Abstract
Nucleotide-binding oligomerization domain (NOD)-containing protein-like receptors (NLRs) are a recently discovered class of innate immune receptors that play a crucial role in initiating the inflammatory response following pathogen recognition. Some NLRs form the framework for cytosolic platforms called inflammasomes, which orchestrate the early inflammatory process via IL-1β activation. Mutations and polymorphisms in NLR-coding genes or in genetic loci encoding inflammasome-related proteins correlate with a variety of autoinflammatory diseases. Moreover, the activity of certain inflammasomes is associated with susceptibility to infections as well as autoimmunity and tumorigenesis. In this review, we will discuss how identifying the genetic characteristics of inflammasomes is assisting our understanding of both autoinflammatory diseases as well as other immune system-driven disorders.
Keywords: autoinflammatory diseases, polymorphisms, NLRs, NLRP3
Introduction
The ability of innate immune cells to sense dangers to the host, such as microbial infections or a disruption in tissue homeostasis, depends upon a plethora of receptor families. The Toll-like receptor (TLR) family is the best-characterized of these,1 but in the last decade new intracellular receptor families have been identified. They include nucleotide-binding oligomerization domain (NOD)-containing protein-like receptors (NLRs), retinoic acid-inducible gene-like receptors and C-type lectin receptors.2, 3, 4 Some members of the NLR family have been deeply investigated for their central role in the formation of functional platforms, named inflammasomes, which mediate the initiation of the inflammatory process through IL-1β and IL-18 secretion.
All these receptors have generated a huge amount of interest recently due to their prominent role in a number of human autoinflammatory diseases. Linkage studies of inherited autoinflammatory conditions identified mutations or polymorphisms in a group of genes named hereditary periodic fever syndromes-associated genes encoding NLR proteins or their inflammasome-related partners. Since then, further genome-wide association studies have characterized specific genetic polymorphisms of NLR genes in relation to autoinflammatory diseases.
Alongside the involvement of NLR inflammasomes with spontaneous inflammatory disorders, other investigations have revealed inflammasome roles in protection against infective agents,5 as well as in the pathogenesis of autoimmunity and tumors.6, 7, 8, 9 How genetic variations influence inflammasome structure and function leading to disease onset is the challenging target for future investigations.
The NLR family
NLRs are fundamental components of the innate immune system that are evolutionarily conserved, having been detected in species as simple as sea urchins. NLR-like proteins functionally and structurally similar to mammalian ones have even been observed in plants.10, 11, 12 This fascinating example of convergent evolution hints at NLR-like structures as an efficient and successful tool of innate immunity against a broad variety of pathogens.
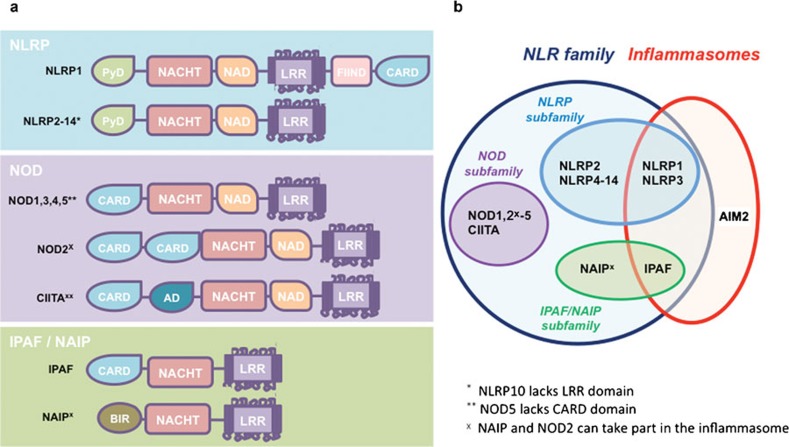
NLRs are cytosolic receptors that are expressed predominately by macrophages and dendritic cells. To date, 23 NLRs have been described in humans and 33 identified in mice. In general, NLRs are composed of three domains: a C-terminal region leucine-rich repeat (LRR), the central nucleotide domain NACHT (also known as NOD) and the N-terminal containing either a caspase recruitment domain (CARD) or pyrin domain (PYD) (Figure 1). LRRs are domains of 20–30 residues forming alpha helix and beta strands, similar to plant disease-resistance genes.13 Indeed, similarly to TLRs, LRR structures in NLRs are important for the recognition of pathogen-associated molecular patterns, although direct evidence of an interaction with known NLR-agonists has not yet been demonstrated. The central domain NACHT shares structural similarities with the central motif of apoptotic protease-activating factor 1, which promotes formation of oligomeric structures. Therefore, an analogous mechanism has been proposed for the formation of the NLR inflammasome platform.14, 15
Figure 1.
NLRs and inflammasomes. (a) Schematic representation of human NLR structures. NLR family comprises the three main subfamilies NLRP, NOD and IPAF/NAIP, characterized by the conserved central oligomerization NACHT domain and the C-terminal LRR domain. The N-terminal CARD or PYD are present in the NOD and NLRP subfamilies, respectively. (b) Among the NLRs, NLRP1, NLRP3 and IPAF participate in the formation of the inflammasome platform. AIM2, not belonging to the NLR family, has also been identified as forming part of a specific inflammasome. AIM2, absent in melanoma 2; CARD, caspase recruitment domain; IPAF, ICE-protease-activating factor; LRR, leucine-rich repeat; NAIP, neuronal apoptosis inhibitory protein; NLR, nucleotide-binding oligomerization domain-containing protein-like receptor; NOD, nucleotide-binding oligomerization domain; PYD, pyrin domain.
The N-terminal CARD and PYD are the key regions linking the NLR to its downstream functions via oligodimerization with an analogous domain harbored by specific adaptor proteins. NLRs with N-terminal domains that contain CARD usually activate signaling cascades leading to transcription of proinflammatory genes (Figure 1). Alternatively an N-terminus PYD will recruit pro-inflammatory caspases.
Based on their molecular structure and function, NLRs can be divided into three subfamilies: NLRP, NOD and ICE-protease-activating factor (IPAF)/neuronal apoptosis inhibitory protein (NAIP) (Figure 1).12 NLRP, also known as NALP (NACHT domain-, LRR- and PYD-containing protein), is the largest subfamily, having 14 members in human and 12 in mouse. The prototypical members are NLRP1 and NLRP3. NLRP3 contains N-terminal PYD, a central NATCH domain responsible for oligomerization and activation, and a NACHT-associated domain followed by the highly conserved LRR region of 171 nucleotides at its C-terminus (Figure 1).16 NLRP1 has a similar structure to NLRP3 except that at the C-terminus of the LRR region, there is a FIIND (domain with function to find) followed by a C-terminal CARD for caspase recruitment. The two also differ in their expression patterns; the NLRP3 gene is strongly expressed in macrophages, dendritic cells, granulocytes and osteoblasts, whereas NLRP1 has a wider expression pattern, including brain, lung, placenta, small intestine, colon, kidney, liver, muscle, testes and epithelial cells. The functions and the tissue and cellular distribution of the other NLRP proteins have not yet been fully characterized.
The NOD family of NLRs includes NOD1–5 and the class II transactivator (Figure 1). An N-terminal CARD is characteristic of the NOD family. NOD1 and NOD2 are the two best-studied members. The natural ligands of NOD1 and NOD2 are moieties of peptidoglycan, a polymer of the glycan chains within bacterial cell walls; specifically, NOD1 detects mesodiaminopimelic acid, while NOD2 recognizes muramyl dipeptide (MDP).17 Upon activation, NOD1 and NOD2 recruit the receptor-interacting protein-2 through homodimerization of CARDs, which leads to the activation of NF-κB.18 NOD1 and NOD2 also engage CARD9, which mediates the activation of mitogen-activated protein kinase cascades responsible for nuclear translocation of Jun and ATF2 transcription factors.19 NF-κB and mitogen-activated protein kinase-mediated pathways trigger a vigorous inflammatory response characterized by secretion of proinflammatory cytokines and chemokines.
The third NLR subfamily is evolutionarily separate from other NLRs and consists of IPAF and NAIP (Figure 1). IPAF contains a CARD, whereas NAIP includes three baculovirus inhibitor domains that are probably involved in inhibition of apoptosis.20 Both IPAF and NAIP are involved in the formation of inflammasomes. Whole bacteria, such as Salmonella typhimurium, Pseudomonas aeruginosa or Legionella pneumophila, as well as bacterial components including flagellin, also promote IPAF oligomerization resulting in recruitment and activation of caspase-1.21 IPAF seems to cooperate with NAIP to induce caspase-1-mediated secretion of IL-1β by macrophages. However, the exact mechanisms underlying IPAF/NAIP detection of bacterial compounds and activation of caspase-1 remain largely unclear.
The inflammasomes
Inflammasomes are the multiprotein cytoplasmic complexes that facilitate activation of proinflammatory caspases.22 To date, three inflammasomes have been described according to the NLR protein they engage: the NLRP1, NLRP3 and IPAF inflammasomes.
NLRP3 is one of the best-characterized inflammasomes. The framework of the NLRP3 inflammasome structure seems to form through homotypic reactions of NLRP3's PYD with the PYD of an adaptor protein called apoptotic-associated speck-like protein containing a caspase recruitment domain (ASC) (Figure 2). ASC then recruits procaspase-1 through CARD–CARD interactions. Similarly, NLRP1 can directly recruit both procaspase-1 and -5 via CARD homotypic interactions. The ability to form inflammasome platforms is not limited to NLRP subfamily members; IPAF can recruit caspase-1 directly via CARD–CARD interactions and therefore the IPAF platform is referred as a third inflammasome prototype.23, 24 There is some evidence that that NAIP can assemble with IPAF to form heterocomplex inflammasomes.25, 26 NLRP1 can also associate with NOD2 to mediate caspase-1-dependent IL-1β secretion in response to Bacillus anthracis and MDP.27, 28
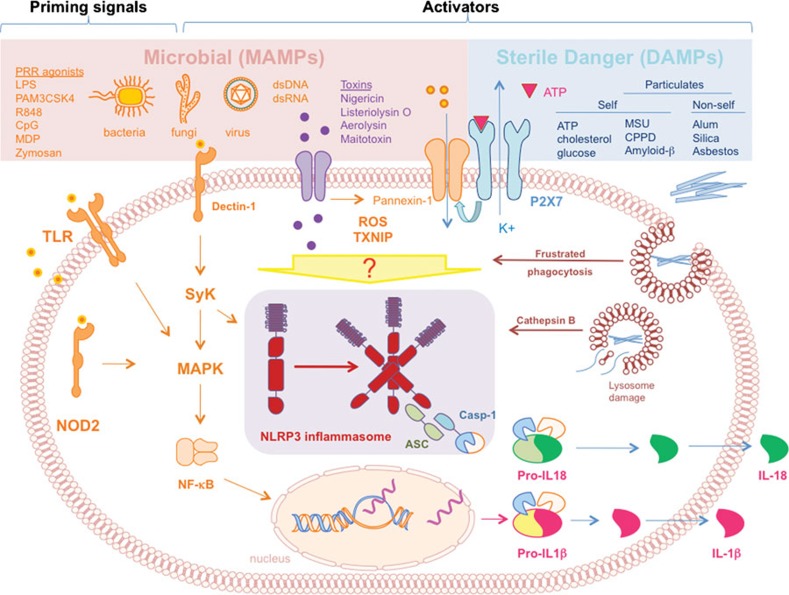
Figure 2.
NLRP3 inflammasome activation pathways. The assembly of the NLRP3 inflammasome in innate cells is induced by several different stimuli, including MAMPs (pink box) or sterile DAMPs (light blue box). A first priming signal delivered predominantly by MAMPs, such as LPS, is required to promote IL-1β transcription via NF-κB translocation to the nucleus. Inflammasome oligomerization and activation can be induced by bacterial and fungal moieties, pore forming toxins, or viral dsDNA and RNA. Upon binding to the P2X7 receptor, ATP also triggers inflammasome formation through K+ efflux and opening of the pannexin-1 channel. Particulates, such as MSU, fibrillar amyloid β, CPPD, silica, asbestos and alum, are phagocytosed and activate the inflammasome. MSU can also promote inflammasome formation via frustrated phagocytosis. ROS formation induced by certain inflammasome activators also mediates inflammasome assembly via recruitment of TXNIP. Inflammasome oligomerization results in ASC-mediated caspase-1 activation responsible for cleavage and release of IL-1β and IL-18 cytokines. ASC, apoptotic-associated speck like protein containing a caspase recruitment domain; CPPD, calcium pyrophosphate dihydrate; DAMP, danger-associated molecular pattern; LPS, lipopolysaccharide; MDP, muramyl dipeptide; MAMP, microbial-associated molecular pattern; MSU, monosodium urate; ROS, reactive oxygen species; TXNIP, thioredoxin-interacting protein; Casp-1, caspase-1.
Recently a fourth type of inflammasome has been identified, involving the protein absent in melanoma 2 (AIM2) (Figure 1).29 Despite the lack of NACHT domain, AIM2 can undergo oligodimerization and recruit ASC via PYD in response to double stranded cytosolic DNA through its C-terminal HIN domain. AIM2 is generating a strong interest as a sensor of DNA and therefore its putative involvement in viral surveillance, as well as onset of autoimmunity against self-DNA, as seen in systemic lupus erythematosus.30, 31, 32 Since AIM2 does not belong to the NLR family, it will not be discussed further.
Activators of the NLRP3 inflammasome
Similar to the TLR family, much work has focused on identifying the stimuli recognized by NLRs. Activation of the inflammasomes can be triggered by exposure to whole live bacterial, fungal or viral pathogens (Salmonella typhimurium, Shigella flexneri, Legionella pneumonia, Pseudomonas aeruginosa, Candida albicans, Saccharomyces cerevisiae, Sendai virus, adenovirus and influenza virus),30, 33, 34, 35 or their components (MDP flagellin),36, 37, 38 but also by toxins required for pathogen entry into the host cell, such as membrane-disrupting and pore-forming compounds (maitotoxin, nigericin, aereolysis, anthrax lethal toxin and alpha-toxin of Staphylococcus aureus) (Figure 2).39, 40, 41, 42, 43 Interestingly, NLRP3 also contributes to the ability of dendritic cells and macrophages to detect endogenous danger molecules released by dying cells. NLRP3 can signal the presence of adenosine triphosphate (ATP), crystals of monosodium urate (MSU) or calcium pyrophosphate crystals,39, 44 and other large particulates of non-microbial origin, such as alum, silica and asbestos (Figure 2).45, 46, 47 The NLRP3 inflammasome has also been proposed as a sensor of adenoviral DNA, double-stranded RNA and viral RNA.30, 35
Proposed mechanisms of inflammasome activation
The NLRP3 inflammasome is the best-characterized, but the precise mechanism of NLRP3 activation is still debated. At steady state, inflammasome components are present in the cytosol and their assembly is prevented by auto-inhibitory mechanisms mediated by chaperone proteins.48 Inflammasome assembly is promoted by the application of specific stimuli. However there is no evidence that inflammasome-activating molecules would interact directly, as specific ligands, with the NLR proteins themselves. Moreover, it is still puzzling how many heterogeneous molecules, which differ broadly in biological properties and structures, could trigger NLRs, such as NLRP3. Multiple mechanisms of NLRP3 activation have been proposed, which may also cooperate in a unique and complex way (Figure 2).
Using cell-free systems it was shown that the NLRP3 inflammasome can spontaneously assemble if potassium levels are lowered below the physiological intracellular concentration of 70 mM.22, 49, 50 This observation suggested that inflammasome formation might also be triggered following disruption of cellular integrity that would similarly cause potassium levels to drop. Therefore, it was suggested that NLRP3 activators might act indirectly through pathways able to lower potassium concentration. This mechanism has been demonstrated for inflammasome activation driven by extracellular ATP, which induces inflammasome assembly and ASC-mediated IL-1β secretion.33, 51 ATP binds to the purinergic receptor P2X7, resulting in the opening of the protein channel pannexin-1.52 This allows potassium efflux, inflammasome activation and ATP release.53 However, it has been proposed that the pore formation following pannexin-1 recruitment could allow cytosolic access to microbial products, such as lipopolysaccharide (LPS), which can directly activate the inflammasome.52 Although it is unclear whether the pore width would be sufficient to allow entrance of such microbial-derived molecules.
The inflammasome activators MSU, alum, asbestos and silica possess a crystal or particulate shape. Inhibition of cytoskeletal rearrangement with colchicine or cytochalasin D blocks IL-1β secretion.44 These observations led to the second model of NLRP3 activation. The attempts to engulf large particulates by phagocytes via a mechanism known as frustrated phagocytosis can generate NADPH-dependent formation of reactive oxygen species (ROS).45, 46, 54 Recently, the thioredoxin-interacting protein was identified as an NLRP3-binding protein linking ROS generation to assembly and activation of the NLRP3 inflammasome.55 In support of this model, antioxidants and NAPDH inhibitors prevent inflammasome assembly.34, 46, 47, 49, 54, 56 Although ROS production is required for NLRP3 activation, several ROS-inducing agents do not promote inflammasome formation.57, 58
A third mechanism of NLRP3 activation proposed by Hornung and colleagues was that, in the case of large particulates, inflammasome activation occurs as a result of lysosomal swelling and leakage.57 In contrast to the hypothesis of frustrated phagocytosis, this model proposes that upon cellular uptake silica and aluminum salts cause lysosome acidification with consequent release of acidic contents into the cytosol. Indeed, inhibition of the lysosomal protease cathepsin B leads to a substantial decrease in activation of the NLRP3 inflammasome induced by silica.57
Inflammasome-activated cytokines: IL-1β and IL-18
The main outcome of NLRP3 inflammasome activation is the recruitment of caspase-1, inducing the cleavage and secretion of the proinflammatory cytokines IL-1β and IL-18. IL-1 family members are potent modulators of both innate and adaptive immunity and are important for host protection against infections. Notably, IL-1β is involved in the early pathogenesis and the sustained severity of a broad pattern of diseases, including many arthritic diseases and septic shock.59 Blocking the IL-1R is a successful treatment for several immune-related disorders.60, 61
IL-1β production is controlled by a two-step mechanism. TLR or NOD agonists transcribe the immature form, pro-IL-1β, via the NF-κB pathway followed by caspase-1-mediated cleavage of pro-IL-1β upon inflammasome activation.59 The NF-κB pathway is also required for transcription of NLRP3.62 To bypass this limitation, most in vitro studies have used inflammasome agonists in combination with other pathogen-associated molecular patterns, most often LPS, to ‘license' inflammasome formation and activation.33, 44, 57, 63, 64, 65 It is important to consider, however, that LPS used as priming signal in several studies could contribute to NLR activation through additional mechanisms beyond pro-IL-1β and NLRP3 transcription.
NLRP3 activation also leads to IL-18 maturation, although this cytokine can also be activated by other caspase-1-independent mechanisms.66, 67 The immature form of IL-18 is constitutively expressed in some cells but only those with an active inflammasome, such as macrophages, dendritic cells and Kupffer cells, have the ability to release the mature form.68
In the last decade, a number of groups have focused on the study of inflammasomes as initiators of immune disorders mediated by IL-1β and IL-18. Thus, many genetic associations between members of NLR family and immune diseases have been discovered. Moreover, a correlation between NLR members and non-immune mediated disorders has also been proposed, suggesting that this protein family has as yet undefined functions crucial for the balanced physiology of the organism. For these reasons, involvement of genetic mutations of inflammasomes in several diseases will be discussed in detail with a particular focus on IL-1-mediated disorders.
Genetics of inflammasomes in autoinflammatory diseases
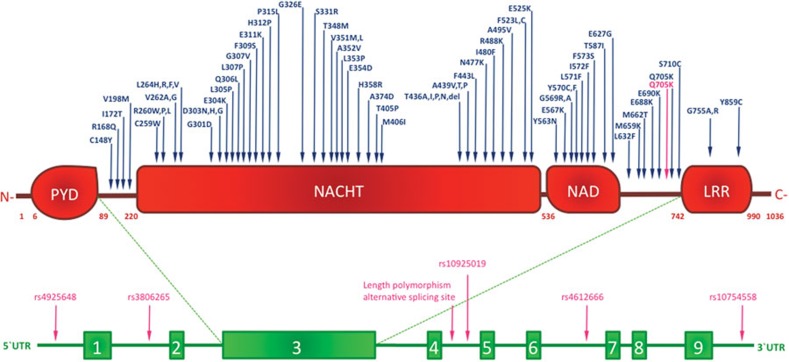
NLRP3 mutations are associated with a group of rare hereditary autoinflammatory diseases called cryopyrin-associated periodic syndromes (CAPS), which includes familial cold auto-inflammatory syndrome (FCAS), Muckle–Wells syndrome (MWS), and chronic infantile neurological, cutaneous, and articular (CINCA) syndrome, also known as neonatal onset multisystem inflammatory disorder.69 These pathologies are listed in order of increasing disease severity and life-threatening potential, and are characterized by recurrent episodes of fever, rush, arthropathies and various degrees of neurological complications.70, 71, 72 Genetic studies of CAPS identified more than 90 disease-associated genetic variants of the NLRP3 gene, the majority of which are autosomal dominant missense point mutations located in exon 3, encoding the NATCH domain (Figure 3 and Table 1).73, 74 Despite the familial recurrence, some genetic variants inducing CAPS phenotypes result from de novo mutations. Interestingly, identical de novo mutations were independently found in different patients and often overlapped with hereditary mutations, suggesting the presence of hot-spot loci within the NLRP3 gene that exhibit high mutation susceptibility.75
Figure 3.
Mutations and polymorphisms in the NLRP3 gene are associated with diseases. Schematic representation of residue substitutions in NLRP3 protein (upper panel), or polymorphysms in NLRP3 gene intronic regions (lower panel). Single residue-allelic variants associated with cryopyrin-associated periodic syndromes or with other NLRP3-related diseases are highlighted in blue and pink, respectively. LRR, leucine-rich repeat; NAD, NACHT-associated domain; PYD, pyrin domain.
Table 1. Inflammasome-related genes correlating with human diseases.
| Disease | Gene | Allele | Mechanism | Ref. |
|---|---|---|---|---|
| MWS | NLRP3 | V198M, R260L, R260W, V262G, L264V, D303N, E311K, H312P, T348M, A352V, A439T, I480F, A495V, F523C(t>g), E567K, G569R, S710C | Spontaneous inflammasome assembly,IL-1β hyperproduction | 69, 71, 72, 74, 75 |
| FCAS | NLRP3 | V198M, C259W, G301D, L353P, T436A, A439V, R488K, E525K, Y563N, E627G, M659K | Spontaneous inflammasome assembly,IL-1β hyperproduction | 71, 72, 74, 75 |
| CINCA syndrome | NLRP3 | C148Y, R168Q, I172T, V198M, R260P, R260W, V262A, V262G, L264H,R,F, D303N, D303H, D303G, E304K, Q306L, G307V, F309S, P315L, G326E, S331R, V351M,L, E354D, H358R, A374D, T405P, M406I, T436I,P,N, T436del, A439P, F443L, N477K, I480F, F523L(c>a), F523L(c>g), G569A, Y570C,F, L571F, I572F, F573S, T587I, L632F, M662T, E688K, E690K, G755A, G755R, Y859C | Spontaneous inflammasome assembly,IL-1β hyperproduction,necrosis-like cell death? | 69, 74, 75, 76, 81, 83 |
| FCAS-like diseases | NLRP12 | R284X | Non-canonical NF-κB activation | 90, 91 |
| Familial Mediterranean fever-like syndrome | PYPAF1 | R554X | Non-canonical NF-κB activation | 97 |
| HIV infection | NLRP3 | SNP 3′ UTR rs10754558 G | Unknown | 99 |
| C. albicans infection (vulvar vestibulitis) | NLRP3 | Intron 4, alternative splicing site | Reduced inflammasome activity? Impaired IL-1β production? | 103 |
| Mycoplasm infection (infertility) | NLRP3 | Intron 4, alternative splicing site | Reduced inflammasome activity?Impaired IL-1β production? | 104 |
| C. trachomatis infection (tubal pathology) | NLRP3 | rs12065526 G>A | Unknown | 105 |
| Crohn's disease | NLRP3 | Q705K (+ C10X CARD8 gene)SNP 5′ UTR rs4925648 C/TSNP rs10925019 C/T | IL-1-mediated? | 85, 86, 87, 88 |
| Psoriatic juvenile idiopathic arthritis | NLRP3 | SNP rs3806265 | IL-1-mediated? | 117 |
| Rheumatoid arthritis | NLRP3 | Q705K (+ C10X CARD8 gene) | IL-1-mediated? | 120 |
| Food-induced anaphylaxis | NLRP3 | SNP rs4612666SNP rs10754558 | IL-1-mediated? | 123 |
| Aspirin-induced asthma | NLRP3 | SNP rs4612666 | IL-1-mediated? | 123 |
| Urticaria | NLRP3 | ? | IL-1β hyperproduction in the skin? | 125 |
| Vitiligo | NLRP1 | 5′ UTR ?3′ UTR rs6502867 | IL-1-mediated autoimmunity?Aberrant apoptosome formation? | 126, 127 |
| Autoimmune Addison's disease | NLRP1 | rs12150220 | IL-1-mediated autoimmunity? | 128 |
| Type 1 diabetes | NLRP1 | rs12150220 | IL-1-mediated autoimmunity? | 128 |
| Type 2 diabetes | NLRP3 | ? | Activation of NLRP3 by glucose? | 133 |
| Hypertension | NLRP3 | Intronic mutation | Activation of NLRP3 by cholesterol? | 131 |
| Alzheimer's disease | IL-1β | ? | Activation of NLRP3 by amyloid β? | 136, 138 |
| Cancer | NLRP3 | ? | IL-1β-mediated inflammation? | 8 |
Abbreviations: CINCA, chronic infantile neurological, cutaneous and articular; FCAS, familial cold autoinflammatory syndrome; MWS, Muckle–Wells syndrome.
It is presently unclear how NLRP3 point mutations affect the functional properties of the inflammasome at a molecular level, leading to the high expressivity and penetrance observed in several CAPS-related genetic variances. Exon 3 missense mutations result in continuous IL-1β secretion caused by constitutive inflammasome assembly and activation (Figure 3). NLRP3 hyperactivation bypasses the need for an NLR agonist. Indeed, blood monocytes from CINCA patients spontaneously release IL-1β upon stimulation by LPS alone without any inflammasome activator.76, 77, 78 This hypothesis is also supported by knock-in mouse models carrying the point mutations R258W and A350V in the exon 3 of NLRP3 gene, which correspond respectively to the human NLRP3 R260W and A352V mutations associated with MWS (Figure 3 and Table 1).75, 77, 79 R258W and A350V mutations result in spontaneous IL-1β release due to aberrant inflammasome structural conformation and hyperactivation in the absence of NLRP3 agonists. Intriguingly, mouse macrophages carrying the L351P mutation, analogous to the human L353P mutation in FCAS patients, showed increased IL-1β secretion when incubated at lower temperatures, mimicking the cold-sensitive phenotype observed in FCAS subjects.79 The pathologies seen in knock-in mouse models of CAPS-associated mutations are similar to human disease syndromes, suggesting that IL-1β hyperactivation is sufficient to initiate and sustain several important inflammatory events. Indeed, therapies blocking the action of IL-1β using an IL-1R antagonist or humanized anti-IL-1β monoclonal antibody are successfully used to treat CAPS and other IL-1-mediated disorders.
Several CAPS-associated mutations and polymorphisms have been identified, but only a few have been analyzed for their effect on the molecular and cellular function. Using an in silico modeling prediction of the NACHT domain structure, Neven and coworkers investigated how the single residue substitutions in NLRP3 frequently associated with CAPS could affect secondary and tertiary protein configurations.75 According to the putative tertiary structure, all the substitutions clustered close to the nucleotide binding cleft, suggesting that nucleotide binding site, located in proximity of the NACHT domain in a specific site outside the structure core, could be crucial for NLRP3 oligomerization. Hence, mutations affecting this region were also suggested to influence the quaternary structure determining aberrant inflammasome formation.75
Mutations in the C-terminal region of the NACHT domain correlate with the most severe CINCA syndrome phenotypes and are associated with significant neurological impairment (Figure 3 and Table 1). LPS-treated monocytes expressing the Y570C mutation showed an increased necrotic-like cell death associated with lysosomal leakage and other cellular metabolic alterations.80, 81 Moreover, monocytes transfected with Y570C exhibited higher NF-κB activity compared to healthy controls.82, 83
A second important aspect is that some CINCA patients with milder symptoms apparently lack NLRP3 mutations but carry a latent low-level mosaicism. This implies that some missense mutations in the NLRP3 gene can exert a dominant phenotype leading to CAPS even when NLRP3 is expressed by less than 25% of the cells in mosaic patients.81, 84 This could be due to the positive feedback loop sustained by the IL-1/IL-1R axis.
Recently disease-correlated missense mutations in exons other than exon 3 have been identified. Exon 6 encodes the LRR domain and the Y859C mutation has been associated with a different CAPS phenotype lacking the typical urticarial rush, while other symptoms were overlapping with late on-set phenotypes of MWS and CINCA (Figure 3).83 Similar to mutations in the NACHT domain, Y859C mutations lead to spontaneous inflammasome formation, ASC recruitment and IL-1β secretion. Remarkably, treatment of patients carrying the Y859C mutation with an IL-1R antagonist was effective, suggesting that a complete screening of the whole NLRP3 gene should be recommended, especially for patients showing non-classical autoinflammatory symptoms.83
Two additional NLRP3 variants, G755R and G755A of exon 4, also result in a CAPS phenotype probably through enhancement of NF-κB activation (Figure 3 and Table 1). However, the role of NLRP3 in regulating the NF-κB pathway is still widely debated. A hint comes from studying the homology among NLRs. Indeed, the missense mutation R334W in the NOD2 gene associated with Crohn's disease exhibits a strong positional and structural analogy to R260W in the NLRP3 gene, which results in aberrant NF-κB activation. Interestingly, an association between Crohn's disease and NLRP3 polymorphisms leading to impaired IL-1β secretion has been proposed.85, 86, 87 However, recent studies in a larger patient group failed to replicate this observation.88 Further work is needed to clarify the possible association of inflammasome genetics to multifactorial diseases.
Although several missense mutations in NLRP3 inflammasomes strongly correlate with CAPS, a significant number of patients with CAPS-like phenotype do not present any mutation in NLRP3 gene.75, 89 Some autoinflammatory syndromes resembling CAPS have been correlated with mutations in inflammasome or IL-1 pathway regulators. NLRP12 mutations are associated with hereditary periodic fever syndromes, and while NLRP12 shares strong homology with NLRP3, it is not involved in inflammasome formation but regulates the NF-κB cascade.90, 91 A nonsense mutation causing defective splicing and introducing a premature stop codon in NLRP12 causes a dominant phenotype due to the lack of regulation of the inflammatory pathway.
Despite its typical inflammasome-promoting structure, another member of NLRP family, NLRP7, acts as a negative regulator of caspase-1 activation.92, 93 Mutations in NLRP7 have mainly been associated with aberrant development of the reproductive tract, such as endometrial cancer tissues and familial recurrent hydatidiform moles.94
Mutations in genes encoding pyrin and proline-serine-threonine phosphatase-interacting protein 1 are associated with two autoinflammatory diseases with hereditary periodic fever syndrome-like symptoms: familial mediterranean fever and pyogenic arthritis pyoderma gangrenosum and acne syndrome, respectively. Pyrin and proline–serine–threonine phosphatase-interacting protein 1 interacts with and modulates pyrin function, which affects inflammasome activity. However, it is still unknown whether pyrin acts as an ASC activator, inducing inflammasome-independent IL-1β release or as negative inflammasome regulator able to dampen NLRP3 function by competing for ASC.95, 96 In both hypotheses pyrin and proline–serine–threonine phosphatase-interacting protein 1 mutations are thought to induce dysregulation of IL-1β secretion.97
A similar hypothesis was formulated for caspase-12, which is expressed in several mammals but commonly present as a null allele in humans, where a mutation leads to a truncated non-functional form of the protein. Caspase-12 is a negative regulator of the NF-κB pathway and for this reason it can increase susceptibility to sepsis following infection. It is likely that in humans the null allele of caspase-12 has been selected for under evolutionary pressure in areas subject to persistent infections. However, in some populations of Africa, South Asia and South America, a caspase-12 mutation causing expression of a full-length protein is responsible for a dominant negative autoinflammatory phenotype.
Genetics of inflammasome-related non-autoinflammatory disorders
Besides auto-inflammation, IL-1β is involved in the pathogenesis of other immune-related disorders ranging from autoimmunity to pathogen clearance and tumorigenesis. Therefore, it follows that mutations in NLR-proteins and inflammasome-related components may also be implicated in this broad spectrum of conditions.
Infections
The involvement of the inflammasome in controlling bacterial infections has mainly been linked to its central role in IL-1β production, which is required for effective pathogen clearance. In this regard, the control of Mycobacterium tuberculosis (MTB) infection by innate cells through inflammasome-derived IL-1β can be taken as an example. IL-1R-deficient mice are more susceptible to MTB infection due to defective bacterial clearance. Moreover, IL-1β is thought to kill MTB bacilli by promoting phagosome maturation. The MTB gene Zmp1, which encodes a Zn2+ metalloprotease, may suppress inflammasome activity in the host macrophage, supporting the idea of a key role for the inflammasome in this system.98 However, while there are no data yet to link inflammasome genetics in the host to MTB susceptibility, there is an association between NLRP3 genetics and immune protection against viruses. Recent studies have suggested a role of inflammasomes in the recognition of viral DNA and RNA during infection.30, 35 For example, NLRP3-deficient mice exhibited limited protection to influenza A infection. Furthermore, an association between NLRP3 polymorphisms and HIV-1 infection has been described; a lower frequency of the 3' UTR SNP rs10754558 G allele was found within HIV-seropositive patients compared to controls.99 Interestingly, such association was seen in infected adults as well as in vertically infected children, suggesting that mutations in NLRP3-mediated immune response could impair protection against HIV-1, independent of the route of transmission. However, the precise mechanism of NLRP3-mediated control of HIV-1 has not yet been revealed. Although several independent studies have uncovered a protective function of the inflammasome during viral infections35, 100, 101, others observed that inflammasome activation by certain viruses was mainly crucial for innate and healing response but not for virus control and adaptive immune activation.102 Thus, the exact mechanism of virus recognition and the possible influence of genetics of inflammasomes on protection from viruses remain unclear.
NLRP3 has also been associated with the control of fungal infections.34 An intronic polymorphism in NLRP3 that causes production of a shorter form of the protein is linked to candidiasis-mediated vulvar vestibulitis.103 In addition, the same splicing variant of NLRP3 correlated with susceptibility to mycoplasma infection causing infertility.104 The NLRP3 polymorphism rs12065526 G>A was identified as a risk factor for the development of tubal pathology symptoms induced by Chlamydia trachomatis infection (Figure 3).105
All these association studies suggest a crucial role for NLRP3 in immune surveillance against a broad spectrum of different infective agents invading the host through the mucosal barriers.
Adaptive immunity-mediated diseases
The boundaries between autoimmunity and autoinflammation are not clearly defined for many immune disorders and the adaptive components in autoinflammation, as well as the innate component in autoimmunity, may have been underestimated.106 A growing body of evidence suggests that inflammasome activation could also drive adaptive immunity and the correlation between inflammasome dysfunction and autoimmunity has become the subject of intense investigation.
In a transgenic mouse model of CAPS, excessive IL-1β secretion resulted in NLRP3-dependent pathogenic activation of a specific IL-17-secreting T-cell subset, namely, Th17 cells.77 Nonetheless, another study excluded an involvement of adaptive immunity in a very similar model of CAPS.79 IL-1β plays an important role in activating and sustaining the Th17-mediated response, especially in humans.107, 108, 109, 110 Considering the central role of Th17 cells in autoimmune diseases including rheumatoid arthritis (RA) and psoriasis, mutations in inflammasome-encoding genes may result in a broad spectrum of Th17 cell-mediated autoimmune disorders, through deregulation of IL-1β secretion.107, 111, 112, 113, 114, 115, 116 Genetic variants in several inflammatory genes, including NLRP3, are associated with two forms of autoimmune arthritis: juvenile idiopathic arthritis and RA, characterized by Th17 cells.117, 118 Indeed, increased expression of NLRP3 was observed in the synovium of RA patients compared to subjects suffering from non-autoimmune osteoarthritis.119 Moreover, in a Swedish cohort, RA susceptibility and severity are significantly influenced by the combination of the Q705K variant of NLRP3, already known for its low-penetrance in FCAS, and the C10X variant of caspase recruitment domain family, member 8.120 Notably, these two polymorphisms are highly recurrent: Q705K polymorphism in NLRP3 was found in 6.5% of the Caucasian population and CARD-8 gene polymorphism C10X is present in 40%.121, 122 However, their penetrance in the pathogenesis of immune-immune mediated disorders is very low, and leads to susceptibility to autoimmunity only when in combination.
Polymorphisms in the NLRP3 gene also correlate with different forms of allergies, including susceptibility to food-induced anaphylaxis, aspirin-induced asthma and urticaria.123, 124 Urticarial reactions are recurrent clinical manifestations in the continuum of CAPS, and skin disorders are a phenotypic feature of knock-in mice carrying NLRP3 hypermutations.77 Moreover, mast cells from CAPS patients express NLRP3 and constitutively secrete IL-1β.124, 125 Therefore, we can speculate that genetics of inflammasome components could be involved in the onset of other types of dermatitis and skin-related allergies, as well as asthma.124
NLRP1 is expressed not only by innate cells but also by T cells, and therefore has a role in adaptive immunity as well as innate. A strong correlation links some intronic variants of NLRP1 with vitiligo and vitiligo-associated autoimmune disorders (Table 1).126, 127 In addition, new associations of NLRP1 SNPs with other autoimmune disorders such as Addison's disease, type 1 diabetes, multiple sclerosis and RA have been identified, even though a genome-wide study does not support all of these associations.128, 129, 130. However, as well as its role in inflammasomes, NLRP1 also contributes to the formation of the apoptosome via caspase-2 and -9 recruitment. For this reason, the genetics of NLRP1 could be associated with susceptibility to autoimmune disorders where apoptosis plays a crucial role.
Chronic tissue damage disorders
NLRP3 is now considered as an intracellular sensor of tissue damage. Environmental threats (silica and asbestos) and endogenous signals (ATP and urate crystals) together with genetic predisposition can significantly contribute to inflammasome-related disorders. Indeed, silica and asbestos leads to chronic pulmonary inflammation, and urate crystals are responsible for gout. A clear correlation between gout and NLRP3 polymorphism has not yet been described. However, hypertension, which affects 57% of gouty patients, has been correlated to a specific intron 4 polymorphism of the NLRP3 gene causing a dominant expression of the transcript.131
IL-1β can promote pathogenesis of disorders associated with tissue damage, in which hyperinflammatory responses worsen the disease rather than promote its resolution. For example, inflammasome-mediated IL-1β overproduction is involved in the pathogenesis of type 2 diabetes, liver damage and muscular dystrophy.132, 133, 134 Inflammasome-mediated processes were also observed in cancer, but the association between the inflammasome/IL-1β axis and cancer development is ambiguous. In some studies the inflammasome seems to promote inflammation-mediated tumor development, also confirmed in lung mesotheliomas caused by silica and asbestos.8, 47 However, in vivo animal models of cancer showed that ATP from dying cells promoted production of IL-1β and interferon-γ-secreting CD8+ T cells able to eliminate the tumor.9 Remarkably, supporting this finding, breast cancer patients with defects in the ATP receptor P2X7 have a higher probability of developing metastases.135 To date, no mutation in NLRP3 has been correlated to tumor susceptibility.
Finally, IL-1β polymorphisms are associated with Alzheimer's disease, a neurological disorder characterized by fibrillar peptide amyloid β accumulation in the brain.136, 137 Notably, amyloid β has been identified as a NLRP3 activator.138 Considering the pleiotropic functions of IL-1β in the nervous system, these observations underline the importance of studying inflammasome genetics in the context of disorders that apparently do not involve the immune system.
Concluding remarks
The genetics of inflammasomes have been deeply investigated in relation to autoinflammation, where missense point mutations in NLR-coding genes determine with high penetrance the pathogenesis of autoinflammatory processes (Figure 4). This discovery allowed better characterization of the genotype of patients exhibiting autoinflammatory symptoms, in order to design a specific anti-IL-1β therapy. Moreover, transgenic animal models bearing these pathological mutations helped in defining the molecular mechanisms mediating aberrant inflammasome activation. However, several genome-wide screenings have recently uncovered the association between genetic variants of NLRs and other diseases beyond autoinflammation. These findings suggest that specific polymorphisms in NLR genes could contribute, albeit with lower penetrance, to the susceptibility to more common multifactorial diseases such as autoimmune disorders and cancer (Figure 4). The study of inflammasome genetics is also important to help in the diagnostic process and in the design of effective therapeutic strategies. Further investigations aimed to define the genetics of inflammasomes in human diseases are urgently needed to reveal the potent mechanisms mediating the pathology of rare autoinflammatory disorders, but also to depict the complexity of widespread multifactorial diseases, where NLR polymorphisms may play an important hidden role.
Figure 4.
Proposed ranking of human diseases based on penetrance and frequency of inflammasome polymorphisms. Missense point mutations in genes encoding inflammasome components exhibit high penetrance and contribute to the pathogenic autoinflammation, leading to severe but rare autoinflammatory syndromes, such as CAPS. Allelic variants of inflammasome encoding genes in combination with other genetic polymorphisms affecting immune cells lead to more common autoinflammatory-like syndromes, including Crohn's diseases, RA and JIA. Multifactorial disorders highly diffused in the human population, such as infections, autoimmunity, cancer, cardiovascular and neurological diseases, might be associated with polymorphisms in inflammasome-encoding genes of low and variable penetrance. CAPS, cryopyrin-associated periodic syndrome; JIA, juvenile idiopathic arthritis; RA, rheumatoid arthritis.
Acknowledgments
We thank Lucy Robinson for critically reviewing the manuscript. This work was supported by Agency for Science, Technology and Research (A*STAR) of Singapore.
References
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Franchi L, Warner N, Viani K, Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vallejo JJ, van Kooyk Y. Endogenous ligands for C-type lectin receptors: the true regulators of immune homeostasis. Immunol Rev. 2009;230:22–37. doi: 10.1111/j.1600-065X.2009.00786.x. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD. Central roles of NLRs and inflammasomes in viral infection. Nat Rev Immunol. 2010;10:688–698. doi: 10.1038/nri2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, TeKippe EM, Woodford RMT, Uronis JM, Holl EK, Rogers AB, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti TD. IL-18 production downstream of the NLRP3 inflammasome confers protection against colorectal tumor formation. J Immunol. 2010;185:4912–4920. doi: 10.4049/jimmunol.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Liu W, Luo Y, Tanaka A, Cai X, Norris DA, et al. Constitutively active inflammasome in human melanoma cells mediating autoinflammation via caspase-1 processing and secretion of interleukin-1beta. J Biol Chem. 2010;285:6477–6488. doi: 10.1074/jbc.M109.064907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C, et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat Med. 2009;15:1170–1178. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin EV, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- Proell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS One. 2008;3:e2119. doi: 10.1371/journal.pone.0002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bella J, Hindle KL, McEwan PA, Lovell SC. The leucine-rich repeat structure. Cell Mol Life Sci. 2008;65:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Tschopp J. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 2004;117:561–574. doi: 10.1016/j.cell.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Faustin B, Lartigue L, Bruey JM, Luciano F, Sergienko E, Bailly-Maitre B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Martinon F, Gaide O, Petrilli V, Mayor A, Tschopp J. NALP inflammasomes: a central role in innate immunity. Semin Immunopathol. 2007;29:213–229. doi: 10.1007/s00281-007-0079-y. [DOI] [PubMed] [Google Scholar]
- McDonald C, Inohara N, Nunez G. Peptidoglycan signaling in innate immunity and inflammatory disease. J Biol Chem. 2005;280:20177–20180. doi: 10.1074/jbc.R500001200. [DOI] [PubMed] [Google Scholar]
- Tattoli I, Travassos LH, Carneiro LA, Magalhaes JG, Girardin SE. The Nodosome: Nod1 and Nod2 control bacterial infections and inflammation. Semin Immunopathol. 2007;29:289–301. doi: 10.1007/s00281-007-0083-2. [DOI] [PubMed] [Google Scholar]
- Hsu YM, Zhang Y, You Y, Wang D, Li H, Duramad O, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8:198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, et al. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- Case CL, Shin S, Roy CR. Asc and Ipaf Inflammasomes direct distinct pathways for caspase-1 activation in response to Legionella pneumophila. Infect Immun. 2009;77:1981–1991. doi: 10.1128/IAI.01382-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- Miao EA, Andersen-Nissen E, Warren SE, Aderem A. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin Immunopathol. 2007;29:275–288. doi: 10.1007/s00281-007-0078-z. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Lamkanfi M, Kanneganti TD. NOD-like receptor (NLR) signaling beyond the inflammasome. Eur J Immunol. 2010;40:624–627. doi: 10.1002/eji.200940211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni DS, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, Vance RE, et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, Vance RE, et al. A NOD2–NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Natl Acad Sci USA. 2008;105:7803–7808. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferwerda G, Ali SR, McGillivray S, Tseng PH, Mariathasan S, Humke EW, et al. Engagement of NOD2 has a dual effect on proIL-1beta mRNA transcription and secretion of bioactive IL-1beta. Eur J Immunol. 2008;38:184–191. doi: 10.1002/eji.200737103. [DOI] [PubMed] [Google Scholar]
- Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T, Alnemri ES, et al. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross J, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Muruve DA, Tschopp J. Innate immunity: cytoplasmic DNA sensing by the AIM2 inflammasome. Curr Biol. 2009;19:R262–R265. doi: 10.1016/j.cub.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- Gross O, Poeck H, Bscheider M, Dostert C, Hannesschläger N, Endres S, et al. Syk kinase signalling couples to the NLRP3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–436. doi: 10.1038/nature07965. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- Pan Q, Mathison J, Fearns C, Kravchenko VV, Da Silva Correia J, Hoffman HM, et al. MDP-induced interleukin-1beta processing requires NOD2 and CIAS1/NALP3. . J Leukoc Biol. 2007;82:177–183. doi: 10.1189/jlb.1006627. [DOI] [PubMed] [Google Scholar]
- Martinon F, Agostini L, Meylan E, Tschopp J. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr Biol. 2004;14:1929–1934. doi: 10.1016/j.cub.2004.10.027. [DOI] [PubMed] [Google Scholar]
- Lightfield KL, Persson J, Brubaker SW, Witte CE, von Moltke J, Dunipace EA, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- Craven RR, Gao X, Allen IC, Gris D, Bubeck Wardenburg J, McElvania-Tekippe E, et al. Staphylococcus aureus alpha-hemolysin activates the NLRP3-inflammasome in human and mouse monocytic cells. PLoS One. 2009;4:e7446. doi: 10.1371/journal.pone.0007446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurcel L, Abrami L, Girardin S, Tschopp J, van der Goot FG. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 2006;126:1135–1145. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- Ozoren N, Masumoto J, Franchi L, Kanneganti TD, Body-Malapel M, Erturk I, et al. Distinct roles of TLR2 and the adaptor ASC in IL-1beta/IL-18 secretion in response to Listeria monocytogenes. J Immunol. 2006;176:4337–4342. doi: 10.4049/jimmunol.176.7.4337. [DOI] [PubMed] [Google Scholar]
- Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- Eisenbarth SC, Colegio OR, O'Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C, PÊtrilli V, van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassel SL, Eisenbarth SC, Iyer SS, Sadler JJ, Colegio OR, Tephly LA, et al. The Nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci USA. 2008;105:9035–9040. doi: 10.1073/pnas.0803933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor A, Martinon F, de Smedt T, Petrilli V, Tschopp J. A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat Immunol. 2007;8:497–503. doi: 10.1038/ni1459. [DOI] [PubMed] [Google Scholar]
- Petrilli V, Papin S, Dostert C Mayor A, Martinon F, Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- Kahlenberg JM, Dubyak GR. Mechanisms of caspase-1 activation by P2X7 receptor-mediated K+ release. Am J Physiol Cell Physiol. 2004;286:C1100–C1108. doi: 10.1152/ajpcell.00494.2003. [DOI] [PubMed] [Google Scholar]
- Laliberte RE, Eggler J, Gabel CA. ATP treatment of human monocytes promotes caspase-1 maturation and externalization. J Biol Chem. 1999;274:36944–36951. doi: 10.1074/jbc.274.52.36944. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Lamkanfi M, Kim YG, Chen G, Park JH, Franchi L, et al. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity. 2007;26:433–443. doi: 10.1016/j.immuni.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Locovei S, Bao L, Dahl G. Pannexin 1 in erythrocytes: function without a gap. Proc Natl Acad Sci USA. 2006;103:7655–7659. doi: 10.1073/pnas.0601037103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz CM, Rinna A, Forman HJ, Ventura AL, Persechini PM, Ojcius OM, et al. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J Biol Chem. 2007;282:2871–2879. doi: 10.1074/jbc.M608083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- Shio MT, Eisenbarth SC, Savaria M, Vinet AF, Bellemare MJ, Harder KW, et al. Malarial hemozoin activates the NLRP3 inflammasome through Lyn and Syk kinases. PLoS Pathog. 2009;5:e1000559. doi: 10.1371/journal.ppat.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutault K, Alderman C, Chain BM, Katz DR. Reactive oxygen species activate human peripheral blood dendritic cells. Free Radic Biol Med. 1999;26:232–238. doi: 10.1016/s0891-5849(98)00194-4. [DOI] [PubMed] [Google Scholar]
- Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- Hawkins PN, Lachmann HJ, McDermott MF. Interleukin-1-receptor antagonist in the Muckle–Wells syndrome. N Engl J Med. 2003;348:2583–2584. doi: 10.1056/NEJM200306193482523. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Shi Y, Hearn A, Fitzgerald K, Golenbock D, Reed G, et al. MyD88-dependent IL-1 receptor signaling is essential for gouty inflammation stimulated by monosodium urate crystals. J Clin Invest. 2006;116:2262–2271. doi: 10.1172/JCI28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarda G, Dostert C, Staehli F, Cabalzar K, Castillo R, Tardivel A, et al. T cells dampen innate immune responses through inhibition of NLRP1 and NLRP3 inflammasomes. Nature. 2009;460:269–273. doi: 10.1038/nature08100. [DOI] [PubMed] [Google Scholar]
- Kool M, Petrilli V, de Smedt T, Rolaz A, Hammad H, van Nimwegen M, et al. Cutting edge: alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J Immunol. 2008;181:3755–3759. doi: 10.4049/jimmunol.181.6.3755. [DOI] [PubMed] [Google Scholar]
- Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichten-Berger GS, Grant EP, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- Sugawara S, Uehara A, Nochi T, Yamaguchi T, Ueda H, Sugiyama A, et al. Neutrophil proteinase 3-mediated induction of bioactive IL-18 secretion by human oral epithelial cells. J Immunol. 2001;167:6568–6575. doi: 10.4049/jimmunol.167.11.6568. [DOI] [PubMed] [Google Scholar]
- Puren AJ, Fantuzzi G, Dinarello CA. Gene expression, synthesis, and secretion of interleukin 18 and interleukin 1beta are differentially regulated in human blood mononuclear cells and mouse spleen cells. Proc Natl Acad Sci USA. 1999;96:2256–2261. doi: 10.1073/pnas.96.5.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattorno M, Tassi S, Carta S, Delfino L, Ferlito F, Pelagatti MA, et al. Pattern of interleukin-1beta secretion in response to lipopolysaccharide and ATP before and after interleukin-1 blockade in patients with CIAS1 mutations. Arthritis Rheum. 2007;56:3138–3148. doi: 10.1002/art.22842. [DOI] [PubMed] [Google Scholar]
- Shinkai K, McCalmont TH, Leslie KS. Cryopyrin-associated periodic syndromes and autoinflammation. Clin Exp Dermatol. 2008;33:1–9. doi: 10.1111/j.1365-2230.2007.02540.x. [DOI] [PubMed] [Google Scholar]
- Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle–Wells syndrome. Nat Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aganna E, Martinon F, Hawkins PN, Ross JB, Swan DC, Booth DR, et al. Association of mutations in the NALP3/CIAS1/PYPAF1 gene with a broad phenotype including recurrent fever, cold sensitivity, sensorineural deafness, and AA amyloidosis. Arthritis Rheum. 2002;46:2445–2452. doi: 10.1002/art.10509. [DOI] [PubMed] [Google Scholar]
- Touitou I, Lesage S, McDermott M, Cuisset L, Hoffman H, Dode C, et al. Infevers: an evolving mutation database for auto-inflammatory syndromes. Hum Mutat. 2004;24:194–198. doi: 10.1002/humu.20080. [DOI] [PubMed] [Google Scholar]
- Masters SL, Simon A, Aksentijevich I, Kastner DL. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease (*) Annu Rev Immunol. 2009;27:621–668. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neven B, Callebaut I, Prieur AM, Feldmann J, Bodemer C, Lepore L, et al. Molecular basis of the spectral expression of CIAS1 mutations associated with phagocytic cell-mediated autoinflammatory disorders CINCA/NOMID, MWS, and FCU. Blood. 2004;103:2809–2815. doi: 10.1182/blood-2003-07-2531. [DOI] [PubMed] [Google Scholar]
- Goldbach-Mansky R, Dailey NJ, Canna SW, Gelabert A, Jones J, Rubin BI, et al. Neonatal-onset multisystem inflammatory disease responsive to interleukin-1beta inhibition. N Engl J Med. 2006;355:581–592. doi: 10.1056/NEJMoa055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, Zhang F, Fuss I, Kitani A, Strober W. A mutation in the NLRP3 gene causing inflammasome hyperactivation potentiates Th17 cell-dominant immune responses. Immunity. 2009;30:860–874. doi: 10.1016/j.immuni.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksentijevich I, Remmers EF, Goldbach-Mansky R, Reiff A, Kastner DL.Mutational analysis in neonatal-onset multisystem inflammatory disease: comment on the articles by Frenkel et al and Saito et al. Arthritis Rheum 2006542703–2704.author reply 2704–2705. [DOI] [PubMed] [Google Scholar]
- Brydges SD, Mueller JL, McGeough MD, Pena CA, Misaghi A, Gandhi C, et al. Inflammasome-mediated disease animal models reveal roles for innate but not adaptive immunity. Immunity. 2009;30:875–887. doi: 10.1016/j.immuni.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa A, Kambe N, Saito M, Nishikomori R, Tanizaki H, Kanazawa N, et al. Disease-associated mutations in CIAS1 induce cathepsin B-dependent rapid cell death of human THP-1 monocytic cells. Blood. 2007;109:2903–2911. doi: 10.1182/blood-2006-07-033597. [DOI] [PubMed] [Google Scholar]
- Saito M, Nishikomori R, Kambe N, Fujisawa A, Tanizaki H, Takeichi K, et al. Disease-associated CIAS1 mutations induce monocyte death, revealing low-level mosaicism in mutation-negative cryopyrin-associated periodic syndrome patients. Blood. 2008;111:2132–2141. doi: 10.1182/blood-2007-06-094201. [DOI] [PubMed] [Google Scholar]
- Kambe N, Satoh T, Tanizaki H, Fujisawa A, Saito MK, Nishikomori R.Enhanced NF-kappaB activation with an inflammasome activator correlates with activity of autoinflammatory disease associated with NLRP3 mutations outside of exon 3: comment on the article by Jeru et al. Arthritis Rheum 2010623123–3124.author reply 3124–3125 [DOI] [PubMed] [Google Scholar]
- Jeru I, Marlin S, Le Borgne G, Cochet E, Normand S, Duquesnoy P, et al. Functional consequences of a germline mutation in the leucine-rich repeat domain of NLRP3 identified in an atypical autoinflammatory disorder. Arthritis Rheum. 2010;62:1176–1185. doi: 10.1002/art.27326. [DOI] [PubMed] [Google Scholar]
- Arostegui JI, Lopez Saldaña MD, Pascal M, Clemente D, Aymerich M, Balaguer F, et al. A somatic NLRP3 mutation as a cause of a sporadic case of chronic infantile neurologic, cutaneous, articular syndrome/neonatal-onset multisystem inflammatory disease: novel evidence of the role of low-level mosaicism as the pathophysiologic mechanism underlying mendelian inherited diseases. Arthritis Rheum. 2010;62:1158–1166. doi: 10.1002/art.27342. [DOI] [PubMed] [Google Scholar]
- Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, et al. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat Genet. 2009;41:71–76. doi: 10.1038/ng285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JR, Cooney RM, Clarke G, , Beckly J, , Geremia A, , Pathan S, et al. The genetics of NOD-like receptors in Crohn's disease. Tissue Antigens. 2010;76:48–56. doi: 10.1111/j.1399-0039.2010.01470.x. [DOI] [PubMed] [Google Scholar]
- Schoultz I, Verma D, Halfvarsson J, Torkvist L, Fredrikson M, Sjöqvist U, et al. Combined polymorphisms in genes encoding the inflammasome components NALP3 and CARD8 confer susceptibility to Crohn's disease in Swedish men. Am J Gastroenterol. 2009;104:1180–1188. doi: 10.1038/ajg.2009.29. [DOI] [PubMed] [Google Scholar]
- Lewis GJ, Massey DC, Zhang H, Bredin F, Tremelling M, Lee JC, et al. Genetic association between NLRP3 variants and Crohn's disease does not replicate in a large UK panel. Inflamm Bowel Dis 2010 Oct 25. [Epub ahead of print]DOI: 10.1002/ibd.21499. [DOI] [PubMed]
- Aksentijevich I, Nowak M, Mallah M, Chae JJ, Watford WT, Hofmann SR, et al. De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): a new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum. 2002;46:3340–3348. doi: 10.1002/art.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeru I, Duquesnoy P, Fernandes-Alnemri T, Cochet E, Yu JW, Lackmy-Port-Lis M, et al. Mutations in NALP12 cause hereditary periodic fever syndromes. Proc Natl Acad Sci USA. 2008;105:1614–1619. doi: 10.1073/pnas.0708616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lich JD, Williams KL, Moore CB, Arthur JC, Davis BK, Taxman DJ, et al. Monarch-1 suppresses non-canonical NF-kappaB activation and p52-dependent chemokine expression in monocytes. J Immunol. 2007;178:1256–1260. doi: 10.4049/jimmunol.178.3.1256. [DOI] [PubMed] [Google Scholar]
- Pinheiro AS, Proell M, Eibl C, Page R, Schwarzenbacher R, Peti W, et al. Three-dimensional structure of the NLRP7 pyrin domain: insight into pyrin–pyrin-mediated effector domain signaling in innate immunity. J Biol Chem. 2010;285:27402–27410. doi: 10.1074/jbc.M110.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Wang Y, Hasegawa M, Imamura R, Suda T. PYPAF3, a PYRIN-containing APAF-1-like protein, is a feedback regulator of caspase-1-dependent interleukin-1beta secretion. J Biol Chem. 2005;280:21720–21725. doi: 10.1074/jbc.M410057200. [DOI] [PubMed] [Google Scholar]
- Deveault C, Qian JH, Chebaro W, Ao A, Gilbert L, Mehio A. NLRP7 mutations in women with diploid androgenetic and triploid moles: a proposed mechanism for mole formation. Hum Mol Genet. 2009;18:888–897. doi: 10.1093/hmg/ddn418. [DOI] [PubMed] [Google Scholar]
- Yu JW, Fernandes-Alnemri T, Datta P, Wu J, Juliana C, Solorzano L, et al. Pyrin activates the ASC pyroptosome in response to engagement by autoinflammatory PSTPIP1 mutants. Mol Cell. 2007;28:214–227. doi: 10.1016/j.molcel.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae JJ, Komarow HD, Cheng J, Wood G, Raben N, Liu PP, et al. Targeted disruption of pyrin, the FMF protein, causes heightened sensitivity to endotoxin and a defect in macrophage apoptosis. Mol Cell. 2003;11:591–604. doi: 10.1016/s1097-2765(03)00056-x. [DOI] [PubMed] [Google Scholar]
- Jeru I, Hayrapetyan H, Duquesnoy P, Sarkisian T, Amselem S. PYPAF1 nonsense mutation in a patient with an unusual autoinflammatory syndrome: role of PYPAF1 in inflammation. Arthritis Rheum. 2006;54:508–514. doi: 10.1002/art.21618. [DOI] [PubMed] [Google Scholar]
- Master SS, Rampini SK, Davis AS, Keller C, Ehlers S, Springer B, et al. Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe. 2008;3:224–232. doi: 10.1016/j.chom.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontillo A, Brandao LA, Guimaraes RL, Segat L, Athanasakis E, Crovella S. A 3'UTR SNP in NLRP3 gene is associated with susceptibility to HIV-1 infection. J Acquir Immune Defic Syndr. 2010;54:236–240. doi: 10.1097/QAI.0b013e3181dd17d4. [DOI] [PubMed] [Google Scholar]
- Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PG, Dash P, Aldridge JR, Jr, Ellebedy AH, Reynolds C, Funk AJ, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Sagie A, Prus D, Linhares IM, Lavy Y, Ledger WJ, Witkin SS. Polymorphism in a gene coding for the inflammasome component NALP3 and recurrent vulvovaginal candidiasis in women with vulvar vestibulitis syndrome. Am J Obstet Gynecol. 2009;200:303–306. doi: 10.1016/j.ajog.2008.10.039. [DOI] [PubMed] [Google Scholar]
- Witkin SS, Bierhals K, Linhares I, Normand N, Dieterle S, Neuer A. Genetic polymorphism in an inflammasome component, cervical mycoplasma detection and female infertility in women undergoing in vitro fertilization. J Reprod Immunol. 2010;84:171–175. doi: 10.1016/j.jri.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Wang W, Stassen FR, Surcel HMÖ, hman H, Tiitinen A, Paavonen J, et al. Analyses of polymorphisms in the inflammasome-associated NLRP3 and miRNA-146A genes in the susceptibility to and tubal pathology of Chlamydia trachomatis infection. Drugs Today (Barc) 2009;45 Suppl B:95–103. [PubMed] [Google Scholar]
- McGonagle D, McDermott MF. A proposed classification of the immunological diseases. PLoS Med. 2006;3:e297. doi: 10.1371/journal.pmed.0030297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton C, Brereton C, Keogh B, Mills KH, Lavelle EC. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J Exp Med. 2006;203:1685–1691. doi: 10.1084/jem.20060285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, Kang HS, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–587. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol. 2007;8:942–949. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- van Beelen AJ, Zelinkova Z, Taanman-Kueter EW, Muller FJ, Hommes DW, Zaat SA, et al. Stimulation of the intracellular bacterial sensor NOD2 programs dendritic cells to promote interleukin-17 production in human memory T cells. Immunity. 2007;27:660–669. doi: 10.1016/j.immuni.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Nistala K, Moncrieffe H, Newton KR, Varsani H, Hunter P, Wedderburn LR. Interleukin-17-producing T cells are enriched in the joints of children with arthritis, but have a reciprocal relationship to regulatory T cell numbers. Arthritis Rheum. 2008;58:875–887. doi: 10.1002/art.23291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzartos JS, Friese MA, Craner MJ, Palace J, Newcombe J, Esiri MM, et al. Interleukin-17 production in central nervous system-infiltrating T cells and glial cells is associated with active disease in multiple sclerosis. Am J Pathol. 2008;172:146–155. doi: 10.2353/ajpath.2008.070690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernis AB. Th17 cells in rheumatoid arthritis and systemic lupus erythematosus. J Intern Med. 2009;265:644–652. doi: 10.1111/j.1365-2796.2009.02099.x. [DOI] [PubMed] [Google Scholar]
- Lubberts E. Th17 cytokines and arthritis. Semin Immunopathol. 2010;32:43–53. doi: 10.1007/s00281-009-0189-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg WB, Miossec P. IL-17 as a future therapeutic target for rheumatoid arthritis. Nat Rev Rheumatol. 2009;5:549–553. doi: 10.1038/nrrheum.2009.179. [DOI] [PubMed] [Google Scholar]
- Evans HG, Gullick NJ, Kelly S, Pitzalis C, Lord GM, Kirkham BW, et al. In vivo activated monocytes from the site of inflammation in humans specifically promote Th17 responses. Proc Natl Acad Sci USA. 2009;106:6232–6237. doi: 10.1073/pnas.0808144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day TG, Ramanan AV, Hinks A, Lamb R, Packham J, Wise C, et al. Autoinflammatory genes and susceptibility to psoriatic juvenile idiopathic arthritis. Arthritis Rheum. 2008;58:2142–2146. doi: 10.1002/art.23604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, Misra R, Aggarwal A. Interleukin 17 levels are increased in juvenile idiopathic arthritis synovial fluid and induce synovial fibroblasts to produce proinflammatory cytokines and matrix metalloproteinases. J Rheumatol. 2008;35:515–519. [PubMed] [Google Scholar]
- Rosengren S, Hoffman HM, Bugbee W, Boyle DL. Expression and regulation of cryopyrin and related proteins in rheumatoid arthritis synovium. Ann Rheum Dis. 2005;64:708–714. doi: 10.1136/ard.2004.025577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastbom A, Verma D, Eriksson P, Skogh T, Wingren G, Soderkvist P. Genetic variation in proteins of the cryopyrin inflammasome influences susceptibility and severity of rheumatoid arthritis (the Swedish TIRA project) Rheumatology (Oxford) 2008;47:415–417. doi: 10.1093/rheumatology/kem372. [DOI] [PubMed] [Google Scholar]
- Hoffman HM, Gregory SG, Mueller JL, Tresierras M, Broide DH, Kolodner RD. Fine structure mapping of CIAS1: identification of an ancestral haplotype and a common FCAS mutation, L353P. . Hum Genet. 2003;112:209–216. doi: 10.1007/s00439-002-0860-x. [DOI] [PubMed] [Google Scholar]
- McGovern DP, Butler H, Ahmad T, Paolucci M, van Heel DA, Negoro K, et al. TUCAN (CARD8) genetic variants and inflammatory bowel disease. Gastroenterology. 2006;131:1190–1196. doi: 10.1053/j.gastro.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Hitomi Y, Ebisawa M, Tomikawa M, Imai T, Komata T, Hirota T, et al. Associations of functional NLRP3 polymorphisms with susceptibility to food-induced anaphylaxis and aspirin-induced asthma. J Allergy Clin Immunol. 2009;124:779–785. doi: 10.1016/j.jaci.2009.07.044. [DOI] [PubMed] [Google Scholar]
- Kambe N, Nakamura Y, Saito M, Nishikomori R. The inflammasome, an innate immunity guardian, participates in skin urticarial reactions and contact hypersensitivity. Allergol Int. 2010;59:105–113. doi: 10.2332/allergolint.09-RAI-0160. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kambe N, Saito M, Nishikomori R, Kim YG, Murakami M, et al. Mast cells mediate neutrophil recruitment and vascular leakage through the NLRP3 inflammasome in histamine-independent urticaria. J Exp Med. 2009;206:1037–1046. doi: 10.1084/jem.20082179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Mailloux CM, Gowan K, Riccardi SL, LaBerge G, Bennett DC, et al. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007;356:1216–1225. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- Jin Y, Birlea SA, Fain PR, Spritz RA. Genetic variations in NALP1 are associated with generalized vitiligo in a Romanian population. J Invest Dermatol. 2007;127:2558–2562. doi: 10.1038/sj.jid.5700953. [DOI] [PubMed] [Google Scholar]
- Magitta NF, Boe Wolff AS, Johansson S, Skinningsrud B, Lie BA, Myhr KM, et al. A coding polymorphism in NALP1 confers risk for autoimmune Addison's disease and type 1 diabetes. Genes Immun. 2009;10:120–124. doi: 10.1038/gene.2008.85. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi T, Kumada M, Kamesaki T, Okuda H, Munkhtulga L, Yanagisawa Y, et al. An intronic variable number of tandem repeat polymorphisms of the cold-induced autoinflammatory syndrome 1 (CIAS1) gene modifies gene expression and is associated with essential hypertension. Eur J Hum Genet. 2006;14:1295–1305. doi: 10.1038/sj.ejhg.5201698. [DOI] [PubMed] [Google Scholar]
- Rawat R, Cohen TV, Ampong B, Francia D, Henriques-Pons A, Hoffman EP, et al. Inflammasome up-regulation and activation in dysferlin-deficient skeletal muscle. Am J Pathol. 2010;176:2891–2900. doi: 10.2353/ajpath.2010.090058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters SL, Dunne A, Subramanian SL, Hull RL, Tannahill GM, Sharp FA, et al. Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1beta in type 2 diabetes. Nat Immunol. 2010;11:897–904. doi: 10.1038/ni.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H, Imamura M, Fujimoto J, Nakanishi K. The TLR4/TRIF-mediated activation of NLRP3 inflammasome underlies endotoxin-induced liver injury in mice. Gastroenterol Res Pract. 2010;2010:641865. doi: 10.1155/2010/641865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu LQ, Li FY, Zhao L, Liu Y, Chu Q, Zang XX, et al. Association of XIAP and P2X7 receptor expression with lymph node metastasis in papillary thyroid carcinoma. Endocrine. 2010;38:276–282. doi: 10.1007/s12020-010-9384-7. [DOI] [PubMed] [Google Scholar]
- Ravaglia G, Paola F, Maioli F, Martelli M, Montesi F, Bastagli L, et al. Interleukin-1beta and interleukin-6 gene polymorphisms as risk factors for AD: a prospective study. Exp Gerontol. 2006;41:85–92. doi: 10.1016/j.exger.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Forlenza OV, Diniz BS, Nunes PV, Memória CM, Yassuda MS, Gattaz WF, et al. Increased serum IL-1beta level in Alzheimer's disease and mild cognitive impairment. Dement Geriatr Cogn Disord. 2009;28:507–512. doi: 10.1159/000255051. [DOI] [PubMed] [Google Scholar]
- Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]