDA in the kidneys of mice exposed to ASB. The homogenates of kidneys from mice treated with or without ASB iv administration for 7 d were used to measure MDA content. n=10. Mean±SD. aP>0.05, cP<0.01 vs control, t-test.
Figure 2.
Specific activity changes in SOD in the kidneys of mice exposed to ASB. The homogenates of kidneys from mice treated with or without ASB iv administration for 7 d were used to measure the SOD specific activity. n=10. Mean±SD. aP>0.05, cP<0.01 vs control, t-test.
Differentially expressed renal proteins
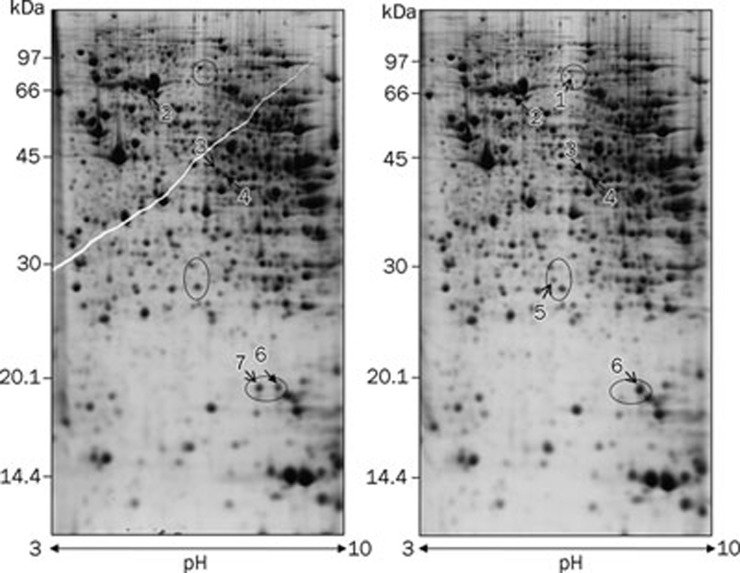
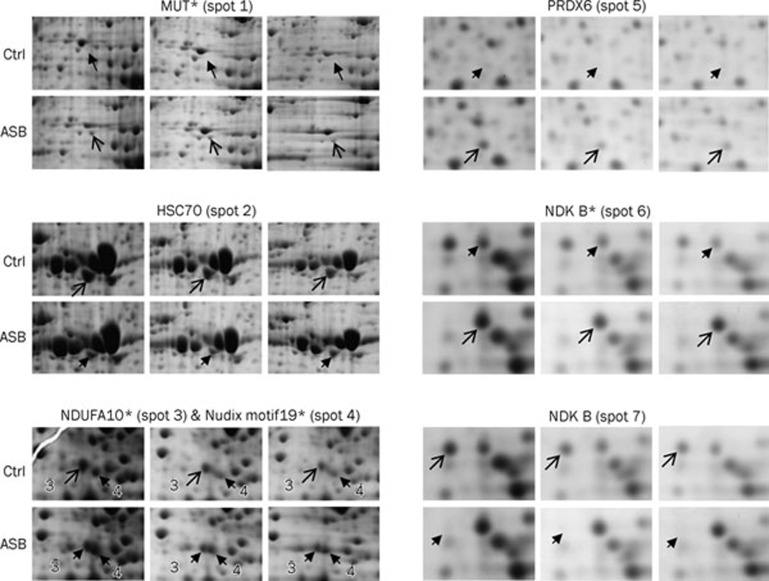
Because only the high dose of ASB caused significant changes in renal function and redox status, this study focused on the proteomic analysis of the kidney after treatment with a high dose of ASB. Approximately 500 protein spots were visualized in each 2-D gel. Representative 2-D gels of both the high-dose group and the control group are illustrated in Figure 3, in which the proteins that were differentially expressed in the ASB group are numbered. In total, seven differentially expressed protein spots in the 2-D gel were identified by MALDI TOF/TOF MS (Table 1). Because two protein spots (spots 6 and 7) were identified as the same protein, nucleoside diphosphate kinase B (NDK B), six proteins were actually regulated by ASB administration. The individual changes in the seven spots were magnified (Figure 4). Of the seven protein spots, three were upregulated, including methylmalonyl-Coenzyme A mutase (MUT, spot 1), peroxiredoxin-6 (PRDX6, spot 5) and NDK B (spot 6). The other three spots, heat shock cognate 71 kDa protein (HSC70, spot 2), mitochondrial NADH dehydrogenase 1 alpha subcomplex subunit 10 (NDUFA10, spot 3) and NDK B (spot 7) were downregulated. The remaining spot (spot 4), which was identified as nucleoside diphosphate-linked moiety X motif 19 (Nudix motif19), displayed an acidic shift with a slight molecular weight increase in the ASB group, suggesting an ASB-induced post-translational modification (PTM). The increase in MUT (spot 1) was also due to an ASB-induced PTM because it showed an acidic shift relative to its theoretical pI and was found only in ASB group gels. In contrast, both NDUFA10 (spot 3) and NDK B (spot 6) exhibited different pIs from their theoretical values, even in the control group (Table 1), suggesting an acidic isoform of NDUFA10 and a basic isoform of NDK B, respectively. Because they were present in both the ASB group and the control group, NDUFA10 (spot 3) and NDK B (spot 6) were considered to be the respective isoforms under physiological conditions. Interestingly, the two spots of NDK B were regulated oppositely: the basic isoform (spot 6) was upregulated, whereas the spot exhibiting the theoretical pI (spot 7) was downregulated, suggesting that ASB treatment shifted NDK B to its basic isoform.
Figure 3.
Representative 2-D gel images of kidney proteins isolated from mice with (right) or without (left) ASB treatment. For each sample, 400 μg of total protein extracted from the mouse kidney was loaded onto a 13-cm IPG strip for isoelectric focusing and separated by SDS-PAGE (12.5%) with CBB staining. The differentially expressed spots detected by Image Master 2D Platinum 7.0 are numbered and summarized in Table 1. Open arrows indicate spots with greater abundance between the corresponding gels.
Table 1. Differentially expressed proteins in the kidney of mouse treated with ASB identified by TOF/TOF MS.
| Spot | Protein | Gene | Accession (UnifrotKB) | Functional classification | MW* (kDa) | pI theoretical | pI estimated | Mascot score | Sequence coverage | ASB effect | Fold change |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Methylmalonyl-Coenzyme A mutase (MUT) | Mut | Q3UJU1 | Amino acid degradation; propanoate metabolism | 82791.6 | 6.45 | 5.95 | 158 | 25% | ↑ | 4.9 |
| 5 | Peroxiredoxin-6 (PRDX6) | Prdx6 | O08709 | Response to ROS | 24871 | 5.7 | 5.7 | 351 | 40% | ↑ | 4.6 |
| 6 | Nucleoside diphosphate kinase B (NDK B) | Nme2 | Q01768 | Nucleotide metabolism | 17351.9 | 6.97 | 7.48 | 184 | 59% | ↑ | 1.9 |
| 2 | Heat shock cognate 71 kDa protein (HSC70) | Hspa8, Hsc70 | P63017 | Stress response | 68736.2 | 5.37 | 5.4 | 225 | 21% | ↓ | 2.3 |
| 3 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subuint 10, mitochondrial (NDUFA10) | Ndufa10 | Q99LC3 | Oxidative phosphorylation | 40577.7 | 7.63 | 6.1 | 72 | 23% | ↓ | 1.3 |
| 7 | Nucleoside diphosphate kinase B (NDK B) | Nme2 | Q01768 | Nucleotide metabolism | 17351.9 | 6.97 | 6.97 | 252 | 46% | ↓ | 6.2 |
| 4 | Nucleoside diphosphate-linked moiety X motif 19, mitochondrial (Nudix motif19, RP2p) | Nudt19, D7Rp2e | P11930 | Fatty acid oxidation | 40294.4 | 6.22 | 6.18 | 261 | 16% | shift | – |
*Molecular weights estimated from the experiment data in this study are all similar to the theoretic one.
Figure 4.
Magnified images of the altered proteins in the mouse kidney induced by ASB treatment. The spots numbered in Figure 3 are shown in the corresponding regions of the individual gels (n=3). The spots with pI values that differ from the theoretical values are marked with asterisks. Open arrows indicate spots with greater abundance between the corresponding gels.
Discussion
In the present study, we used differential proteomic analysis to identify the proteins regulated by ASB in the mouse kidney. Seven spots, which were identified as six proteins, were differentially expressed and/or post-translationally modified after ASB treatment and identified by MALDI TOF/TOF MS (Table 1). This is report to demonstrate an alteration in kidney protein expression following ASB-induced renal injury.
Protein involved in cellular energetics
Nudix motif19 is a member of the Nudix hydrolase superfamily and was recently identified as a CoA diphosphatase involved in fatty acid β-oxidation14. Nudix motif19 is highly expressed in the mouse kidney at both the protein15 and mRNA levels16, suggesting it has an important role in the kidney. The slight PTM of Nudix motif19 induced by ASB reflects a disturbance in fatty acid catabolism, which could lead to catabolite accumulation and result in the inhibition of fatty acid β-oxidation and a reduction in energy supply.
MUT is strictly a mitochondrial enzyme. It catalyzes the isomerization of methylmalonyl-coenzyme A (CoA) to succinyl-CoA17 and is the key enzyme for the transfer of the catabolites of branched-chain amino acids and odd-chain fatty acids into the tricarboxylic acid cycle18. A knockout of Mut in the mouse results in tubulointerstitial renal disease in response to respiratory chain dysfunction19. A similar interstitial nephritis was triggered by ASB in our previous study10. The MUT dysfunction resulting from the ASB-induced acidic shift in pI in this study may be similar to the inactivation caused by Mut deletion. Therefore, the ASB-induced nephrotoxicity is likely due to respiratory chain dysfunction.
NDUFA10 is a subunit of mitochondrial respiration chain complex I (NADH: ubiquinone oxidoreductase)20. PTMs regulate the activity and the interactive capacity of complex I subunits21. The phosphorylation of NDUFA10 in bovine heart has been proposed to influence the affinity of NADH binding and the activity of complex I22. In this study, one of the acidic isoforms of NDUFA10 was more highly expressed in the control group, in agreement with its important role in the electron transport chain of intact mitochondria. The ASB group displayed a relative reduction in this isoform, suggesting that ASB partially inhibits PTM and leads to functional decline. Complex I is the major source of superoxide23, and impairment of complex I causes oxidative damage that is a part of the progression of different pathologies, including Parkinson's disease24. Although further study is needed to identify the PTM of NDUFA10 in the mouse kidney, the partial inhibition of the PTM of NDUFA10 upon treatment with ASB suggests aberrant superoxide generation by complex I. Excessive endogenous ROS can contribute to acute renal injury13. Hence, the ASB-induced changes in the PTM of NDUFA10 might contribute to ASB nephrotoxicity.
NDK B belongs to the nm23 gene family and encodes nucleoside diphosphate kinase (NDK), which has multiple functions that are involved not only in cell energy conversion but also in many cellular processes25. In this study, NDK B was identified from two spots in the control group and one spot in the ASB group due to the ASB-induced basic shift in NDK B, suggesting a change in the function of NDK B. Both NDK B and NDK A are modified after H2O2 treatment26, and Cys109 of NDK A was identified as the site of oxidative modification by ROS27. Because Cys109 is also present in NDK B and no other PTM of NDK B has been reported, the basic isoform of NDK B also seems to be subject to oxidative modification under physiological conditions, which would support the existence of a higher ROS level in the kidney than in other tissues13. Altered NDK expression is associated with the inhibition of cellular proliferation and apoptosis28, and NDK B overexpression reduces H2O2-induced cell death in BAF3 cells29. Furthermore, a recent proteomic analysis revealed that NDK B (pI 7.6) was upregulated in neuronal nitric oxide synthase (nNOS)-knockout mice due to an increased level of superoxide30. Therefore, the increase in the basic isoform of NDK B (pI 7.48) in our proteomic analysis may enable its antioxidative function against superoxide triggered by ASB, which is partially supported by the increased MDA content in the ASB group.
The above proteins were all modified by ASB at the post-translational level and are all mitochondrial proteins except NDK B, a dual-localized mitochondrial protein15. Their modification by ASB reflects a decline or loss of mitochondrial activity in cellular energy metabolism, especially in cell respiration. Taken together, the alterations in the protein profile suggest that the mitochondrion is likely the primary target of ASB.
Proteins involved in the stress response
HSC70 is a required chaperone-mediated protein folding cofactor that belongs to the heat shock protein (HSP) 70 family. Unlike its cognate protein HSP70, which is induced only under stress, HSC70 is constitutively expressed and is involved in other housekeeping functions, including cell cycle regulation and the stress response. Depletion of the HSC70 pool has been linked to a block in the activation of caspases that inhibit apoptosis signaling31, 32. Attenuated HSC70 expression was also observed in ochratoxin A-induced cell death associated with ROS generation33. Therefore, the decrease in HSC70 after ASB administration indicates that apoptosis signaling was activated in renal cells in response to oxidative stress. In addition, PRDX6, an antioxidant enzyme, was upregulated by ASB. PRDX6 belongs to the peroxiredoxin family, and its overexpression in mice or transfected cells reduces cellular H2O2 levels and decreases oxidative stress-induced apoptosis34. The regulation of stress-responsive proteins suggests a correlation between oxidative stress and the renal toxicity of ASB that occurs via cell apoptosis. This proposal is supported by the alterations in the biochemical index, including an increase in the MDA content and a decrease in SOD activity, which suggest that a redox imbalance occurs in the mouse kidney after ASB treatment.
Upregulation of PRDX6 by superoxide
Although PRDX6 is frequently reported to increase in vitro in response to cytotoxic substrates, such as glucose and H2O2, most efforts have failed to detect a corresponding increase in vivo35, 36. Several mechanisms for the in vivo upregulation of PRDX6 have been described, including the high level of superoxide caused by a lack of NO to scavenge superoxide in the nNOS-knockout mouse30, the excessive ROS generated by complex I due to increased glutathione oxidation after ischemia-reperfusion37, and the high level of endogenous ROS resulting from the markedly reduced GSH level in a CFTR-defective mouse lung38. Complex I is the major source of superoxide23. Furthermore, superoxide production by complex I increases in response to oxidation of the mitochondrial GSH pool39. From these previous studies, it has been suggested that PRDX6, which likely belongs to a core cellular antioxidative defense, is upregulated by superoxide generated by complex I. However, not all ROS inducers increase the in vivo expression of PRDX6. Paraquat fails to upregulate PRDX6 in the mouse lung38. A similar result was observed in the rat kidney after ip injection of chloroform40. To our knowledge, this is report of a reagent that can upregulate PRDX6 in the kidney in vivo.
In summary, we have identified changes in six renal mouse proteins after exposure to ASB, all of which were related to cellular energetics or the oxidative stress response. The modification of MUT and NDUFA10 is related to the dysfunction of the mitochondrial respiratory chain, and the regulation of NDK B and PRDX6 is associated with abnormal superoxide generation. Our results suggest that oxidative stress in response to superoxide production in the mitochondria plays an important role in the renal toxicity induced by ASB treatment. The proteomics data in this study provide new insights into the biochemical pathways involved in ASB nephrotoxicity, which will aid in the future reduction of ADRs to LBZ in clinical applications.
Author contribution
Hong LU, Xin-yue ZHANG, and Li-ying ZHU designed the experiments; Hong LU, Xin-yue ZHANG, and Yan-quan ZHOU performed the experiments; Li-ying ZHU and Hong LU performed data analysis and prepared the manuscript; Xin WEN revised the manuscript.
Acknowledgments
This work was supported by the Science Foundation of Zhejiang Provincial Education Administration Y201016256. We thank Dr Yue-zhi TAO and the members of his laboratory at Zhejiang Academy of Agricultural Sciences for their full support. We also thank Dr Jin-mei ZHOU for critically reading the manuscript.
References
- Xu C, Chou GX, Wang ZT. A new diterpene from the leaves of Andrographis paniculata Nees. Fitoterapia. 2010;81:610–3. doi: 10.1016/j.fitote.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Rao YK, Vimalamma G, Rao CV, Tzeng YM. Flavonoids and andrographolides from Andrographis paniculata. Phytochemistry. 2004;65:2317–21. doi: 10.1016/j.phytochem.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Chao WW, Lin BF. Isolation and identification of bioactive compounds in Andrographis paniculata (Chuanxinlian) Chin Med. 2010;5:17. doi: 10.1186/1749-8546-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Ma Q, Yu L, Meng YJ. The clinical applications and adverse reaction of Lianbizhi-injection. China Medical Herald. 2008;5:97. [Google Scholar]
- State Food and Drug Administration [homepage on the Internet]. Beijing: The Administration; c2000–2005 [updated 2005 Apr 12; cited 2010 Nov 30]. Adverse Drug Reaction Information Bulletin vol 8; [about 1 screen]. Available from: : http://www.sda.gov.cn/WS01/CL0078/11280.html .
- Cai WP, Zhou HX, Zhu YQ, Yan F, Huang HH. The report of 10 cases of Lianbizhi-induced acute interstitial nephritis. Journal of Qiqihar Medical College. 2002;23:297. [Google Scholar]
- Zhao JW, Ni ZH, Cao LO, Mou S, Fang W, Zhang QY. Analysis of clinical and pathological of Lianbizhi injection-induced acute renal failure. Chinese Journal of Integrated Traditional and Western Nephrology. 2005;9:529–31. [Google Scholar]
- Wang HM, Du WM, Wang XY, Xu JL, Wang TC, Jin HL. Analysis of adverse reaction/event report in 70 cases of Lianbizhi-injection. Chin J Clin Pharm. 2007;16:252–4. [Google Scholar]
- Hu ZH, Wu CQ, Wang QJ, Wang QX, Luo YW, Yang BH, et al. Toxic effects of two kinds of Lianbizhi injections on rats. Adverse Drug Reactions Journal. 2010;12:10–6. [Google Scholar]
- Lu H, Zhang XY, Zhou YQ, Jin SS. Toxic actions of andrographolide sodium bisulfite on kidney of mice and rabbits. Chin J Pharmacol Toxicol. 2010;24:1–5. [Google Scholar]
- Merrick BA, Witzmann FA. The role of toxicoproteomics in assessing organ specific toxicity. EXS. 2009;99:367–400. doi: 10.1007/978-3-7643-8336-7_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongboonkerd V, Chutipongtanate S, Kanlaya R, Songtawee N, Sinchaikul S, Parichatikanond P, et al. Proteomic identification of alterations in metabolic enzymes and signaling proteins in hypokalemic nephropathy. Proteomics. 2006;6:2273–85. doi: 10.1002/pmic.200500530. [DOI] [PubMed] [Google Scholar]
- Nath KA, Norby SM. Reactive oxygen species and acute renal failure. Am J Med. 2000;109:665–78. doi: 10.1016/s0002-9343(00)00612-4. [DOI] [PubMed] [Google Scholar]
- Ofman R, Speijer D, Leen R, Wanders Ronald JA. Proteomic analysis of mouse kidney peroxisomes: identification of RP2p as a peroxisomal nudix hydrolase with acyl-CoA diphosphatase activity. Biochem J. 2006;393:537–43. doi: 10.1042/BJ20050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini D, Calvo S, Chang B, Sheth S, Vafai S, Ong S, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–23. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasmi L, McLennan AG. The mouse Nudt7 gene encodes a peroxisomal nudix hydrolase specific for coenzyme A and its derivatives. Biochem J. 2001;357:33–8. doi: 10.1042/0264-6021:3570033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton WA, Gravel RA, Rosenblatt DS.Disorders of propionate and methylmalonate metabolismIn: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, et al, editors. The metabolic and molecular bases of inherited disease. New York: McGraw-Hill; 2001p 2165–92. [Google Scholar]
- Chandler RJ, Sloan J, Fu H, Tsai M, Stabler S, Allen R, et al. Metabolic phenotype of methylmalonic acidemia in mice and humans: the role of skeletal muscle. BMC Med Genet. 2007;8:64. doi: 10.1186/1471-2350-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler RJ, Zerfas PM, Shanske S, Sloan J, Hoffmann V, DiMauro S, et al. Mitochondrial dysfunction in mut methylmalonic acidemia. FASEB J. 2009;23:1252–61. doi: 10.1096/fj.08-121848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J, Fearnley IM, Shannon RJ, Hirst J, Walker JE. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol Cell Proteomics. 2003;2:117–26. doi: 10.1074/mcp.M300014-MCP200. [DOI] [PubMed] [Google Scholar]
- Muñoz J, Fernández-Irigoyen J, Santamaría E, Parbel A, Obeso J, Corrales FJ. Mass spectrometric characterization of mitochondrial complex I NDUFA10 variants. Proteomics. 2008;8:1898–908. doi: 10.1002/pmic.200701085. [DOI] [PubMed] [Google Scholar]
- Schulenberg B. Analysis of steady-state protein phosphorylation in mitochondria using a novel fluorescent phosphosensor dye. J Biol Chem. 2003;278:27251–5. doi: 10.1074/jbc.C300189200. [DOI] [PubMed] [Google Scholar]
- Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29:222–30. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- Schilling B, Aggeler R, Schulenberg B, Murray J, Row R, Capaldi R, et al. Mass spectrometric identification of a novel phosphorylation site in subunit NDUFA10 of bovine mitochondrial complex I. FEBS Lett. 2005;579:2485–90. doi: 10.1016/j.febslet.2005.03.061. [DOI] [PubMed] [Google Scholar]
- Mehta A, Orchard S. Nucleoside diphosphate kinase (NDPK, NM23, AWD): recent regulatory advances in endocytosis, metastasis, psoriasis, insulin release, fetal erythroid lineage and heart failure; translational medicine exemplified. Mol Cell Biochem. 2009;329:3–15. doi: 10.1007/s11010-009-0114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song EJ, Kim YS, Chung JY, Kim E, Chae SK, Lee KJ. Oxidative modification of nucleoside diphosphate kinase and its identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Biochemistry. 2000;39:10090–7. doi: 10.1021/bi000267a. [DOI] [PubMed] [Google Scholar]
- Lee E, Jeong J, Kim SE, Song EJ, Kang SW, Lee KJ, et al. Multiple functions of nm23-h1 are regulated by oxido-reduction system. PLoS ONE. 2009;4:e7949. doi: 10.1371/journal.pone.0007949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi D, Lacombe ML, Paggi MG. nm23: unraveling its biological function in cell differentiation. J Cell Physiol. 2000;182:144–9. doi: 10.1002/(SICI)1097-4652(200002)182:2<144::AID-JCP2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Arnaud-Dabernat S, Masse K, Smani M, Peuchant E, Landry M, Bourbon PM, et al. Nm23-M2/NDP kinase B induces endogenous c-myc and nm23-M1/NDP kinase A overexpression in BAF3 cells. Both NDP kinases protect the cells from oxidative stress–induced death. Exp Cell Res. 2004;301:293–304. doi: 10.1016/j.yexcr.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Da Silva-Azevedo L, Jähne S, Hoffmann C, Stalder D, Heller M, Pries AR, et al. Up-regulation of the peroxiredoxin-6 related metabolism of reactive oxygen species in skeletal muscle of mice lacking neuronal nitric oxide synthase. J Physiol. 2009;587:655–68. doi: 10.1113/jphysiol.2008.164947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin B, Hartnett KA, Erhardt JA, Legos JJ, White RF, Barone FC, et al. Caspase 3 activation is essential for neuroprotection in preconditioning. Proc Natl Acad Sci U S A. 2003;100:715–20. doi: 10.1073/pnas.0232966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JE, Zeiger SLH, Hettinger JC, Brooks JD, Holt B, Morrow JD, et al. Essential role of the redox-sensitive kinase p66shc in determining energetic and oxidative status and cell fate in neuronal preconditioning. J Neurosci. 2010;30:5242–52. doi: 10.1523/JNEUROSCI.6366-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Cong WT, Bang Y, Lee SN, Yoon CS, Kwack SJ, et al. Proteome response to ochratoxin A-induced apoptotic cell death in mouse hippocampal HT22 cells. Neurotoxicology. 2009;30:666–76. doi: 10.1016/j.neuro.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Walsh B, Pearl A, Suchy S, Tartaglio J, Visco K, Phelan SA. Overexpression of Prdx6 and resistance to peroxide-induced death in Hepa1–6 cells: Prdx suppression increases apoptosis. Redox Rep. 2009;14:275–84. doi: 10.1179/135100009X12525712409652. [DOI] [PubMed] [Google Scholar]
- Morrison J. Effect of high glucose on gene expression in mesangial cells: upregulation of the thiol pathway is an adaptational response. Physiol Genomics. 2004;17:271–82. doi: 10.1152/physiolgenomics.00031.2004. [DOI] [PubMed] [Google Scholar]
- Lee CK, Kim HJ, Lee YR, So HH, Park HJ, Won KJ, et al. Analysis of peroxiredoxin decreasing oxidative stress in hypertensive aortic smooth muscle. Biochim Biophys Acta. 2007;1774:848–55. doi: 10.1016/j.bbapap.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Eismann T, Huber N, Shin T, Kuboki S, Galloway E, Wyder M, et al. Peroxiredoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G266–74. doi: 10.1152/ajpgi.90583.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudel S, Kelly M, Fritsch J, Nguyen-Khoa T, Thérond P, Couturier M, et al. Peroxiredoxin 6 fails to limit phospholipid peroxidation in lung from cftr-knockout mice subjected to oxidative challenge. PLoS ONE. 2009;4:e6075. doi: 10.1371/journal.pone.0006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ER, Hurrell F, Shannon RJ, Lin TK, Hirst J, Murphy MP. Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J Biol Chem. 2003;278:19603–10. doi: 10.1074/jbc.M209359200. [DOI] [PubMed] [Google Scholar]
- Fujii T, Fujii J, Taniguchi N. Augmented expression of peroxiredoxin VI in rat lung and kidney after birth implies an antioxidative role. Eur J Biochem. 2001;268:218–25. doi: 10.1046/j.1432-1033.2001.01843.x. [DOI] [PubMed] [Google Scholar]