Abstract
The odorant receptors (ORs) provide our main gateway to sensing the world of volatile chemicals. This involves a complex encoding process in which multiple ORs, each of which detects its own set of odorants, work as an ensemble to produce a distributed activation code that is presumably unique to each odorant. One marked challenge to decoding the olfactory code is OR deorphanization, the identification of a set of activating odorants for a particular receptor. Here, we survey various methods used to try to express defined ORs of interest. We also suggest strategies for selecting odorants for test panels to evaluate the functional expression of an OR. Integrating these tools, while retaining awareness of their idiosyncratic limitations, can provide a multi-tiered approach to OR deorphanization, spanning the initial discovery of a ligand to vetting that ligand in a physiologically relevant setting.
Enormous progress has been made in identifying ligands for G protein–coupled receptors (GPCRs) since the 5HT1A serotonin and D2 dopamine receptors were identified as such 25 years ago (Bunzow et al., 1988; Fargin et al., 1988). There is, however, one notable and conspicuous exception: the odorant receptors (ORs). ORs comprise nearly 50% of the ∼800 GPCRs in humans, yet ∼90% of human ORs remain orphan receptors with unknown ligands. This is surprising when you consider that more than half of all non-olfactory GPCRs have been deorphaned to date. Furthermore, unlike most non-olfactory GPCRs, our olfactory perceptual experience means deorphanization strategies for ORs are not necessarily burdened by a lack of knowledge of relevant ligands (Civelli et al., 2013). The molecular mechanisms of OR activity have also been well described (e.g., Firestein, 2001). It is unlikely that fundamental differences in molecular function are the bottleneck in OR deorphanization, as ORs appear to use the same basic molecular mechanisms as other class A GPCRs in terms of ligand binding and transduction (Bockaert and Pin, 1999; Gether, 2000). Regardless, ORs are “poorly behaved” when it comes to functional assays.
Yet large-scale OR deorphanization is needed to elucidate basic principles of odor coding. Deorphanization will allow investigators to probe how OR activity is influenced by specific physicochemical properties of odorant molecules (i.e., develop odorant structure–activity relationships) and ultimately how this information is processed in the brain to elicit an “odor image” that can influence human or animal behavior by providing information on such aspects as perceived odor intensity, odor detection threshold, and valence. As Jerome Lettvin so eloquently and simply put it for the visual system with his seminal 1959 paper “What the Frog’s Eye Tells the Frog’s Brain,” the olfactory field must first know “what the nose knows” to understand what it tells the brain (Lettvin et al., 1959). This Review focuses on recent advances in OR deorphanization, from both the standpoint of receptor expression and that of ligand assay panel selection, highlighting some of the caveats that abound in this continuing but challenging endeavor.
Methods of deorphanization
Deorphanization using olfactory sensory neurons (OSNs).
The most significant impediment to deorphaning an OR of interest has been the lack of a suitable cell-based heterologous expression system. The first three assays discussed circumvent this block by exploiting the OSN itself as the expression system. Using OSNs decreases the difficulty of expressing functional exogenous ORs because the cellular machinery, whatever that may be, is already poised to process ORs. Moreover, the receptive field of the OR (that is, the set of ligands capable of activating the OR and the relative potency of such ligands) is presumed to be of high fidelity because the OR is coupled to its native transduction cascade. There are tradeoffs in terms of the difficulty of the methodology and the throughput rate, but the techniques of adenoviral infection, gene targeting, and imaging of the dorsal olfactory bulb (OB) have added substantially to our knowledge of select ORs.
The earliest approach to studying a defined OR was to use a recombinant adenovirus to deliver exogenous genes into OSNs (Zhao et al., 1998; Touhara et al., 1999). In this approach, the virus is constructed with a constitutively active promoter that drives overexpression of an OR and a green fluorescent protein (GFP) marker. An internal ribosome entry site (IRES) sequence sandwiched between the exogenous OR and GFP produces the receptor and GFP as separate proteins in infected cells (Potter et al., 2001). Introducing exogenous ORs into a native cellular environment permits proper trafficking and expression of untagged, unmodified receptors on the plasma membrane. Using the adenoviral technique, ligands for rat OR-I7—the first OR to be deorphaned—were discovered by recording OR activity ex vivo, using an electro-olfactogram (a moderate throughput screening methodology in which odorants are delivered in vapor phase to the intact olfactory epithelium) (Zhao et al., 1998). The adenoviral technique, coupled to calcium imaging of infected OSNs from dissociated olfactory epithelium (a low throughput ex vivo screening methodology in which odorants are delivered in aqueous phase to dispersed cells), was also used to confirm a ligand for mouse MOR23 (Touhara et al., 1999).
Gene-targeting strategies in mice provide another way to deorphan ORs in their native cell type. In this approach, a fluorescent marker protein sequence is inserted adjacent to the endogenous locus of a specific OR. Once again, an IRES sequence is used to mark cells expressing the particular OR. Like the adenoviral approach, the OR being gene targeted is typically left untagged and unmodified. But unlike the adenovirus approach, in which the exogenous and endogenous ORs are coexpressed in the same OSN, the gene-targeting approach permits expression of only the single OR. Two highly related mouse ORs, M71 and M72, with 96% amino acid identity, were deorphaned through gene targeting, using ex vivo calcium imaging of labeled M71 or M72 OSNs from dissociated olfactory epithelia (Bozza et al., 2002; Feinstein et al., 2004).
Fully in vivo expression systems for ORs in OSNs have now been developed by coupling gene-targeting and OB recording techniques. This allows for realistic vapor-phase odorant delivery where the presence of mucus is preserved and odorants can be delivered under physiological conditions. The olfactory system is kept intact, from cell–cell contacts in the olfactory epithelium all the way to axonal projections to the brain. Further, odorants are delivered in the vapor phase. Although it is difficult to know the precise concentration and even the final form of an odorant that reaches the olfactory neurons because of the influence of an air–water or mucus interphase (Nagashima and Touhara, 2010), vapor-phase delivery provides a more realistic physicochemical environment. In contrast, in vitro and ex vivo (e.g., calcium imaging of dissociated olfactory epithelium) assays typically require odorants to be delivered in the water phase. Although water-phase odorant delivery may not exactly recapitulate the native physicochemical environment, it does allow for more accurate assessment of the concentrations of odorant delivered. Indeed, even higher odorant concentrations may be achieved in the water phase depending on the vapor pressure of the odorant. This may be one reason for the observed discrepancies in receptor profiles observed across the different assay systems (see below; Oka et al., 2006). Thus, although a particularly technically challenging technique, in vivo assays provide the most realistic albeit perhaps a less well defined olfactory environment in which to perform functional studies.
The in vivo recording method takes advantage of the stereotyped topography of OSN projections between the olfactory epithelium and the OB. Sensory neurons expressing a specific OR coalesce in roughly two stereotyped locations of the OB called glomeruli, one located laterally and one medially (Ressler et al., 1994; Vassar et al., 1994; Mombaerts et al., 1996). These neuropil conglomerates are composed largely of the axons of OSNs synapsing onto dendrites of mitral and tufted neurons, with these second-order neurons then projecting axons forward to cortical regions of the brain (Haberly and Price, 1977; Shepherd, 2006; Ghosh et al., 2011). Glomeruli located on the dorsal surface of the OB are accessible with minimal surgical manipulation, making them amenable to in vivo optical imaging recordings of OSN activity. Several imaging techniques have been used in the dorsal bulb to detect odorant-induced activity of OSNs of unknown OR identity. These include recording intrinsic signals (Rubin and Katz, 1999; Uchida et al., 2000), using the genetically encoded activity marker synapto-pHluorin (Bozza et al., 2004), applying calcium-indicator dyes (Oka et al., 2006), and using optogenetic reporters for calcium and membrane voltage (Storace, D.A., L.B. Cohen, and U. Sung. 2013. Association for Chemoreception Sciences Annual Meeting. Abstr. P109). Although beneficial as an in vivo technique, imaging the dorsal surface of the OB only provides a partial picture of receptor activity because it can only report on the subset of OSNs that project to the dorsal glomeruli. Although receptor activity has been recorded from other areas of the OB (Igarashi and Mori, 2005; Shirasu et al., 2014), this requires substantial surgical manipulation and still suffers from sampling only a small subpopulation of receptor types.
More recently, specific ORs have been gene targeted so that identified ORs can be screened in the dorsal OB (Belluscio et al., 2002; Bozza et al., 2002). Here, a fluorescent marker (e.g., tauGFP) is inserted in the locus just downstream of an OR that directs axons to the dorsal OB (such as M71). The resultant GFP-labeled glomerulus for that specific OR can thus be identified while functional activity can be viewed using intrinsic activity or a fluorescent activity indicator of a different wavelength. An extension of this technique expands the range of ORs that can be tested by replacing the dorsal bulb–targeting OR sequences with OR sequences that would normally direct axons elsewhere (as in the I7→M71 locus swap). The OSNs that express the replacement OR now form glomeruli located in the dorsal OB and are therefore amenable to dorsal OB imaging techniques (Soucy et al., 2009; Tsuboi et al., 2011). This gene-targeted approach in which dorsal-projecting OR loci are replaced by presumably any target OR sequence currently allows for the highest throughput of ligand screening among OSN-based systems, and it also allows odorants to be screened in the more physiological vapor phase. However, producing such a mouse line is both expensive and slow.
Contributions from OSN expression systems.
Native expression systems, in which defined ORs are expressed in OSNs, have yielded important insights into fundamentals of the olfactory code. Rat OR-I7 and mouse M71/M72 provide good examples. OR-I7 has been screened with panels of diverse and panels of only subtly different chemicals, thus providing in-depth structure–activity relationship insight for this receptor (Araneda et al., 2000). The initial electro-olfactogram recordings from OR-I7 adenovirus-infected regions of the epithelium revealed that, among a diverse panel of 74 test odorants spanning multiple chemical functional groups, only saturated aliphatic n-aldehydes with simple backbone chains of 7–10 carbons activated OR-I7. Within this range, the eight-carbon octanal was the most potent. Octanal was then used as the lead compound in a further electro-olfactogram screen in which a medicinal chemistry approach was undertaken. 90 different compounds with subtle and systematic changes in functional group, carbon chain length, degree of saturation, and type of side-chain substitution were tested (Araneda et al., 2000). This screen revealed that OR-I7 could tolerate particular patterns of unsaturation and branching but not others. This work provided the first thorough characterization of an OR’s molecular receptive field and identified citral, a partial agonist of OR-I7, as the first known modulator of any OR’s activity.
Screening OSNs from M71 or M72 IRES tauGFP gene-targeted animals (Bozza et al., 2002; Feinstein et al., 2004) provided an opportunity to investigate how closely related receptors parse odorants. These receptors differ by only 11 amino acids. In initial calcium imaging of labeled OSNs from dissociated olfactory epithelia, the two ORs both seemed narrowly tuned to acetophenone. However, later patch-clamp recordings from the dendritic knobs of labeled OSNs revealed several receptor-discriminating ligands from among a large screen of 330 compounds (Zhang et al., 2012). How minor changes in OR amino acid composition can affect response profiles, and the implications for how confidently one can predict receptive fields based on overall sequence similarity, is an area open to active investigation.
Dose–response relationships (EC50s) of GFP-labeled M71 OSNs in the dissociated olfactory epithelium preparations display a surprising level of variability. For instance, the EC50 for acetophenone differed by more than an order of magnitude in different experiments (Bozza, et al., 2002). This variability is also evident in ex vivo patch recordings of intact epithelia, as seen in the response of the mouse MOR23 receptor to Lyral (Grosmaitre et al., 2006), suggesting that it is not merely a consequence of the dissociation method. Thus, it may be an inherent biological property of OSNs. Although this phenomenon has yet to be explored in in vivo systems, such biological variability calls into question how odorant concentration is encoded when OSNs expressing the same OR can nevertheless have quite different sensitivities.
An important aspect to consider when comparing multiple expression systems is fidelity, and the OSN-based expression systems show good cross-validation. In vivo functional imaging of GFP-labeled OR-I7 glomeruli (Belluscio et al., 2002) has confirmed ex vivo electro-olfactogram and OSN calcium imaging results that indicate octanal is a potent OR-I7 ligand.
Deorphanization using heterologous expression systems.
Although they most faithfully recapitulate the receptive field, OSN-based expression systems are relatively low throughput. Moreover, they have been the principal mode of extended investigation for only seven ORs: rat OR-I7 (Zhao et al., 1998), mouse OR-I7 (Bozza et al., 2002), MOR23 (Touhara et al., 1999), MOR-EG (Kajiya et al., 2001), MOR-EV (Kajiya et al., 2001), M71 (Bozza et al., 2002), and M72 (Feinstein et al., 2004). Although the main goal of high throughput compound screening is to identify more comprehensively the set of odorants that make up a given OR’s receptive field, in time this process can lead to a compendium of ORs that share a given ligand (Malnic, 2007; Touhara, 2007; Krautwurst, 2008; Reisert and Restrepo, 2009). Large scale, heterologous-based screening thus holds the promise of providing a more substantive window onto the olfactory combinatorial code.
A particularly challenging problem, however, is that many ORs are retained in the ER of heterologous cells. Failing to traffic to the cell surface, the ORs are unable to interact with the odorant (Lu et al., 2003, 2004). Models of a two-step process of intracellular OR trafficking in OSNs tend to focus primarily on the role of OSN-specific forward trafficking proteins (McClintock and Sammeta, 2003). These include assistive proteins such as receptor-transporting protein (RTP)1, RTP2, and receptor expression–enhancing protein (REEP)1 (Saito et al., 2004), and certain non-olfactory GPCRs that may act as heterodimer partners (Bush and Hall, 2008). One relatively unexplored possibility, however, is the inverse scenario in which heterologous cells possess or overexpress one or more negative trafficking factor(s). Such factors may retain ORs in the ER because of protein misfolding or because they fail to recognize even a properly folded OR as appropriate export cargo. In support of this possibility, ER retention by the organelle’s quality control machinery has been documented for several non-olfactory GPCRs (Kim and Arvan, 1998; Ellgaard and Helenius, 2003; van Anken and Braakman, 2005). The OR trafficking complex in the OSN may thus involve the presence of OSN-specific forward trafficking factors or the absence of specific ER quality-control genes, both of which are largely unknown. A more thorough understanding of the apparently unique protein trafficking environment in OSNs could suggest modifications to help alleviate the bottleneck of functional OR expression in heterologous cells.
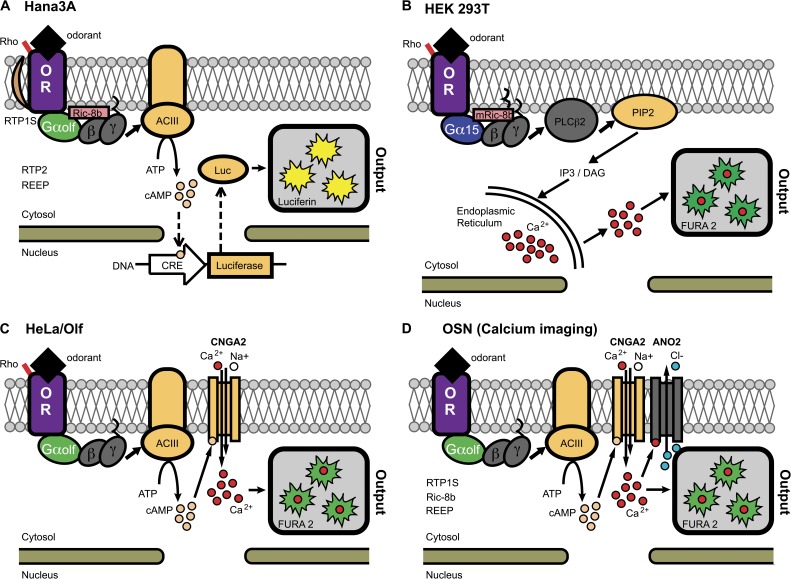
Some modified cell lines do provide an acceptable environment for functionally expressing at least certain ORs. Although systems such as Xenopus laevis oocytes (Katada et al., 2003; Abaffy et al., 2006, 2007), the insect Sf9 cell line (Matarazzo et al., 2005), yeast (Minic et al., 2005), even budding baculovirus (Mitsui et al., 2012) have been used, the mainstay of deorphanization through heterologous expression has been two human cell lines, HEK293 and HeLa (Fig. 1). HEK293 has been a reliable vehicle for studying mOR-EG (Kajiya et al., 2001; Katada et al., 2003, 2005; Oka et al., 2004a,b, 2006; Baud et al., 2011), making mOR-EG a staple receptor for pharmacology studies and comparison with other heterologous expression systems. mOR-EG was identified through single-cell RT-PCR (Kajiya et al., 2001) of eugenol-responding OSNs from dissociated mouse olfactory epithelia. Rather strikingly (in retrospect), mOR-EG could be functionally expressed directly in HEK293 cells, without cofactors or even an N-terminal tag on the OR. Although a non-olfactory G protein, Gα15, is the OR coupling partner in HEK293 systems, mOR-EG still responded to the same ligands in the in vitro assay as those identified in the original ex vivo OSN screening (Kajiya et al., 2001). HEK293 cells have also been used to express the human ORs hOR17-4 (Spehr et al., 2003), hOR17-40 (Wetzel et al., 1999), and OR51E2 (Shepard et al., 2013), as well as the rat OR-I7 (Krautwurst et al., 1998), and the mouse ORs OR-I7 (Krautwurst et al., 1998) and Olfr78 (Shepard et al., 2013), although some of these latter cases involved the addition of a tag or leader peptide sequence to the N terminus of the OR sequence.
Figure 1.
OR signal transduction and functional readout of the main in vitro assay systems (A–C) and ex vivo dissociated OSNs (D). Salient differences between these assays include the use of N-terminal amino acid tags (e.g., Rho) and trafficking proteins (e.g., RTP1S) to promote cell surface expression, Gαolf or Gα15 G proteins that trigger calcium ion (Ca2+) flux or cAMP production upon ligand binding, and Ric8b or myristoylated Ric8B (mRic8b) proteins that attenuate G protein signaling. RTP2 and REEP are present in Hana3A cells but appear to only marginally improve functional OR expression. RTP1S, REEP, and Ric8b mRNAs are expressed in OSNs, but there is a paucity of evidence for their function there. ACIII, adenylyl cylcase type III; ANO2, anoctamin 2 calcium-activated chloride channel; CNGA2, cyclic nucleotide–gated ion channel A2; CRE, cAMP response element; Luc, luciferase protein; OR, odorant receptor.
The most common functional readout for ORs expressed in HEK293T cells coupled to Gα15 is an intracellular increase in calcium, monitored using fluorescent calcium-binding dyes such as fura-2. This enables a fast readout, as the aqueous-delivered odorants only need to be applied transiently. But because odorants are typically applied sequentially (as opposed to concurrent application using a fluorescent imaging plate reader [FLIPR] assay, as will be discussed later), the overall time course of the experiment is prolonged and only a limited number of odorants can be tested on the same cell preparation. Such imaging thus represents only a low to mid-throughput approach.
The most widely used in vitro OR assay uses the Hana3A cell line, an extensively engineered system derived from HEK293T cells (Saito et al., 2004). The Hana3A cell line stably expresses the canonical OSN G protein α subunit Gαolf, bringing the signal coupling closer to an in vivo situation. Hana3A also stably expresses three OSN OR trafficking cofactors (RTP1, RTP2, and REEP) that have been shown to enhance cell surface expression of a subset of tested ORs (Saito et al., 2004). In addition, coexpression of an alternative splice variant of RTP1 called RTP1S (Wu et al., 2012), the guanine nucleotide exchange factor Ric8B (Von Dannecker et al., 2005, 2006; Kerr et al., 2008), and the type 3 muscarinic acetylcholine receptor (Li and Matsunami, 2011) indicates that all three appear to work synergistically to enhance OR functionality without altering ligand specificity (Zhuang and Matsunami, 2007).
In addition to this collection of cofactors, optimal cell surface expression often requires modification of the OR itself. In the Hana3A system, as in nearly all other in vitro OR assays, fusing the first 20 amino acids of rhodopsin (Rho tag) or other short noncleavable tags (e.g., Flag, c-myc, or HA) to the receptor’s N terminus is common and typically necessary for optimal functional expression. But it is possible that such permanent tags modify OR response profiles (Zhuang and Matsunami, 2007). Recently, a cleavable 17–amino acid leucine-rich N-terminal signal peptide (Lucy tag) was shown to promote cell surface OR expression in HEK293T cells (Shepard et al., 2013). The Lucy tag appears to operate synergistically with RTP1S, Ric8b, and a Flag-Rho tag. Strikingly, all 15 of the ORs tested with Lucy could be visualized on the cell surface by immunostaining. The use of cleavable tags like Lucy, where little or no tag remains on the mature OR, should help improve the in vivo relevance of screening data derived from in vitro systems.
The functional readout from Hana3A-based systems comes from pairing Gαolf to an endogenously expressed adenylate cyclase to yield odorant-induced cAMP production. The increase in cAMP is monitored via a cAMP response element (CRE)-driven luciferase reporter, yielding light as the output (Katada et al., 2003; Saito et al., 2004). A potential drawback, however, is that when used with a standard luminometer plate reader, measurable and consistent odorant-dependent luciferase production in the CRE luciferase assay can require up to four hours of odorant stimulation. The potential for OR adaptation during such prolonged odorant exposure is unknown.
An alternative to these two main HEK293T-based systems is the HeLa/Olf expression system. This has been used successfully to deorphan ORs without added trafficking cofactors, although an N-terminal Rho tag on the OR is still typically used (Shirokova et al., 2005). The HeLa/Olf system includes Gαolf and an adenylate cyclase like Hana-3A but differs in its functional readout. For the HeLa/Olf system, the odorant-induced cAMP activates an exogenously expressed variant of the CNG channel (CNGA2). cAMP binding to CNGA2 stimulates calcium influx, which can be detected using a fluorescent calcium–sensitive dye (e.g., Calcium 5). This calcium change could, in theory, be monitored for sequential applications of odorants as in the HEK assay. But in investigations using HeLa/Olf, the primary strategy has instead been to record receptor activity in real time with a FLIPR. FLIPR assays, which can be run in a multi-well format, are a widely used strategy for high throughput screening of GPCRs in the pharmaceutical industry (Emkey and Rankl, 2009). The HeLa/Olf system thus allows for reduced odorant exposure times compared with luciferase reporter systems.
Caveats of heterologous expression systems.
Heterologous expression systems have enabled the deorphanization and extended characterization of ∼95 mouse and 41 human ORs (Mainland et al., 2014) as of the beginning of 2014. Although still this means that only ∼8 and ∼10% of each species’ OR repertoire, respectively, have been deorphaned, this contribution by in vitro studies stands in marked contrast to the mere seven ORs primarily studied using OSN-based methods and confirmed in separate studies over this same period (i.e., rat OR-I7 [Zhao et al., 1998], mouse OR-I7 [Bozza et al., 2002], MOR23 [Touhara et al., 1999], MOR-EG [Kajiya et al., 2001], MOR-EV [Kajiya et al., 2001], M71 [Bozza et al., 2002], and M72 [Feinstein et al., 2004]). Thus, heterologous expression assays are the undisputed primary drivers of deorphanization. But several cautionary cases of assay-dependent bias should be considered when interpreting OR response profiles obtained in vitro.
One cautionary case involves the G protein identity. Some GPCRs can couple with more than one G protein α subunit. Recent evidence suggests that when a given GPCR is docked to a particular G protein α subunit, a certain subset of chemicals may serve as ligands, but when docked to an alternate G protein α subunit, the functional set of ligands may change. Thus, some ligands preferentially signal through a particular pathway (Sivertsen et al., 2013). Such stimulus signaling bias has been reported for ORs in heterologous systems and can alter the resulting odorant response profile (Shirokova et al., 2005; Oka et al., 2006; Hamana et al., 2010). Shirokova et al. (2005) showed that the noncanonical Gα15, the G protein commonly used in HEK-based systems, can alter the odorant specificities and pharmacological profile of ORs. For example, when ORs6 was coupled to Gα15, octanoic acid was an agonist. But when ORs6 was coupled to Gαolf, octanoic acid not only failed to activate the receptor, but it actually served as a competitive antagonist. The same study by Shirokova et al. (2005) also illustrated how the Gα15 G protein displays “permissiveness”; 7 of the 14 ligands for mouse receptor olfr43 were agonists in only the Gα15 but not the Gαolf background, suggesting that Gα15 may enable more indiscriminate ligand binding. Although Gα15 assays may thus provide a good measure of ligand-binding potential, they may not reveal the whole story with regard to receptor function. Ligand efficacy, which includes the activity of the entire receptor–G protein complex and downstream signaling cascade, is a more relevant measure of receptor activity. This suggests that assays involving the canonical G protein, Gαolf, may more accurately measure ligand efficacy and may therefore provide a better gauge of the OR’s discriminatory capacity. Although the inconsistencies depending on coupling agent make comparisons across studies difficult, they may lead us to a better understanding of how the G protein–OR complex acts as a unit in determining OR receptive fields.
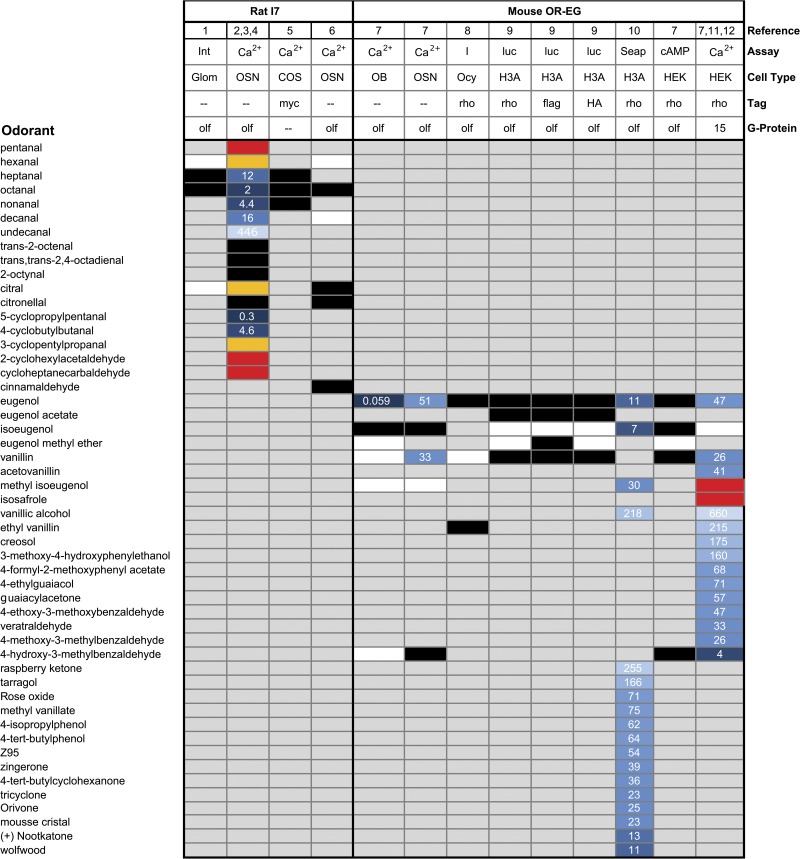
Although the influence of the assay on the reported receptive fields has been documented for rat OR-I7, (Mombaerts, 2004), MOR29B (Tsuboi et al., 2011), and the trace amine-associated receptor TAAR4 (Zhang et al., 2013), this bias has been most thoroughly explored for mOR-EG (Fig. 2). Oka et al. (2006) compared the specificity and sensitivity of the mOR-EG response profile in vitro in HEK293 cells coupled to either Gα15 or Gαolf, ex vivo by calcium imaging of OSNs from the dissociated olfactory epithelium of a mOR-EG–labeled transgenic mouse, and in vivo by intrinsic imaging of GFP-labeled glomeruli in this same transgenic mouse line. The response profile of mOR-EG to seven odorants in HEK293 and dissociated OSNs was nearly identical, although some difference was observed depending on the G protein used in the HEK293 assay. In particular, the downstream coupling partner affected the ability of isoeugenol to serve as an agonist. But the most unexpected results came from the in vivo assay. Vanillin and 4H3MBA, delivered in water phase, were strong mOR-EG agonists in both HEK293 cells coexpressing Gαolf and dissociated OSNs (which also express Gαolf). Yet when OSN activity was measured in the vapor phase at the level of the glomeruli, very modest or no responses to these odorants was obtained. This was not caused by a general decrease in sensitivity in the in vivo method of recording. Indeed, an ∼1,000 times increase in the sensitivity to eugenol was observed with in vivo glomerular imaging compared with the other assay systems. Intriguingly, the responses of mOR-EG glomeruli to vanillin before and after washout of the nasal cavity suggest that the olfactory mucus may account for part of the observed difference. Thus, caution extends even to ex vivo work with OSNs from dissociated olfactory epithelia; the presence of mucus and a fully intact in vivo system may better define the physiologically relevant boundaries of a receptive field. But until it is resolved how odorants may partition into the mucus, and how this affects their effective concentration, a continued benefit of ex vivo work is that the aqueous concentrations can be well controlled. This in turn provides an accurate view of which odorants can bind a given OR and enables the development of structure–activity relationships.
Figure 2.
Response profiles of rat OR-I7 and mouse OR-EG expressed in various assay systems. The response profiles of a given OR can vary depending on the assay system. For rat OR-I7 and mouse mOR-EG, tested but inactive compounds are not included in this figure unless the compound showed agonist or antagonist activity in at least one of the assays shown. For rat OR-I7, only select agonists are reported. See Araneda et al. (2000) for a more complete listing of OR-I7 agonists. Blue boxes, heat map of reported EC50 values; darkest blue, <0.1 µM; lightest blue, 660 µM; black boxes, agonist but no EC50 reported; orange boxes, partial agonist; red boxes, antagonist; white boxes, tested but inactive; Int, intrinsic imaging; Ca2+, calcium imaging; I, current; luc, luciferase; Seap, secreted alkaline phosphatase; OB, olfactory bulb; OSN, olfactory sensory neuron; COS, African green monkey cells; Ocy, Xenopus oocytes; H3A, Hana3A cells; HEK, human embryonic kidney cells. References: 1, Belluscio et al. (2002); 2, Araneda et al. (2004); 3, Peterlin et al. (2008); 4, Kurland et al. (2010); 5, Levasseur et al. (2003); 6, Bozza et al. (2002); 7, Oka et al. (2006); 8, Repicky and Luetje (2009); 9, Zhuang and Matsunami (2007); 10, Baud et al. (2011); 11, Katada et al. (2005); 12, Oka et al. (2004a,b).
Contributions from heterologous expression systems.
Despite these caveats, the throughput capacity of heterologously expressed ORs offers a valuable window to OR biology. For example, mOR-EG’s extensive characterization in the HEK293 system (Katada et al., 2003, 2005; Oka et al., 2004a,b) and the ability to readily express even mutated mOR-EG receptors (Katada et al., 2005; Kato et al., 2008) paved the way for detailed analysis of the relationship between agonist and antagonist ligands. From screening with blends, the odorants methyl isoeugenol, isosafrole, and dimerized isoeugenol were identified as antagonists of mOR-EG as expressed in HEK293 cells with Gα15 (Oka et al., 2004a,b). These results, along with trends among identified agonists and antagonists for other ORs (Araneda et al., 2000; Sanz et al., 2005; Shirokova et al., 2005; Jacquier et al., 2006; Abaffy et al., 2007; Peterlin et al., 2008), suggest that most OR antagonists are likely to be structurally related to the agonist compounds, be odorants themselves, and exhibit competitive antagonism. But it was through a large structure–activity relationship study coupled with point mutagenesis that a mechanism for antagonism in mOR-EG was proposed and tested (Katada et al., 2005). Vanillin, a potent agonist, was used as the lead compound in a medicinal chemistry approach that varied the substituents around three positions of vanillin’s benzene ring. 21 additional compounds were screened and 13 were found to activate mOR-EG over a 100-fold span in relative affinity (EC50), starting in the low micromolar range. By combining these structure–activity relationship results with the results from point mutations and computational homology modeling, the mOR-EG odorant-binding site was predicted. It consisted of a hydrophobic pocket formed by portions of transmembrane domains III, V, and VI. Nine amino acids within the presumptive binding pocket were shown to be involved in receptor activation. They propose that there are critical electronic interactions between a phenyl ring in the binding pocket and the particular double-bond pattern in the tail of isoeugenol and related antagonists. A simple shift of the double bond by one position was sufficient to turn an isoeugenol-like antagonist into a eugenol-like agonist. Thus, this electrostatic interaction between ligand and receptor may be the switch that filters agonists from antagonists for this OR.
Heterologous expression systems can also provide deep insight about the combinatorial code, as is evident in the work of Saito et al. (2009). Using the Hana3A cell-line approach, this group produced the largest single dataset to date, in which 93 odorants, spanning a large portion of the chemical space of commercially available odorants, were tested on 62 ORs. The authors then determined the potency of each of the 63 odorants that activated at least one OR. The resulting information-dense matrix revealed intriguing and unexpected coactivation patterns by odorants both across ORs and within the receptive field of individual ORs. Because these ORs are heterologously expressed, these patterns can be probed further. The ability to devise hypotheses about chemical relationships and then revisit the same ORs, to test them with a new panel of compounds, is a great strength of heterologous expression systems.
In silico approaches to deorphanization
The challenges of expressing ORs on the cell surface and of probing them with very large numbers of ligands make computational in silico approaches an attractive alternative for deorphaning ORs. A robust in silico system could conceivably allow virtual high throughput screening of all compounds that possess physicochemical features that make them likely odorants. This would provide a substantial dataset from which to develop a detailed pharmacophore model of the chemical features and their arrangement that is required for activation of the OR in question. A glimpse of such potential was seen for the in vitro deorphaned OR1G1, which was activated by 59 of 95 diverse odorants (Sanz et al., 2005). Having such a large agonist repertoire allowed derivation of the best-defined pharmacophore for an OR to date (Sanz et al., 2008). In silico approaches that build up large datasets of predicted binders and nonbinders may thus help identify the true feature detector capabilities of each OR.
One major in silico approach that lends itself to receptor deorphanization is to construct OR models and dock potential ligands (Singer and Shepherd, 1994; Singer, 2000; Hall et al., 2004; Hummel et al., 2005; Katada et al., 2005; Lai et al., 2005, 2008, 2014; Abaffy et al., 2007; Schmiedeberg et al., 2007; Kurland et al., 2010; Anselmi et al., 2011; Baud et al., 2011; Charlier et al., 2012; Gelis et al., 2012; Launay et al., 2012). OR modeling has typically worked in close concert with a biologically expressed OR, with deorphaning of the OR in the biological expression system preceding development and refinement of an in silico model. Models have primarily been used to help direct site mutagenesis studies, propose rationale for ligand discrimination, and address other questions of receptor biology versus serving as a stand-alone screening system. On the rare occasions where OR models have been used to try to predict novel ligands for a given OR, success rates have been mixed. Hall et al. (2004) built two models of the rat OR-I7 receptor, into which 62 putative ligands were docked and their predicted binding energy was calculated. A high binding energy should correlate with empirical binding. In their first build, no molecule had a binding energy of 30 kcal/mol or higher. When Hall et al. (2004) refined their model-building procedure, they now found odorants that bound in the 30-kcal/mol range, 69% of which were later empirically confirmed to be activating ligands. In silico screening of libraries of ligands has recently seen a resurgence. For MOR42-3, 40 candidates were selected from an initial 574 odorant panel based on docking to a receptor model; 55% of these high scorers were validated in a physiological screen (Bavan et al., 2014). Dynamic modeling has also brought improvements to in silico screening systems, correctly predicting the binding of amyl butyrate to hOR2AG1 and mOR283-2 (Gelis et al., 2012).
ODORactor, a web-based tool, takes a quite different approach to in silico–assisted deorphanization (Liu et al., 2011). OR models approach the combinatorial code from the “one OR is activated by multiple odorants” facet. ODORactor approaches the code from the “one odorant activates multiple ORs” facet, making in silico predictions of the suite of ORs that detect a given ligand. Eschewing model building, ODORactor has data-mined all published deorphaned human and mouse ORs and uses multidimensional analysis and chemoinformatics in its predictions. ODORactor may prove to be a valuable tool when the odorant is of special interest, such as for potential pheromones or other behavior-modulating chemicals (Lin et al., 2005), or odorants of practical (Corcelli et al., 2010; Li et al., 2012) or commercial relevance (Bieri et al., 2004).
Caveats to in silico approaches.
Although in silico approaches show promise, at present they still benefit from validation by biological means. Some of the challenges still to be surmounted by the modeling approach are evident in work on rat OR-I7. Rat OR-I7 has been modeled independently for over a decade (Singer, 2000; Hall et al., 2004; Lai et al., 2005; Khafizov et al., 2007; Kurland et al., 2010) using various technical approaches. However, only one residue is consistently implicated in binding octanal; the remaining residues predicted to line the binding pocket are highly contested. Alignment of ORs to a template GPCR is a key step in certain types of model building. Bovine rhodopsin in its retinal-bound, inactive state was at one time the only mammalian class A GPCR with 3-D structural information (Palczewski et al., 2000). Even recent OR models still often use the rhodopsin template as part of the construction and optimization process. But now an array of other crystallized class A GPCRs in their active state are available. The GPCRautomodel web tool enables users to select among these new templates when building an OR model (Launay et al., 2012). Whether new alignment choices lead to improved receptive field predictions remains to be seen.
Because ODORactor updates its database with each new publication, one thing to be mindful of is that its predictions are dependent on the sum quality of published data. For example, the predictions for mouse ORs made by the current version of ODORactor may be heavily weighted by the receptor and ligand profiles present in the in vitro heterologous expression work of Saito et al. (2009), given that this study contributes nearly half of all currently deorphaned mouse ORs. How ODORactor deals with conflict, given that the details of the receptive field for the same OR can vary depending on the readout system it is coupled to (Fig. 2), also needs to be considered. Finally (provided the OR can be expressed), although the positive hits predicted by ODORactor can be validated, the frequency of false negative predictions by ODORactor cannot be readily discerned. As more ORs outside the initial ODORactor build are deorphaned and compared with ODORactor predictions before and after publication, this will clarify the capabilities and constraints of this intriguing new tool.
Choosing ligands for deorphanization
Wide-scale screens with various rationale.
Once a given OR has been successfully expressed, there remains the question of how to look for that first activating ligand in vast chemical space. One possibility involves a “shotgun” approach where many odorants are simply tried either alone or in blends without clearly disclosed rationale. Testing blends followed by fractionation would seem the obvious choice to increase the number of odorants screened. But there can be concerns about solvent levels in multiple-component blends. Furthermore, this technique can backfire should an antagonist be present in the same fraction as an agonist. Given that many of the currently published antagonists are structurally similar to agonists of the same receptor (Araneda et al., 2000; Oka et al., 2004a,b; Sanz et al., 2005; Shirokova et al., 2005; Jacquier et al., 2006; Abaffy et al., 2007; Peterlin et al., 2008), blends that contain related odorants may be at particular risk of generating false negatives in functional studies. However, interactions can still occur even among unexpected pairings. This was highlighted during the use of the Henkel100 blend to deorphan hOR17-4 (Spehr et al., 2003). The only agonist to emerge from the series of mixes was cyclamal. During the follow-up medicinal chemistry exploration, however, the closely related bourgeonal was found to be active even though the mix containing it had been inactive. By fractionating that mix, it was determined that the structurally unrelated undecanal was the culprit antagonist. Thus, although precautions can be taken to divide seemingly similar components into different mixes, the encoding of odorants by ORs is still not sufficiently understood to assure that mixes will not exhibit interaction effects.
One guide to shotgun-type approaches is the data emerging from calcium imaging of OSNs from dissociated olfactory epithelia of wild-type mice or rats. The identities of the ORs are not known in this method, but because the full OR repertoire is surveyed, this approach can provide useful population-level information of the relative activity elicited by a given odorant. Although ORs are expressed at different frequencies (Khan et al., 2011), such that an odorant that activates many OSNs could conceivably be targeting just one or a few highly expressed ORs, constructing an odorant panel whose components each activate many OSNs with minimal overlap in their coverage remains a reasonable strategy for first-pass, general deorphanization of an OR. Although surveys using blends have shed light on OR repertoire coverage (Nara et al., 2011), more commonly single odorants are used. Surveys with single odorants have highlighted important trends. For example, flexible odorants appear to activate more OSNs than do highly constrained ones (Peterlin et al., 2008), flexible odorants with mid-length chains (between seven to nine carbon backbone) recruit more OSNs than do much shorter or longer ones (Sato et al., 1994; Kaluza and Breer, 2000; Araneda et al., 2004), and acids seem to be poor recruiters relative to alcohols or aldehydes of the same length (Araneda et al., 2004). Attentiveness to such trends during panel design may help maximize the potential for hits.
A second form of guidance can be found in the computational analysis of biologically relevant chemical space. This is the subset of all chemicals, such as those that can be found in larger databases such as PubChem or the Available Chemicals Directory, filtered based on general parameters that typify odorants. Chemical space is the multidimensional arrangement of these putative or known odorants based on their physicochemical descriptors and biological functional similarity. It is organized so that the displacement of two odorants relative to each other is related to the probability of their being co-detected by ORs (closer odorants being more likely to be co-detected). Determination of an appropriately weighted chemical space has helped generate highly accurate predictions of the receptive fields for the mosquito Anopheles gambia (Wang et al., 2010) and the fruit fly Drosophila melanogaster (Boyle et al., 2013). Currently, there are three models for the chemical space as weighted using data from mammals (Haddad et al., 2008; Saito et al., 2009; Mainland et al., 2014). To help navigate and use one model of this space, Haddad et al. (2008) provided lists of common odorants that should comprise broadly dispersed initial screening panels. “Broad” here refers to the diameter of the hypersphere that encloses the odorants in multidimensional space. Models of chemical space are being used retroactively to assess the breadth of coverage of assay panels and the breadth of tuning of OR receptive fields (Saito et al., 2009; Li et al., 2012). However, panels composed by this method have yet to be deployed for initial deorphanization attempts.
Targeted screens that take the receptor into account.
The above approaches to identifying the first ligand emphasize chemical diversity, focusing solely on the odorants. In contrast, the paralogue/orthologue approach uses information about the receptor to guide selection of a set of odorants to test. Paralogues within a species often share elements of their receptive fields, such as seen with M71 and M72 (96% protein identity) (Zhang et al., 2012), and MOR29A and MOR29B (95% protein homology) (Tsuboi et al., 2011). But when using the paralogue approach to testing compounds that are successful ligands of the “reference” OR against the orphan OR, there is the chance that the reference OR might be narrowly tuned. The three members of the large MOR256 family provide a telling, cautionary example. Whereas MOR256-17 responds to a variety of ligands, MOR256-8 and MOR256-22 can detect only a restricted subset of the MOR256-17 receptive field. Furthermore, the receptive fields of MOR256-8 and MOR256-22 show very sparse overlap with each other (Li et al., 2012). Thus, in this case, which OR is chosen as the reference would greatly affect success in deorphaning even related receptors.
As an alternative to comparing paralogues, looking across species to a deorphaned orthologue is often more productive. Some orthologues with very similar receptive fields include mouse and rat OR-I7 (Bozza et al., 2002), and mouse Olfr43 and human OR1A1 and OR1A2 (Schmiedeberg et al., 2007). In a far-ranging survey of 17 rat ORs and their mouse orthologues and 18 chimpanzee ORs and their rhesus macaque orthologues, Adipietro et al. (2012) concluded that orthologues are nearly 2.5 times more likely to share a ligand in common than are paralogues. They also demonstrated that orthologues often share an extended ligand profile, albeit with differences in odorant EC50s. These findings should provide encouragement for those seeking to extend findings from mouse ORs to human ORs.
Although robust, the orthologue approach is limited to a handful of ORs because the OR to be deorphaned must be a specific match. This is opposed to the paralogue approach that requires only family affiliation and thus has greater widespread applicability for deorphaning. But why might comparing paralogues not be as good a predictor in terms of similarity of receptive fields as when orthologues are compared? One contributing factor may be that the current classification of ORs into families is hierarchical in nature and based on full-length sequence. A reorganization based on short motifs with predicted functional relevance may improve performance. The foundations for alternative organizations already exist. Two studies independently identified short motifs distributed throughout the OR (Liu et al., 2003; Zhang et al., 2007). Man et al. (2004) compared select mouse and human orthologues and paralogues to identify a set of 22–amino acid residues, predicted to have special impact on ligand recognition. Saito et al. (2009) identified 15 residues for which 16 optimized physicochemical property descriptors were very good at predicting the response of 62 ORs to a panel of 63 odorants. Any of these motif lists can be used to break free of hierarchical constraints and plot ORs in multidimensional space. The resultant new relationships between ORs may better incorporate known or inferred physiology.
Beyond deorphanization: Integration with the combinatorial code
When a ligand has been found for the expressed OR of interest, a common next step is to use a medicinal chemistry approach to look for additional ligands to help characterize the structure–activity relationship for that OR. For example, a battery of compounds with subtle changes may be used to examine the discriminatory capacity of the receptor. This approach has been used successfully for many receptors, including mOR-EG (Katada et al., 2005) and M72 (Zhang et al., 2012). Both receptors were tested with panels of small aromatics. Because only compounds with scaffolds similar to the initial target are tested, there may be a tendency to consider that a receptor is “narrowly tuned.” But expanding to more diversified test panels can sometimes yield a marked shift in perspective. For example, Baud et al. (2011) tested an assortment of odorants on mOR-EG, and Soucy et al. (2009) tested a collection of compounds on M72. In neither case did these ORs appear to remain so “narrowly tuned”; mOR-EG also responded to aliphatic cyclic ketones and M72 to aliphatic tiglate esters. It can be daunting to probe OR recognition of structurally diverse odorants because such a search is likely to return many inactive compounds. But this type of expanded testing provides valuable information to challenge pharmacophore models. This, in turn, can better clarify what chemical features the OR truly abstracts.
Another strategy to expand beyond an initial lead compound is to challenge the OR with odorants that the larger OR repertoire reports as “biologically similar.” This strategy can apply to subsections of the odorants as well, as in the case of substituting a cyclohexene ring in place of a benzene ring. Such odorants may appear structurally dissimilar, but as the exchanges proposed are empirically based, the approach may be particularly productive. The information on “biological similarity” can be gained from calcium imaging of OSNs from dissociated olfactory epithelia of wild-type mice or rats. Even though the identity of the ORs is not known, the large-scale patterns across the OR repertoire show how frequently two odorants are recognized by the same OSN. This information can be used to generate probability tables. Such tables may aid deorphaning efforts by suggesting what other odorants are likely activators. Knowledge of expected co-recognition rates can also flag when an OR is making a challenging discrimination, thus contributing in a special way to the combinatorial code.
Future perspective
A relatively new approach to deorphanizing human ORs involves genomics, where the presence of naturally occurring genetic variations (single-nucleotide polymorphisms) in individual OR genes are correlated with the relative activities of the ORs in vitro and the measured odor perception in humans (Keller et al., 2007; Menashe et al., 2007; Lunde et al., 2012; McRae et al., 2012; Jaeger et al., 2013; Mainland et al., 2014). This approach has been successful in correlating the activity of individual ORs with measureable perceptual characteristics like odor intensity and pleasantness ratings. These emergent genomics-based OR percept studies may represent a powerful new strategy for OR deorphanization because they can provide a direct link between OR identity and perception. However, inherent to this and all other OR deorphanization strategies is the absolute requirement for a robust in vitro assay to confirm putative ligand–receptor pairings and to probe odorant structure–activity relationships.
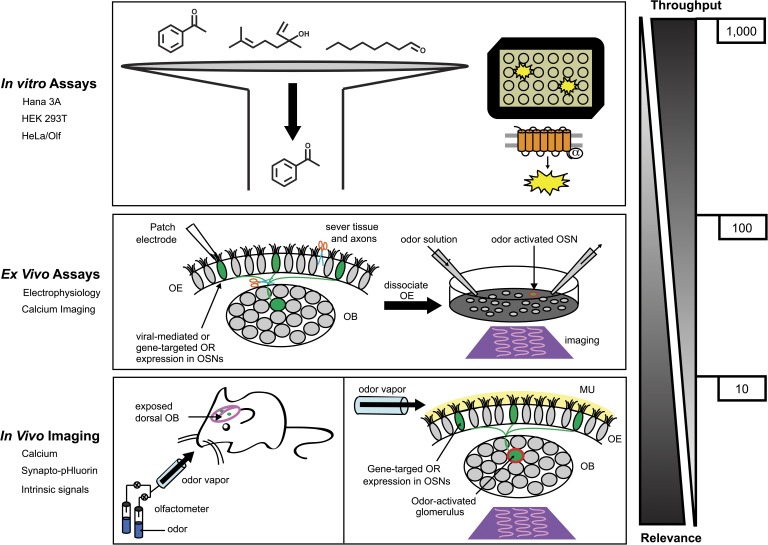
The need for a comprehensive OR assay platform that balances throughput and response accuracy is thus critical for understanding the olfactory combinatorial code. A more robust in vitro platform (Fig. 3) could enable the identification and expression of all ORs contributing to the combinatorial activation code for an odorant or odor mixture (Malnic et al., 1999). The need for such a platform is especially pronounced given that ligands for ∼90% of human ORs remain unknown. Various techniques have been adopted including using native OSNs and an array of heterologous expression systems. But each of these systems has inherent strengths and limitations, and none have all the desired criteria of a suitable assay platform. For now, the optimal screening platform should encompass an initial high throughput chemical screen using in vitro heterologous systems followed by a low throughput in vivo assay for validation. Perhaps future implementation of interdisciplinary strategies that are routinely used by the pharmaceutical industry for the discovery of GPCR agonists and modulators (Inglese et al., 2007; Eggert, 2013) could contribute to more advanced and reliable heterologous OR systems.
Figure 3.
Schematic overview of a comprehensive OR assay platform and the approximate single assay compound throughput. Given the inherent complexity of the combinatorial receptor code and the remaining challenges of current in vitro OR assays, a comprehensive OR assay platform would aim to balance screening throughput and organism relevance by incorporating in vitro, ex vivo, and in vivo assays. A robust in vitro platform would enable the initial screening of thousands of odorant compounds. Agonists or antagonists identified from in vitro systems could then be screened using ex vivo OSN assays (medium throughout and relevance) or in vivo imaging assays (low throughput and high relevance). A salient benefit of in vivo over ex vivo or in vitro assays is that odorants are delivered in the vapor phase to the fully intact peripheral olfactory system (olfactory epithelium and bulb). The main benefit of ex vivo over in vitro assays is the use of OSNs as an expression system for native (i.e., untagged) receptors. OB, olfactory bulb; OE, olfactory epithelium; OSNs, olfactory sensory neurons; MU, olfactory mucus.
Aside from the issue of proper trafficking, another reason for the relative paucity of deorphaned ORs may be that compounds used for screening have limited structural diversity and may not fully span odorant chemical space. This could be a matter of availability as commercial sources possess only limited collections of standard fragrance compounds. Moreover, commercial sources may not provide a sufficient range of subtly different odorants to serve as medicinal chemistry probes, particularly when the lead odorant has a rare or complex chemical scaffold. One relatively untapped source of both diverse and nuanced chemical libraries is the major flavor and fragrance houses. With more than 100 years of experience in chemical synthesis and purification, flavor and fragrance houses possess extraordinary collections of organoleptically defined volatile compounds that could greatly assist in both decoding the combinatorial code at the receptor level and help generate hypotheses of how that code correlates with percept. Olfactory research collaborations between academic and industry partners can be fruitful (e.g., Grabenhorst et al., 2007) and should be strengthened further to provide greater access to diverse and well-characterized odorant libraries. Ultimately, such partnerships may be an important step forward in the quest to unravel the complex and often puzzling relationships between chemical structure, receptor activity, and human perception.
Acknowledgments
The authors would like to thank Ayome Abibi, Huey-Ling Kao, and Patrick Pfister (Firmenich, Inc.) for helpful discussions.
Z. Peterlin and M.E. Rogers are employees of Firmenich, Inc. S. Firestein has ongoing experiments in the field of olfaction with Firmenich, Inc., and consults for Firmenich, Inc., as well. He has no patents with Firmenich, Inc. This Review has no bearing on his interactions with Firmenich, Inc., or vice versa, and it does not alter the authors’ adherence to all of the Journal’s policies on sharing data and materials. The authors have no additional financial interests.
Elizabeth M. Adler served as editor.
Footnotes
Abbreviations used in this paper:
- CRE
- cAMP response element
- FLIPR
- fluorescent imaging plate reader
- GFP
- green fluorescent protein
- GPCR
- G protein–coupled receptor
- IRES
- internal ribosome entry site
- OB
- olfactory bulb
- OR
- odorant receptor
- OSN
- olfactory sensory neuron
- REEP
- receptor expression–enhancing protein
- RTP
- receptor-transporting protein
References
- Abaffy T., Matsunami H., Luetje C.W. 2006. Functional analysis of a mammalian odorant receptor subfamily. J. Neurochem. 97:1506–1518 10.1111/j.1471-4159.2006.03859.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abaffy T., Malhotra A., Luetje C.W. 2007. The molecular basis for ligand specificity in a mouse olfactory receptor: A network of functionally important residues. J. Biol. Chem. 282:1216–1224 10.1074/jbc.M609355200 [DOI] [PubMed] [Google Scholar]
- Adipietro K.A., Mainland J.D., Matsunami H. 2012. Functional evolution of mammalian odorant receptors. PLoS Genet. 8:e1002821 10.1371/journal.pgen.1002821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselmi C., Buonocore A., Centini M., Facino R.M., Hatt H. 2011. The human olfactory receptor 17-40: Requisites for fitting into the binding pocket. Comput. Biol. Chem. 35:159–168 10.1016/j.compbiolchem.2011.04.011 [DOI] [PubMed] [Google Scholar]
- Araneda R.C., Kini A.D., Firestein S. 2000. The molecular receptive range of an odorant receptor. Nat. Neurosci. 3:1248–1255 10.1038/81774 [DOI] [PubMed] [Google Scholar]
- Araneda R.C., Peterlin Z., Zhang X., Chesler A., Firestein S. 2004. A pharmacological profile of the aldehyde receptor repertoire in rat olfactory epithelium. J. Physiol. 555:743–756 10.1113/jphysiol.2003.058040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud O., Etter S., Spreafico M., Bordoli L., Schwede T., Vogel H., Pick H. 2011. The mouse eugenol odorant receptor: Structural and functional plasticity of a broadly tuned odorant binding pocket. Biochemistry. 50:843–853 10.1021/bi1017396 [DOI] [PubMed] [Google Scholar]
- Bavan S., Sherman B., Luetje C.W., Abaffy T. 2014. Discovery of novel ligands for mouse olfactory receptor MOR42-3 using an in silico screening approach and in vitro validation. PLoS One. 9:e92064 10.1371/journal.pone.0092064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belluscio L., Lodovichi C., Feinstein P., Mombaerts P., Katz L.C. 2002. Odorant receptors instruct functional circuitry in the mouse olfactory bulb. Nature. 419:296–300 10.1038/nature01001 [DOI] [PubMed] [Google Scholar]
- Bieri S., Monastyrskaia K., Schilling B. 2004. Olfactory receptor neuron profiling using sandalwood odorants. Chem. Senses. 29:483–487 10.1093/chemse/bjh050 [DOI] [PubMed] [Google Scholar]
- Bockaert J., Pin J.P. 1999. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 18:1723–1729 10.1093/emboj/18.7.1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S.M., McInally S., Ray A. 2013. Expanding the olfactory code by in silico decoding of odor-receptor chemical space. Elife. 2:e01120 10.7554/eLife.01120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza T., Feinstein P., Zheng C., Mombaerts P. 2002. Odorant receptor expression defines functional units in the mouse olfactory system. J. Neurosci. 22:3033–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza T., McGann J.P., Mombaerts P., Wachowiak M. 2004. In vivo imaging of neuronal activity by targeted expression of a genetically encoded probe in the mouse. Neuron. 42:9–21 10.1016/S0896-6273(04)00144-8 [DOI] [PubMed] [Google Scholar]
- Bunzow J.R., Van Tol H.H., Grandy D.K., Albert P., Salon J., Christie M., Machida C.A., Neve K.A., Civelli O. 1988. Cloning and expression of a rat D2 dopamine receptor cDNA. Nature. 336:783–787 10.1038/336783a0 [DOI] [PubMed] [Google Scholar]
- Bush C.F., Hall R.A. 2008. Olfactory receptor trafficking to the plasma membrane. Cell. Mol. Life Sci. 65:2289–2295 10.1007/s00018-008-8028-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier L., Topin J., Ronin C., Kim S.K., Goddard W.A., III, Efremov R., Golebiowski J. 2012. How broadly tuned olfactory receptors equally recognize their agonists. Human OR1G1 as a test case. Cell. Mol. Life Sci. 69:4205–4213 10.1007/s00018-012-1116-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelli O., Reinscheid R.K., Zhang Y., Wang Z., Fredriksson R., Schiöth H.B. 2013. G protein-coupled receptor deorphanizations. Annu. Rev. Pharmacol. Toxicol. 53:127–146 10.1146/annurev-pharmtox-010611-134548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcelli A., Lobasso S., Lopalco P., Dibattista M., Araneda R., Peterlin Z., Firestein S. 2010. Detection of explosives by olfactory sensory neurons. J. Hazard. Mater. 175:1096–1100 10.1016/j.jhazmat.2009.10.054 [DOI] [PubMed] [Google Scholar]
- Eggert U.S. 2013. The why and how of phenotypic small-molecule screens. Nat. Chem. Biol. 9:206–209 10.1038/nchembio.1206 [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181–191 10.1038/nrm1052 [DOI] [PubMed] [Google Scholar]
- Emkey R., Rankl N.B. 2009. Screening G protein-coupled receptors: Measurement of intracellular calcium using the fluorometric imaging plate reader. Methods Mol. Biol. 565:145–158 10.1007/978-1-60327-258-2_7 [DOI] [PubMed] [Google Scholar]
- Fargin A., Raymond J.R., Lohse M.J., Kobilka B.K., Caron M.G., Lefkowitz R.J. 1988. The genomic clone G-21 which resembles a β-adrenergic receptor sequence encodes the 5-HT1A receptor. Nature. 335:358–360 10.1038/335358a0 [DOI] [PubMed] [Google Scholar]
- Feinstein P., Bozza T., Rodriguez I., Vassalli A., Mombaerts P. 2004. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the β2 adrenergic receptor. Cell. 117:833–846 10.1016/j.cell.2004.05.013 [DOI] [PubMed] [Google Scholar]
- Firestein S. 2001. How the olfactory system makes sense of scents. Nature. 413:211–218 10.1038/35093026 [DOI] [PubMed] [Google Scholar]
- Gelis L., Wolf S., Hatt H., Neuhaus E.M., Gerwert K. 2012. Prediction of a ligand-binding niche within a human olfactory receptor by combining site-directed mutagenesis with dynamic homology modeling. Angew. Chem. Int. Ed. Engl. 51:1274–1278 10.1002/anie.201103980 [DOI] [PubMed] [Google Scholar]
- Gether U. 2000. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr. Rev. 21:90–113 10.1210/edrv.21.1.0390 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Larson S.D., Hefzi H., Marnoy Z., Cutforth T., Dokka K., Baldwin K.K. 2011. Sensory maps in the olfactory cortex defined by long-range viral tracing of single neurons. Nature. 472:217–220 10.1038/nature09945 [DOI] [PubMed] [Google Scholar]
- Grabenhorst F., Rolls E.T., Margot C., da Silva M.A., Velazco M.I. 2007. How pleasant and unpleasant stimuli combine in different brain regions: Odor mixtures. J. Neurosci. 27:13532–13540 10.1523/JNEUROSCI.3337-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosmaitre X., Vassalli A., Mombaerts P., Shepherd G.M., Ma M. 2006. Odorant responses of olfactory sensory neurons expressing the odorant receptor MOR23: A patch clamp analysis in gene-targeted mice. Proc. Natl. Acad. Sci. USA. 103:1970–1975 10.1073/pnas.0508491103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberly L.B., Price J.L. 1977. The axonal projection patterns of the mitral and tufted cells of the olfactory bulb in the rat. Brain Res. 129:152–157 10.1016/0006-8993(77)90978-7 [DOI] [PubMed] [Google Scholar]
- Haddad R., Khan R., Takahashi Y.K., Mori K., Harel D., Sobel N. 2008. A metric for odorant comparison. Nat. Methods. 5:425–429 10.1038/nmeth.1197 [DOI] [PubMed] [Google Scholar]
- Hall S.E., Floriano W.B., Vaidehi N., Goddard W.A., III 2004. Predicted 3-D structures for mouse I7 and rat I7 olfactory receptors and comparison of predicted odor recognition profiles with experiment. Chem. Senses. 29:595–616 10.1093/chemse/bjh063 [DOI] [PubMed] [Google Scholar]
- Hamana H., Shou-xin L., Breuils L., Hirono J., Sato T. 2010. Heterologous functional expression system for odorant receptors. J. Neurosci. Methods. 185:213–220 10.1016/j.jneumeth.2009.09.024 [DOI] [PubMed] [Google Scholar]
- Hummel P., Vaidehi N., Floriano W.B., Hall S.E., Goddard W.A., III 2005. Test of the Binding Threshold Hypothesis for olfactory receptors: Explanation of the differential binding of ketones to the mouse and human orthologs of olfactory receptor 912-93. Protein Sci. 14:703–710 10.1110/ps.041119705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K.M., Mori K. 2005. Spatial representation of hydrocarbon odorants in the ventrolateral zones of the rat olfactory bulb. J. Neurophysiol. 93:1007–1019 10.1152/jn.00873.2004 [DOI] [PubMed] [Google Scholar]
- Inglese J., Johnson R.L., Simeonov A., Xia M., Zheng W., Austin C.P., Auld D.S. 2007. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 3:466–479 10.1038/nchembio.2007.17 [DOI] [PubMed] [Google Scholar]
- Jacquier V., Pick H., Vogel H. 2006. Characterization of an extended receptive ligand repertoire of the human olfactory receptor OR17-40 comprising structurally related compounds. J. Neurochem. 97:537–544 10.1111/j.1471-4159.2006.03771.x [DOI] [PubMed] [Google Scholar]
- Jaeger S.R., McRae J.F., Bava C.M., Beresford M.K., Hunter D., Jia Y., Chheang S.L., Jin D., Peng M., Gamble J.C., et al. 2013. A Mendelian trait for olfactory sensitivity affects odor experience and food selection. Curr. Biol. 23:1601–1605 10.1016/j.cub.2013.07.030 [DOI] [PubMed] [Google Scholar]
- Kajiya K., Inaki K., Tanaka M., Haga T., Kataoka H., Touhara K. 2001. Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants. J. Neurosci. 21:6018–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaluza J.F., Breer H. 2000. Responsiveness of olfactory neurons to distinct aliphatic aldehydes. J. Exp. Biol. 203:927–933 [DOI] [PubMed] [Google Scholar]
- Katada S., Nakagawa T., Kataoka H., Touhara K. 2003. Odorant response assays for a heterologously expressed olfactory receptor. Biochem. Biophys. Res. Commun. 305:964–969 10.1016/S0006-291X(03)00863-5 [DOI] [PubMed] [Google Scholar]
- Katada S., Hirokawa T., Oka Y., Suwa M., Touhara K. 2005. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: Mapping the odorant-binding site. J. Neurosci. 25:1806–1815 10.1523/JNEUROSCI.4723-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Katada S., Touhara K. 2008. Amino acids involved in conformational dynamics and G protein coupling of an odorant receptor: targeting gain-of-function mutation. J. Neurochem. 107:1261–1270 10.1111/j.1471-4159.2008.05693.x [DOI] [PubMed] [Google Scholar]
- Keller A., Zhuang H., Chi Q., Vosshall L.B., Matsunami H. 2007. Genetic variation in a human odorant receptor alters odour perception. Nature. 449:468–472 10.1038/nature06162 [DOI] [PubMed] [Google Scholar]
- Kerr D.S., Von Dannecker L.E., Davalos M., Michaloski J.S., Malnic B. 2008. Ric-8B interacts with Gαolf and Gγ13 and co-localizes with Gαolf, Gβ1 and Gγ13 in the cilia of olfactory sensory neurons. Mol. Cell. Neurosci. 38:341–348 10.1016/j.mcn.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Khafizov K., Anselmi C., Menini A., Carloni P. 2007. Ligand specificity of odorant receptors. J. Mol. Model. 13:401–409 10.1007/s00894-006-0160-9 [DOI] [PubMed] [Google Scholar]
- Khan M., Vaes E., Mombaerts P. 2011. Regulation of the probability of mouse odorant receptor gene choice. Cell. 147:907–921 10.1016/j.cell.2011.09.049 [DOI] [PubMed] [Google Scholar]
- Kim P.S., Arvan P. 1998. Endocrinopathies in the family of endoplasmic reticulum (ER) storage diseases: Disorders of protein trafficking and the role of ER molecular chaperones. Endocr. Rev. 19:173–202 [DOI] [PubMed] [Google Scholar]
- Krautwurst D. 2008. Human olfactory receptor families and their odorants. Chem. Biodivers. 5:842–852 10.1002/cbdv.200890099 [DOI] [PubMed] [Google Scholar]
- Krautwurst D., Yau K.W., Reed R.R. 1998. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 95:917–926 10.1016/S0092-8674(00)81716-X [DOI] [PubMed] [Google Scholar]
- Kurland M.D., Newcomer M.B., Peterlin Z., Ryan K., Firestein S., Batista V.S. 2010. Discrimination of saturated aldehydes by the rat I7 olfactory receptor. Biochemistry. 49:6302–6304 10.1021/bi100976w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai P.C., Singer M.S., Crasto C.J. 2005. Structural activation pathways from dynamic olfactory receptor-odorant interactions. Chem. Senses. 30:781–792 10.1093/chemse/bji070 [DOI] [PubMed] [Google Scholar]
- Lai P.C., Bahl G., Gremigni M., Matarazzo V., Clot-Faybesse O., Ronin C., Crasto C.J. 2008. An olfactory receptor pseudogene whose function emerged in humans: a case study in the evolution of structure-function in GPCRs. J. Struct. Funct. Genomics. 9:29–40 10.1007/s10969-008-9043-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai P.C., Guida B., Shi J., Crasto C.J. 2014. Preferential binding of an odor within olfactory receptors: A precursor to receptor activation. Chem. Senses. 39:107–123 10.1093/chemse/bjt060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launay G., Téletchéa S., Wade F., Pajot-Augy E., Gibrat J.F., Sanz G. 2012. Automatic modeling of mammalian olfactory receptors and docking of odorants. Protein Eng. Des. Sel. 25:377–386 10.1093/protein/gzs037 [DOI] [PubMed] [Google Scholar]
- Lettvin J.Y., Maturana H.R., McCulloch W.S., Pitts W.H. 1959. What the frog’s eye tells the frog’s brain. Proceedings of the IRE. 47:1940–1951 10.1109/JRPROC.1959.287207 [DOI] [Google Scholar]
- Levasseur G., Persuy M.A., Grebert D., Remy J.J., Salesse R., Pajot-Augy E. 2003. Ligand-specific dose-response of heterologously expressed olfactory receptors. Eur. J. Biochem. 270:2905–2912 10.1046/j.1432-1033.2003.03672.x [DOI] [PubMed] [Google Scholar]
- Li J., Haddad R., Chen S., Santos V., Luetje C.W. 2012. A broadly tuned mouse odorant receptor that detects nitrotoluenes. J. Neurochem. 121:881–890 10.1111/j.1471-4159.2012.07740.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.R., Matsunami H. 2011. Activation state of the M3 muscarinic acetylcholine receptor modulates mammalian odorant receptor signaling. Sci. Signal. 4:ra1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D.Y., Zhang S.Z., Block E., Katz L.C. 2005. Encoding social signals in the mouse main olfactory bulb. Nature. 434:470–477 10.1038/nature03414 [DOI] [PubMed] [Google Scholar]
- Liu A.H., Zhang X., Stolovitzky G.A., Califano A., Firestein S.J. 2003. Motif-based construction of a functional map for mammalian olfactory receptors. Genomics. 81:443–456 10.1016/S0888-7543(03)00022-3 [DOI] [PubMed] [Google Scholar]
- Liu X., Su X., Wang F., Huang Z., Wang Q., Li Z., Zhang R., Wu L., Pan Y., Chen Y., et al. 2011. ODORactor: a web server for deciphering olfactory coding. Bioinformatics. 27:2302–2303 10.1093/bioinformatics/btr385 [DOI] [PubMed] [Google Scholar]
- Lu M., Echeverri F., Moyer B.D. 2003. Endoplasmic reticulum retention, degradation, and aggregation of olfactory G-protein coupled receptors. Traffic. 4:416–433 10.1034/j.1600-0854.2003.00097.x [DOI] [PubMed] [Google Scholar]
- Lu M., Staszewski L., Echeverri F., Xu H., Moyer B.D. 2004. Endoplasmic reticulum degradation impedes olfactory G-protein coupled receptor functional expression. BMC Cell Biol. 5:34 10.1186/1471-2121-5-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde K., Egelandsdal B., Skuterud E., Mainland J.D., Lea T., Hersleth M., Matsunami H. 2012. Genetic variation of an odorant receptor OR7D4 and sensory perception of cooked meat containing androstenone. PLoS ONE. 7:e35259 10.1371/journal.pone.0035259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainland J.D., Keller A., Li Y.R., Zhou T., Trimmer C., Snyder L.L., Moberly A.H., Adipietro K.A., Liu W.L., Zhuang H., et al. 2014. The missense of smell: functional variability in the human odorant receptor repertoire. Nat. Neurosci. 17:114–120 10.1038/nn.3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B. 2007. Searching for the ligands of odorant receptors. Mol. Neurobiol. 35:175–181 10.1007/s12035-007-0013-2 [DOI] [PubMed] [Google Scholar]
- Malnic B., Hirono J., Sato T., Buck L.B. 1999. Combinatorial receptor codes for odors. Cell. 96:713–723 10.1016/S0092-8674(00)80581-4 [DOI] [PubMed] [Google Scholar]
- Man O., Gilad Y., Lancet D. 2004. Prediction of the odorant binding site of olfactory receptor proteins by human-mouse comparisons. Protein Sci. 13:240–254 10.1110/ps.03296404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarazzo V., Clot-Faybesse O., Marcet B., Guiraudie-Capraz G., Atanasova B., Devauchelle G., Cerutti M., Etiévant P., Ronin C. 2005. Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chem. Senses. 30:195–207 10.1093/chemse/bji015 [DOI] [PubMed] [Google Scholar]
- McClintock T.S., Sammeta N. 2003. Trafficking prerogatives of olfactory receptors. Neuroreport. 14:1547–1552 10.1097/00001756-200308260-00001 [DOI] [PubMed] [Google Scholar]
- McRae J.F., Mainland J.D., Jaeger S.R., Adipietro K.A., Matsunami H., Newcomb R.D. 2012. Genetic variation in the odorant receptor OR2J3 is associated with the ability to detect the “grassy” smelling odor, cis-3-hexen-1-ol. Chem. Senses. 37:585–593 10.1093/chemse/bjs049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menashe I., Abaffy T., Hasin Y., Goshen S., Yahalom V., Luetje C.W., Lancet D. 2007. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 5:e284 10.1371/journal.pbio.0050284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minic J., Persuy M.A., Godel E., Aioun J., Connerton I., Salesse R., Pajot-Augy E. 2005. Functional expression of olfactory receptors in yeast and development of a bioassay for odorant screening. FEBS J. 272:524–537 10.1111/j.1742-4658.2004.04494.x [DOI] [PubMed] [Google Scholar]
- Mitsui K., Sakihama T., Takahashi K., Masuda K., Fukuda R., Hamana H., Sato T., Hamakubo T. 2012. Functional reconstitution of olfactory receptor complex on baculovirus. Chem. Senses. 37:837–847 10.1093/chemse/bjs067 [DOI] [PubMed] [Google Scholar]
- Mombaerts P. 2004. Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 5:263–278 10.1038/nrn1365 [DOI] [PubMed] [Google Scholar]
- Mombaerts P., Wang F., Dulac C., Chao S.K., Nemes A., Mendelsohn M., Edmondson J., Axel R. 1996. Visualizing an olfactory sensory map. Cell. 87:675–686 10.1016/S0092-8674(00)81387-2 [DOI] [PubMed] [Google Scholar]
- Nagashima A., Touhara K. 2010. Enzymatic conversion of odorants in nasal mucus affects olfactory glomerular activation patterns and odor perception. J. Neurosci. 30:16391–16398 10.1523/JNEUROSCI.2527-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nara K., Saraiva L.R., Ye X., Buck L.B. 2011. A large-scale analysis of odor coding in the olfactory epithelium. J. Neurosci. 31:9179–9191 10.1523/JNEUROSCI.1282-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y., Nakamura A., Watanabe H., Touhara K. 2004a. An odorant derivative as an antagonist for an olfactory receptor. Chem. Senses. 29:815–822 10.1093/chemse/bjh247 [DOI] [PubMed] [Google Scholar]
- Oka Y., Omura M., Kataoka H., Touhara K. 2004b. Olfactory receptor antagonism between odorants. EMBO J. 23:120–126 10.1038/sj.emboj.7600032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y., Katada S., Omura M., Suwa M., Yoshihara Y., Touhara K. 2006. Odorant receptor map in the mouse olfactory bulb: In vivo sensitivity and specificity of receptor-defined glomeruli. Neuron. 52:857–869 10.1016/j.neuron.2006.10.019 [DOI] [PubMed] [Google Scholar]
- Palczewski K., Kumasaka T., Hori T., Behnke C.A., Motoshima H., Fox B.A., Le Trong I., Teller D.C., Okada T., Stenkamp R.E., et al. 2000. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 289:739–745 10.1126/science.289.5480.739 [DOI] [PubMed] [Google Scholar]
- Peterlin Z., Li Y., Sun G., Shah R., Firestein S., Ryan K. 2008. The importance of odorant conformation to the binding and activation of a representative olfactory receptor. Chem. Biol. 15:1317–1327 10.1016/j.chembiol.2008.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S.M., Zheng C., Koos D.S., Feinstein P., Fraser S.E., Mombaerts P. 2001. Structure and emergence of specific olfactory glomeruli in the mouse. J. Neurosci. 21:9713–9723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisert J., Restrepo D. 2009. Molecular tuning of odorant receptors and its implication for odor signal processing. Chem. Senses. 34:535–545 10.1093/chemse/bjp028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repicky S.E., Luetje C.W. 2009. Molecular receptive range variation among mouse odorant receptors for aliphatic carboxylic acids. J. Neurochem. 109:193–202 10.1111/j.1471-4159.2009.05925.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler K.J., Sullivan S.L., Buck L.B. 1994. Information coding in the olfactory system: Evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 79:1245–1255 10.1016/0092-8674(94)90015-9 [DOI] [PubMed] [Google Scholar]
- Rubin B.D., Katz L.C. 1999. Optical imaging of odorant representations in the mammalian olfactory bulb. Neuron. 23:499–511 10.1016/S0896-6273(00)80803-X [DOI] [PubMed] [Google Scholar]
- Saito H., Kubota M., Roberts R.W., Chi Q., Matsunami H. 2004. RTP family members induce functional expression of mammalian odorant receptors. Cell. 119:679–691 10.1016/j.cell.2004.11.021 [DOI] [PubMed] [Google Scholar]
- Saito H., Chi Q., Zhuang H., Matsunami H., Mainland J.D. 2009. Odor coding by a mammalian receptor repertoire. Sci. Signal. 2:ra9 10.1126/scisignal.2000016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz G., Schlegel C., Pernollet J.C., Briand L. 2005. Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chem. Senses. 30:69–80 10.1093/chemse/bji002 [DOI] [PubMed] [Google Scholar]
- Sanz G., Thomas-Danguin T., Hamdani H., Le Poupon C., Briand L., Pernollet J.C., Guichard E., Tromelin A. 2008. Relationships between molecular structure and perceived odor quality of ligands for a human olfactory receptor. Chem. Senses. 33:639–653 10.1093/chemse/bjn032 [DOI] [PubMed] [Google Scholar]
- Sato T., Hirono J., Tonoike M., Takebayashi M. 1994. Tuning specificities to aliphatic odorants in mouse olfactory receptor neurons and their local distribution. J. Neurophysiol. 72:2980–2989 [DOI] [PubMed] [Google Scholar]
- Schmiedeberg K., Shirokova E., Weber H.P., Schilling B., Meyerhof W., Krautwurst D. 2007. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J. Struct. Biol. 159:400–412 10.1016/j.jsb.2007.04.013 [DOI] [PubMed] [Google Scholar]
- Shepard B.D., Natarajan N., Protzko R.J., Acres O.W., Pluznick J.L. 2013. A cleavable N-terminal signal peptide promotes widespread olfactory receptor surface expression in HEK293T cells. PLoS ONE. 8:e68758 10.1371/journal.pone.0068758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd G.M. 2006. Smell images and the flavour system in the human brain. Nature. 444:316–321 10.1038/nature05405 [DOI] [PubMed] [Google Scholar]
- Shirasu M., Yoshikawa K., Takai Y., Nakashima A., Takeuchi H., Sakano H., Touhara K. 2014. Olfactory receptor and neural pathway responsible for highly selective sensing of musk odors. Neuron. 81:165–178 10.1016/j.neuron.2013.10.021 [DOI] [PubMed] [Google Scholar]
- Shirokova E., Schmiedeberg K., Bedner P., Niessen H., Willecke K., Raguse J.D., Meyerhof W., Krautwurst D. 2005. Identification of specific ligands for orphan olfactory receptors. G protein-dependent agonism and antagonism of odorants. J. Biol. Chem. 280:11807–11815 10.1074/jbc.M411508200 [DOI] [PubMed] [Google Scholar]
- Singer M.S. 2000. Analysis of the molecular basis for octanal interactions in the expressed rat 17 olfactory receptor. Chem. Senses. 25:155–165 10.1093/chemse/25.2.155 [DOI] [PubMed] [Google Scholar]
- Singer M.S., Shepherd G.M. 1994. Molecular modeling of ligand-receptor interactions in the OR5 olfactory receptor. Neuroreport. 5:1297–1300 10.1097/00001756-199406020-00036 [DOI] [PubMed] [Google Scholar]
- Sivertsen B., Holliday N., Madsen A.N., Holst B. 2013. Functionally biased signalling properties of 7TM receptors—opportunities for drug development for the ghrelin receptor. Br. J. Pharmacol. 170:1349–1362 10.1111/bph.12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy E.R., Albeanu D.F., Fantana A.L., Murthy V.N., Meister M. 2009. Precision and diversity in an odor map on the olfactory bulb. Nat. Neurosci. 12:210–220 10.1038/nn.2262 [DOI] [PubMed] [Google Scholar]
- Spehr M., Gisselmann G., Poplawski A., Riffell J.A., Wetzel C.H., Zimmer R.K., Hatt H. 2003. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 299:2054–2058 10.1126/science.1080376 [DOI] [PubMed] [Google Scholar]
- Touhara K. 2007. Deorphanizing vertebrate olfactory receptors: recent advances in odorant-response assays. Neurochem. Int. 51:132–139 10.1016/j.neuint.2007.05.020 [DOI] [PubMed] [Google Scholar]
- Touhara K., Sengoku S., Inaki K., Tsuboi A., Hirono J., Sato T., Sakano H., Haga T. 1999. Functional identification and reconstitution of an odorant receptor in single olfactory neurons. Proc. Natl. Acad. Sci. USA. 96:4040–4045 10.1073/pnas.96.7.4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi A., Imai T., Kato H.K., Matsumoto H., Igarashi K.M., Suzuki M., Mori K., Sakano H. 2011. Two highly homologous mouse odorant receptors encoded by tandemly-linked MOR29A and MOR29B genes respond differently to phenyl ethers. Eur. J. Neurosci. 33:205–213 10.1111/j.1460-9568.2010.07495.x [DOI] [PubMed] [Google Scholar]
- Uchida N., Takahashi Y.K., Tanifuji M., Mori K. 2000. Odor maps in the mammalian olfactory bulb: domain organization and odorant structural features. Nat. Neurosci. 3:1035–1043 10.1038/79857 [DOI] [PubMed] [Google Scholar]
- van Anken E., Braakman I. 2005. Versatility of the endoplasmic reticulum protein folding factory. Crit. Rev. Biochem. Mol. Biol. 40:191–228 10.1080/10409230591008161 [DOI] [PubMed] [Google Scholar]
- Vassar R., Chao S.K., Sitcheran R., Nuñez J.M., Vosshall L.B., Axel R. 1994. Topographic organization of sensory projections to the olfactory bulb. Cell. 79:981–991 10.1016/0092-8674(94)90029-9 [DOI] [PubMed] [Google Scholar]
- Von Dannecker L.E., Mercadante A.F., Malnic B. 2005. Ric-8B, an olfactory putative GTP exchange factor, amplifies signal transduction through the olfactory-specific G-protein Gαolf. J. Neurosci. 25:3793–3800 10.1523/JNEUROSCI.4595-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Dannecker L.E., Mercadante A.F., Malnic B. 2006. Ric-8B promotes functional expression of odorant receptors. Proc. Natl. Acad. Sci. USA. 103:9310–9314 10.1073/pnas.0600697103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Carey A.F., Carlson J.R., Zwiebel L.J. 2010. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA. 107:4418–4423 10.1073/pnas.0913392107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel C.H., Oles M., Wellerdieck C., Kuczkowiak M., Gisselmann G., Hatt H. 1999. Specificity and sensitivity of a human olfactory receptor functionally expressed in human embryonic kidney 293 cells and Xenopus Laevis oocytes. J. Neurosci. 19:7426–7433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Pan Y., Chen G.Q., Matsunami H., Zhuang H. 2012. Receptor-transporting protein 1 short (RTP1S) mediates translocation and activation of odorant receptors by acting through multiple steps. J. Biol. Chem. 287:22287–22294 10.1074/jbc.M112.345884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Huang G., Dewan A., Feinstein P., Bozza T. 2012. Uncoupling stimulus specificity and glomerular position in the mouse olfactory system. Mol. Cell. Neurosci. 51:79–88 10.1016/j.mcn.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Pacifico R., Cawley D., Feinstein P., Bozza T. 2013. Ultrasensitive detection of amines by a trace amine-associated receptor. J. Neurosci. 33:3228–3239 10.1523/JNEUROSCI.4299-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang X., Firestein S. 2007. Comparative genomics of odorant and pheromone receptor genes in rodents. Genomics. 89:441–450 10.1016/j.ygeno.2007.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Ivic L., Otaki J.M., Hashimoto M., Mikoshiba K., Firestein S. 1998. Functional expression of a mammalian odorant receptor. Science. 279:237–242 10.1126/science.279.5348.237 [DOI] [PubMed] [Google Scholar]
- Zhuang H., Matsunami H. 2007. Synergism of accessory factors in functional expression of mammalian odorant receptors. J. Biol. Chem. 282:15284–15293 10.1074/jbc.M700386200 [DOI] [PubMed] [Google Scholar]