Abstract
Key intracytosolic pattern recognition receptors of innate immunity against bacterial infections are nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs). We elucidated the NOD1 and NOD2-mediated activation of human eosinophils, the principal effector cells for allergic inflammation, upon interacting with human bronchial epithelial BEAS-2B cells in allergic asthma. Eosinophils constitutively expressed NOD1,2 but exhibited nonsignificant responses to release chemokines upon the stimulation by NOD1 ligand γ-D-glutamyl-meso-diaminopimelic acid (iE-DAP) and NOD2 ligand muramyl dipeptide (MDP). However, iE-DAP and MDP could significantly upregulate cell surface expression of CD18 and intercellular adhesion molecule (ICAM)-1 on eosinophils and ICAM-1 on BEAS-2B cells, as well as induce chemokines CCL2 and CXCL8 release in the coculture system (all P<0.05). Both eosinophils and BEAS-2B cells were the main source for CXCL8 and CCL2 release in the coculture system upon iE-DAP or MDP stimulation. Direct interaction between eosinophils and BEAS-2B cells is responsible for CCL2 release, and soluble mediators are implicated in CXCL8 release. ERK and NF-κB play regulatory roles for the expression of adhesion molecules and chemokines in coculture. Treatment with NOD1,2 ligand could induce the subepithelial fibrosis and significantly enhance the serum concentration of total IgE, chemokine CCL5 for eosinophils and T helper type 2 (Th2) cells and asthma Th2 cytokine IL-13 in bronchoalveolar lavage fluid of ovalbumin-sensitized allergic asthmatic mice (all P<0.05). This study provides further evidence of bacterial infection-mediated activation of NOD1,2 in triggering allergic asthma via the activation of eosinophils interacting with bronchial epithelial cells at inflammatory airway.
Keywords: allergy, bronchial epithelial cells, chemokines, eosinophils, signal transduction
Introduction
Eosinophils are the most important inflammatory effector cells infiltrating and accumulating at the site of allergic inflammation, e.g. the airway submucosa, through the adhesion molecules.1 Activated eosinophils release cytotoxic and inflammatory molecules such as major basic protein, eosinophil peroxidase, eosinophilic cationic protein, superoxides, cysteinyl leukotrienes, cytokines and chemokines.1 These molecules together cause epithelial, epidermal damage, bronchoconstriction and mucus secretion, and eventually result in the manifestation of allergic asthma.1,2 Eosinophils are also capable of producing and releasing a variety of proinflammatory cytokines, together with chemokines including CXCL8 and CCL2, which exacerbate allergic inflammation.1 We and other groups have found that the interaction of eosinophils and epithelial cells at the inflammatory sites can result in the elevated induction of inflammatory cytokines, chemokines and eosinophilic cationic protein release.3,4,5,6 Therefore, the contact of eosinophils with bronchial epithelial cells exhibits significant implications in the immunopathology of airway inflammatory diseases such as asthma.3,4,5,6
Moderate pulmonary bacterial infection, such as Chlamydia pneumoniae, can provoke allergic inflammation upon the activation of innate immunity-related dendritic cells by inhaled allergen.7,8,9 The innate immune system recognizes microorganisms through pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) and RIG-I-like receptors which detect conserved microbial components called pathogen-associated molecular patterns. The most widely expressed and well-characterized TLRs recognize microbes on the cell surface and in endosomes; whereas NLRs and RIG-I-like receptors detect bacterial and viral components in the cytosol.10 Among these PRRs, members of the intracellular NLR family including NOD1 and NOD2 have been identified as key mediators in inflammatory and immune responses, by recognizing intracellular microbes and commensal organisms.11,12,13 NOD2 can also function as a cytoplasmic viral PRR by producing of interferon-β.14 Regarding the antibacterial innate immunity, NOD1 and NOD2, can sense the cytosolic presence of the peptidoglycan fragment γ-D-glutamyl-meso-diaminopimelic acid (iE-DAP) in all Gram-negative and certain Gram-positive bacteria, and muramyl dipeptide (MDP) in almost all bacteria, respectively.10,11,15 Bacterial sensor NOD2 can trigger a potent antigen-specific immune response with a T helper type 2 (Th2)-type polarization cytokine profile.16 The crucial pathological roles of NOD1,2 in immune disorders have been proved from the linkage of NOD1,2 polymorphisms with chronic inflammatory Crohn's disease, atopic dermatitis, atopic eczema and allergic asthma.17,18,19 In fact, NOD1-mediated recognition of bacterial products in the skin or at mucosal interfaces may regulate Th2 polarization and IgE concentrations.18 NOD1,2-mediated eosinophil activation has recently been reported.20 To further evaluate the cellular mechanisms of bacterial infection-induced innate immunity-mediated allergic asthma, we further elucidate the in vitro and in vivo immunological effects and the underlying intracellular regulatory mechanisms of NOD1,2-mediated activation of eosinophils upon the interaction with bronchial epithelial cells.
Materials and methods
Materials
NOD1 ligand iE-DAP or its inactive negative control peptide iE-Lys and the NOD2 ligand N-acetylmuramyl-ℓ-alanyl-𝒹-isoglutamine (muramyl dipeptide/MDP) or its inactive negative control MDP D–D isomer were from Invivogen Corp., San Diego, CA, USA.
Endotoxin-free solutions
Cell culture medium was purchased from Life Technologies, Carlsbad, CA, USA, free of detectable lipopolysaccharide (<0.1 EU/ml). All other solutions were prepared using pyrogen-free water and sterile polypropylene plastic ware. No solution contained detectable lipopolysaccharide, as determined by the Limulus amoebocyte lysate assay (sensitivity limit 12 pg/ml; Biowhittaker Inc., Walkersville, MD, USA).
Isolation of human blood eosinophils from buffy coat and eosinophil culture
Fresh human buffy coat obtained from the healthy volunteers of Hong Kong Red Cross Blood Transfusion Service was diluted 1∶2 with phosphate-buffered saline (PBS) at 4 °C and centrifuged using an isotonic Percoll solution (density 1.082 g/ml; GE Healthcare, Piscataway, NJ, USA) for 30 min at 1000g. The eosinophil-rich granulocyte fraction was collected and washed twice with cold PBS containing 2% fetal bovine serum (FBS) (Life Technologies). The cells were then incubated with anti-CD16 magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany) at 4 °C for 45 min and CD16-positive cells were depleted by passing through an LS+ column (Miltenyi Biotec) within a magnetic field. With this preparation, the drop-through fraction contained eosinophils with a purity of at least 99% as assessed by Hemacolor rapid blood smear stain (E Merck Diagnostica, Darmstadt, Germany). The isolated eosinophils were cultured in RPMI-1640 medium (Life Technologies) supplemented with 10% FBS and 20 mM Hepes (Life Technologies). The protocol using human eosinophils purified from human buffy coat was approved by the Clinical Research Ethics Committee of The Chinese University of Hong Kong-New Territories East Cluster Hospitals with written consent from all healthy volunteers of Hong Kong Red Cross Blood Transfusion Service.
Coculture of eosinophils and bronchial epithelial BEAS-2B cells
The human bronchial epithelial cell line (BEAS-2B) transformed by adenovirus 12-SV40 virus hybrid (Ad12SV40) was obtained from the American Type Culture Collection. BEAS-2B cells were grown in Dulbecco's modified Eagle's medium nutrient mixture F12 (DMEM/F12; Life Technologies) with 10% FBS in 6- or 24-well cell culture plates at 37 °C with 5% CO2–95% humidified air until confluence to cell monolayer, and the medium was replaced with RPMI-1640 medium containing 10% FBS (Life Technologies) with or without eosinophils. BEAS-2B cells maintain the characteristics of primary human bronchial epithelial cells including the expression of intercellular adhesion molecule (ICAM)-1 and chemokine CXCL8.21,22,23,24
Real-time quantitative PCR of NOD1 and NOD2 of eosinophils
Briefly, total RNA of eosinophils (1×106) was extracted using TRI-Reagent (Molecular Research Center Inc., Cincinnati, OH, USA) and treated with DNase I to exclude genomic DNA contamination (Ambion Inc., Austin, TX, USA). Extracted RNA was then reversely transcribed into first-strand complementary DNA using first-strand cDNA synthesis kit (Applied Biosystems, Foster City, CA, USA). The quantitative expression of human NOD1 and NOD2 of eosinophils was performed using FastStart Universal SYBR Green Master (ROX) (Roche Applied Science, Indianapolis, IN, USA) with reversed transcribed product, AmpliTaq Gold DNA polymerase (Applied Biosystems) and primers of NOD1, forward 5′-TTCCCTGCTCACTCAGAGCAAAG-3′, and reverse 5′-TAGCACAGCACGAACTTGGAGTCA-3′, NOD2, forward 5′-CGAGGCATCTGCAAGCTCATTGAA-3′, and reverse 5′-GTGCACAGCCGTCAGTCAATTTGT-3′ and GAPDH, forward 5′-ATGGGGAAGGTGAAGGTCG-3′, and reverse 5′- GGGGTCATTGATGGCAACAATA-3′. The relative gene expression was calculated using 2−ΔCt (Ct, NLR-Ct, GAPDH).
Coculture of fixed eosinophils and BEAS-2B cells
Confluent BEAS-2B cells or eosinophils were treated with 1% paraformaldehyde in PBS in ice for 1 h to prevent the release of mediators from cells while preserving the cell membrane integrity to maintain intercellular interaction. After fixation, cells were washed at least 10 times with PBS containing 2% FBS, and fixed or unfixed eosinophils and BEAS-2B cells were cocultured in RPMI-1640 supplemented with 10% FBS.4,5,6
Coculture of eosinophils and BEAS-2B cells in the presence of transwell inserts
To prevent direct interaction between eosinophils and BEAS-2B cells in the coculture, transwell inserts (pore size: 0.4 µM) (BD Biosciences Corp., San Jose, CA, USA) were used to separate these two cells into two compartments. Confluent BEAS-2B cells and eosinophils were cultured together in the presence of transwell inserts, in which eosinophils and BEAS-2B cells were placed in the upper and lower compartment, respectively.4,5,6
Quantitative analysis of cytokines and chemokines
Concentrations of human chemokines CXCL8 and CCL2 in culture supernatant were quantitated with human chemokine cytometric bead array (CBA) kit (BD Pharmingen Corp., San Diego, CA, USA) using four-color FACSCalibur flow cytometer (BD Biosciences Corp.). Murine CCL5 and IL-13 in bronchoalveolar lavage fluid (BALF) was quantitated using mouse cytokine Bio-Plex assay reagent with Bio-Plex 200 suspension array system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).6
Quantitative analysis of total IgE
Murine serum concentrations of total IgE were determined with mouse IgE ELISA kit (BioLegend Corp., San Diego, CA, USA).
Immunofluorescence staining and flow cytometry
To determine the expression of adhesion molecules on the cell surface, cells were washed and resuspended with cold PBS after preceding treatments. After blocking with 2% human pooled serum for 20 min at 4 °C and washing with cold PBS, cells were incubated with mouse antihuman CD18-FITC conjugate (BD Pharmingen Corp.), mouse antihuman ICAM-1/L-selectin (CD62L)-FITC-conjugate (R & D Systems, Minneapolis, MN, USA) or FITC-conjugated mouse IgG1 isotypic control antibody (BD Pharmingen Corp.) for 30 min at 4 °C in the dark. After washing, cells were resuspended in 1% paraformaldehyde as fixative and subjected to flow cytometric analysis.
To determine the intracellular expression of NOD1, NOD2 and phosphorylated signaling molecules, cells were fixed with 4% paraformaldehyde for 10 min at 37 °C after preceding treatments. After centrifugation, cells were permeabilized in ice-cold methanol for 30 min and then stained with mouse antihuman NOD1 antibody (Cell Signaling Technology Inc., Bervely, MA, USA) or mouse antihuman NOD2 antibody (BioLegend Corp.) together with FITC-conjugated goat antimouse IgG (Life Technologies), or mouse FITC-conjugated antiphosphorylated ERK1/2, mouse FITC-conjugated antiphosphorylated IκB-α (Cell Signaling Technology Inc.) or corresponding mouse IgG1 isotypic antibody (BD Pharmingen Corp.) for 30 min at 4 °C in dark. Cells were then washed, resuspended and subjected to analysis. Expression of intracellular NOD1,2 and phosphorylated signaling molecules of 10 000 viable cells was analyzed by flow cytometry (FACSCalibur) as arithmetic mean of mean fluorescence intensity (MFI) plus standard deviation (s.d.) of three independent experiments with the subtraction with appropriate isotypic control.6
Allergic asthmatic mice model
Inbred female BALB/c mice (8 weeks old and 25 g weight) were bred under specific pathogen-free conditions and maintained at laboratory animal services center, The Chinese University of Hong Kong. Allergic airway inflammation was induced using a widely used allergen ovalbumin sensitization and challenge method.25 Briefly, mice were sensitized with intraperitoneal ovalbumin (20 µg) plus aluminum hydroxide powder [2 mg Al (OH)3 in 100 µl PBS] on days 0 and 14, and challenged twice with intranasal 1% ovalbumin aerosol on days 28, 29 and 30. Mice were treated with intravenous (iv) injection with PBS solution or NOD1, 2 ligands (200 µg) on days 29 and 30 (1 h before the first ovalbumin challenge). Mice were then sacrificed on day 33 to collect BALF and serum. The concentration of serum total IgE, and BALF CCL5 and IL-13 was analyzed using mouse IgE ELISA kits and mouse cytokine Bio-Plex assay reagent, respectively. Excised lungs were fixed 48 h at 4 °C with fixative (10% formalin). Fixed lung samples were rinsed in PBS, dehydrated and embedded in paraffin. Sections (4 µm) were stained with periodic acid Schiff to evaluate the general morphology and the presence of mucin-producing goblet cells. The animal protocol was approved by the Animal Experimentation Ethics Committee of The Chinese University of Hong Kong.
Statistical analysis
All data were expressed as mean±s.d. from three independent experiments. The statistical significance of differences was determined by one-way analysis of variance or unpaired t-test. Any difference with P values <0.05 was considered significant. When analysis of variance or unpaired t-test indicated a significant difference, Bonferroni post hoc test was then used to assess the difference between groups. All analysis was performed using the Statistical Package for the Social Sciences (SPSS) statistical software for Windows, version 16.0 (SPSS Inc., Chicago, IL, USA).
Results
Expression of NOD1 and NOD2 in eosinophils
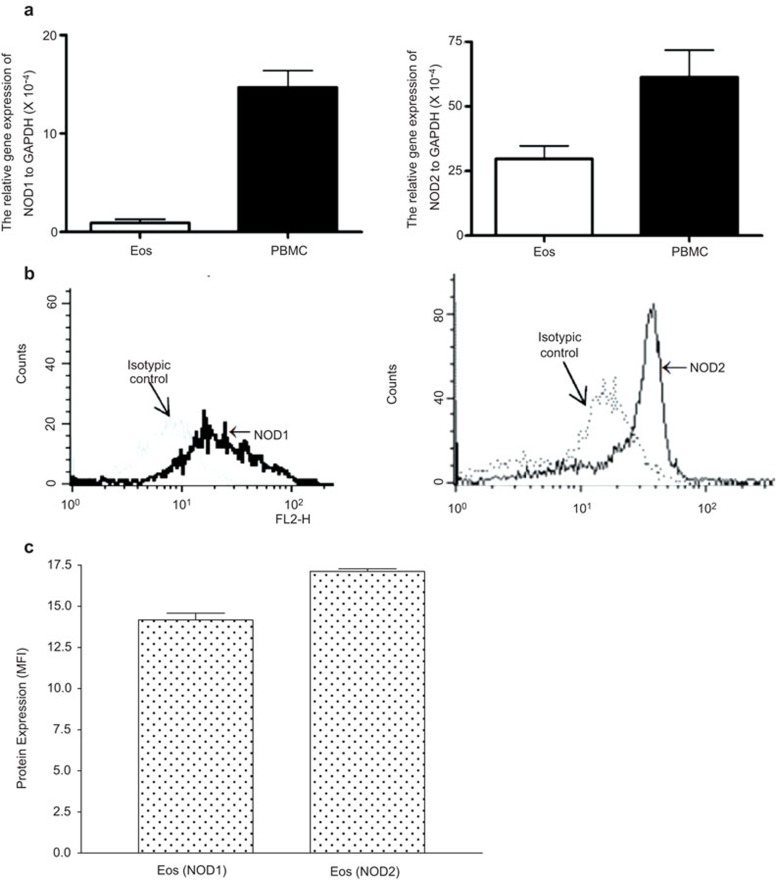
BEAS-2B cells have previously been shown to express NOD1 and NOD2 proteins either constitutively or by induction.26,27 We confirmed the expressions of gene (Figure 1A) and protein (Figure 1B and C) of NOD1,2 in eosinophils using real-time quantitative PCR and flow cytometry which are in concordance with recent published results.20 Human peripheral blood mononuclear cells (PBMCs) were used as positive control of gene expression.
Figure 1.
QPCR and flow cytometry of intracellular expression of NLR components NOD1 and NOD2 in EOS. (a) Equal amount of extracted total RNA from eosinophils and PBMC (5×105 cells) were used for qPCR. The relative gene expression of NOD1 and NOD2 was calculated using 2−ΔCt (Ct, NLR-Ct, GAPDH). (b) Intracellular expression of NOD1,2 in eosinophils (5×105 cells) was determined by flow cytometry. Triplicate experiments were performed and representative histograms are shown. (c) Quantitative results of flow cytometric analysis of intracellular expression of NOD1,2 in eosinophils are presented with arithmetic mean plus s.d. of MFI subtracting appropriate isotypic control of triplicate experiments. EOS, eosinophils; MFI, mean fluorescence intensity; NLR, NOD-like receptor; NOD, nucleotide-binding oligomerization domain; PBMC, peripheral blood mononuclear cell; qPCR, quantitative PCR; s.d., standard deviation.
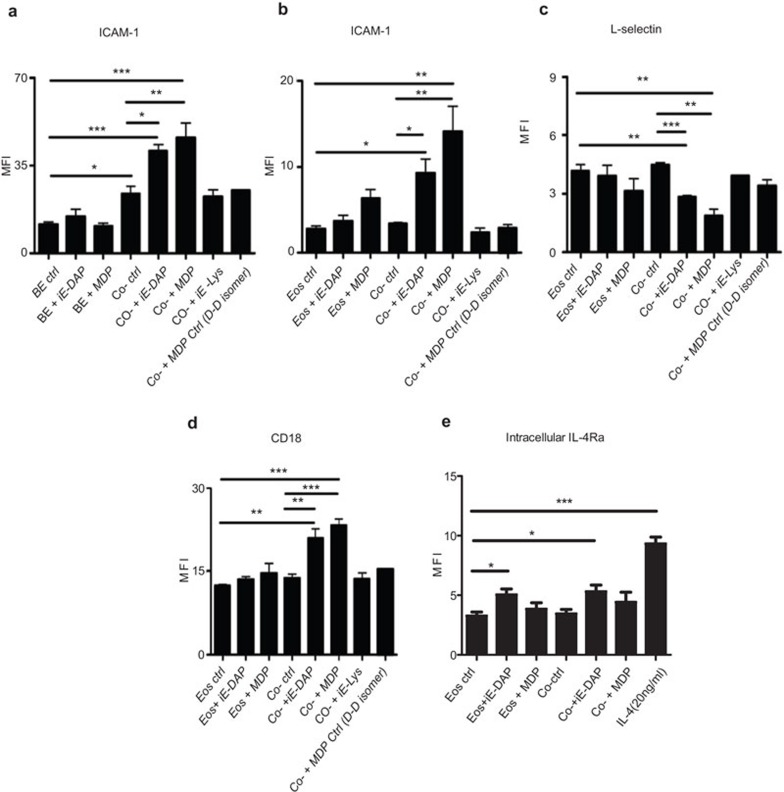
Effects of NOD1 and NOD2 ligands on the surface expression of adhesion molecules and intracellular IL-4 receptor upon the interaction of human eosinophils and BEAS-2B cells
NOD1 ligand iE-DAP or NOD2 ligand MDP (10 µg/ml) stimulation had nonsignificant effects on the surface expression of adhesion molecules on eosinophils or BEAS-2B cells alone (Figure 2A–D), even with high dose up to 100 µg/ml (data not shown). When cocultured together without ligand stimulation, the expression of ICAM-1 on BEAS-2B cells was significantly enhanced, while no significant changes of the expression of CD18, ICAM-1 and L-selectin on eosinophils were observed (Figure 2A–D). However, upon iE-DAP or MDP stimulation in the coculture system, the expression of CD18 and ICAM-1 on eosinophils was significantly upregulated, while L-selectin on eosinophils was significantly downregulated (Figure 2B–D). Moreover, the expression of ICAM-1 on BEAS-2B cells was also further augmented in the coculture system with iE-DAP or MDP stimulation (Figure 2A). The NOD1 ligand iE-DAP-negative control iE-Lys and NOD2 ligand MDP-negative control MDP (D–D isomer) showed no significant effects on the expression of adhesion molecules upon coculture of eosinophils and BEAS-2B cells (Figure 2A–D). In addition, iE-DAP but not coculture could significantly upregulate the intracellular expression of IL-4 receptor-α (ΙL-4Ra) in eosinophils (Figure 2E). We also observed that eosinophils exhibited strong cell surface expression of CCR3 and weak expression of IL-4Ra but NOD1,2 ligands and coculture with BEAS-2B cells did not exhibit any significant effect on the cell surface expression of CCR3 and IL-4Ra on eosinophils (all P>0.05) (data not shown).
Figure 2.
Effect of NOD1 ligand iE-DAP and NOD2 ligand MDP on cell surface expression of adhesion molecules and intracellular expression of IL-4Ra upon the interaction of human eosinophils and BEAS-2B cells. Eosinophils (5×105 cells) and confluent BEAS-2B cells (8×104 cells) were cultured either together or separately with or without iE-DAP, MDP, negative control iE-Lys, MDP Ctrl (D–D isomer) (10 µg/ml) or positive control of IL-4 (20 ng/ml) for 16 h. Surface expression of (a) ICAM-1 on BEAS-2B cells and (b) ICAM-1, (c) L-selectin, (d) CD18 on eosinophils and (e) intracellular IL-4Ra in eosinophils was analyzed by flow cytometry and shown as MFI. MFIs are normalized by subtracting appropriate isotypic control and shown as arithmetic mean plus s.d. of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 when compared between groups denoted by horizontal lines. Ctrl: control; BE: BEAS-2B cells; Eos: eosinophils; Co-: coculture of eosinophils and BEAS-2B cells. ICAM, intercellular adhesion molecule; iE-DAP, γ-D-glutamyl-meso-diaminopimelic acid; IL-4Ra, IL-4 receptor-α MDP, muramyl dipeptide; MFI, mean fluorescence intensity; NOD, nucleotide-binding oligomerization domain; s.d., standard deviation.
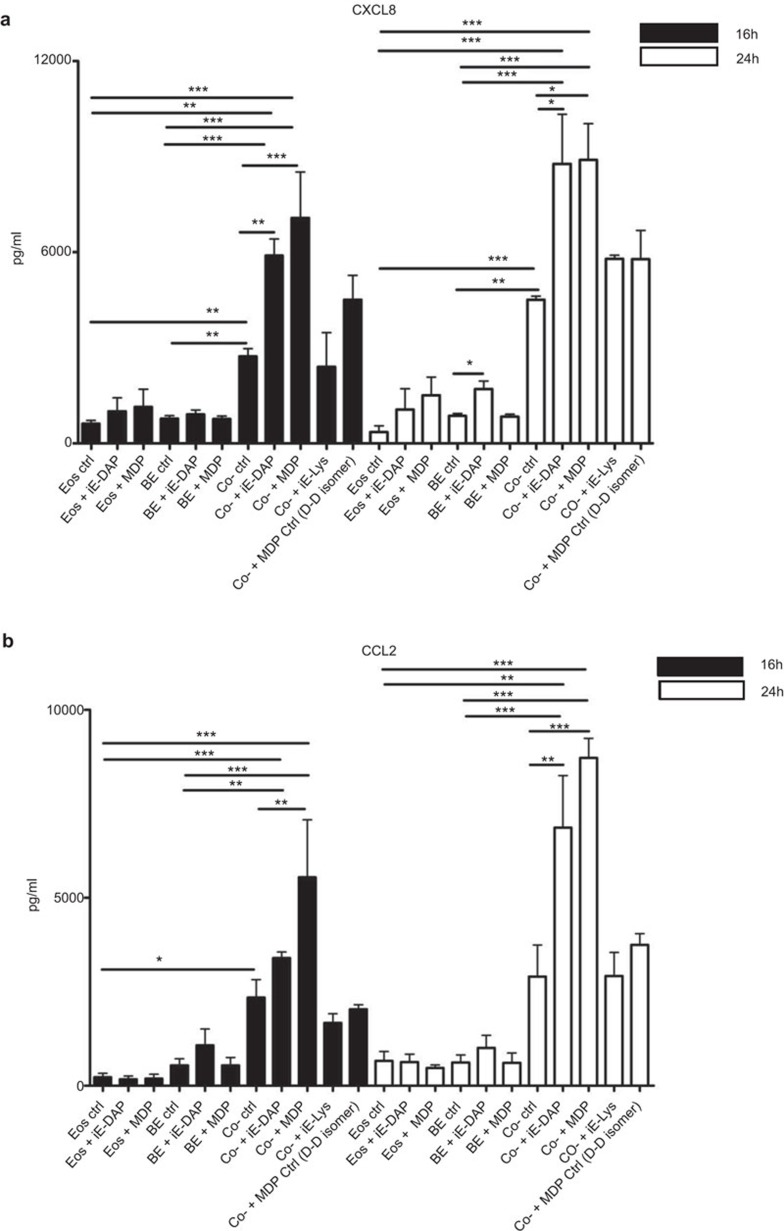
Effects of NOD1 and NOD2 ligands on the cytokine and chemokine release upon the interaction of human eosinophils and BEAS-2B cells
NOD1 ligand iE-DAP (10 µg/ml) alone could significantly induce the release of CXCL8 from BEAS-2B cells at 24 h (Figure 3A), which was consistent with other report,28 while NOD2 ligand MDP (10 µg/ml) alone showed little effects on CXCL8 induction from BEAS-2B cells (Figure 3A). In addition, iE-DAP or MDP (10 µg/ml) alone exhibited no prominent effects on CCL2 and CXCL8 release from human eosinophils (Figure 3A and B). Upon coculture, the levels of CCL2 and CXCL8 were found to be markedly elevated than those of eosinophils alone or BEAS-2B cells alone (Figure 3A and B). Besides, concentrations of CCL2 and CXCL8 were found to be significantly enhanced in the coculture of eosinophils and BEAS-2B cells under the stimulation of iE-DAP or MDP (10 µg/ml). The release of CCL2 and CXCL8 of 24 h culture was higher than those of 16 h (Figure 3A and B). The NOD1 ligand-negative control iE-Lys and NOD2 ligand-negative control MDP (D–D isomer) showed nonsignificant effects on the expression of CCL2 and CXCL8 upon coculture of eosinophils and BEAS-2B cells (Figure 3A and B). We found that NOD1,2 ligands did not exhibit any significant effect on the induction of other chemokines such as CCL5, CXCL9 and CXCL10 from coculture (all P>0.05, data not shown); therefore, we further elucidated the regulatory mechanisms on the induction of CCL2 and CXCL8 in subsequent in vitro experiments, and applied asthmatic mice experiment to evaluate the in vivo effects of NOD1,2 ligands on eosinophils.
Figure 3.
Effect of NOD1 ligand iE-DAP and NOD2 ligand MDP on the release of CXCL8 and CCL2 upon the interaction of human eosinophils and BEAS-2B cells. Eosinophils (5×105 cells) and confluent BEAS-2B (8×104 cells) were cultured either together or separately with or without iE-DAP (10 µg/ml), MDP (10 µg/ml), and negative control iE-Lys (10 µg/ml) and MDP Ctrl (D–D isomer, 10 µg/ml) for 16 and 24 h. Cell-free culture supernatant was collected, and CXCL8 and CCL2 released into the supernatant were measured using CBA. Results are expressed as arithmetic mean plus s.d. of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 when compared between groups denoted by horizontal lines. Ctrl: control; BE: BEAS-2B cells; Eos: eoisnophils; Co-: coculture of eosinophils and BEAS-2B cells. CBA, cytometric bead array; iE-DAP, γ-D-glutamyl-meso-diaminopimelic acid; MDP, muramyl dipeptide; NOD, nucleotide-binding oligomerization domain; s.d., standard deviation.
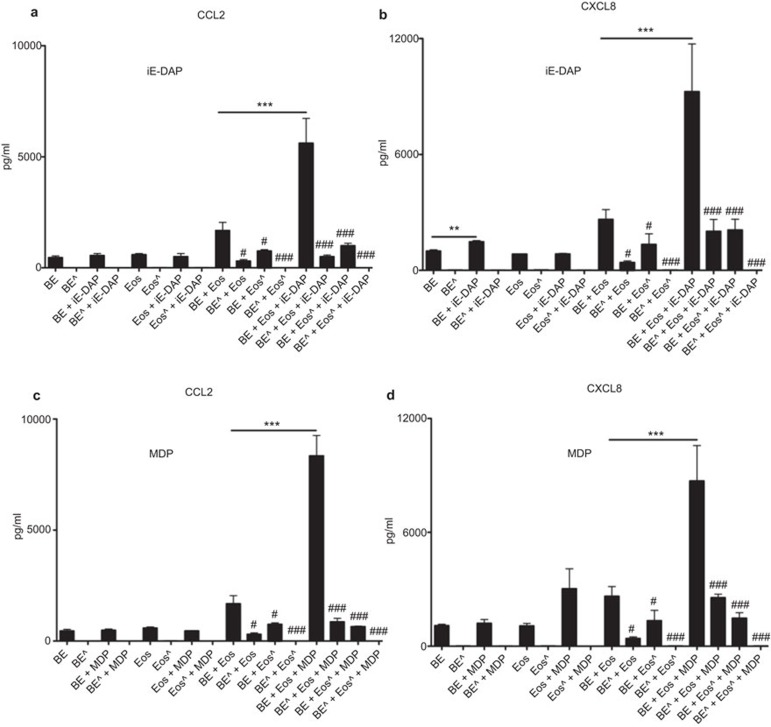
Source of CCL2 and CXCL8 in coculture of human eosinophils and BEAS-2B cells upon NOD1 and NOD2 ligand stimulation
We have previously shown that both eosinophils and BEAS-2B cells can produce CCL2 and CXCL8.4,5,29 To further investigate the source(s) of chemokines CCL2 and CXCL8 released in the coculture system, 1% paraformaldehyde was used to fix eosinophils or BEAS-2B cells to prevent the secretion of cytokines and chemokines, while preserving the cell membrane integrity to maintain the direct intercellular interaction between eosinophils and BEAS-2B cells via the surface adhesion molecules.30 We compared CCL2 and CXCL8 release in the coculture of normal intact cells with the cells fixed with 1% paraformaldehyde. As shown in Figure 4, the coculture of fixed eosinophils and unfixed BEAS-2B cells, and fixed BEAS-2B cells and unfixed eosinophils could almost abrogate the release of chemokines CCL2 and CXCL8 in coculture system with or without NOD1 ligand iE-DAP or NOD2 ligand MDP stimulation. These results suggest that both eosinophils and BEAS-2B cells are the sources of CCL2 and CXCL8 in the coculture system.
Figure 4.
Source of chemokine CCL2 and CXCL8 in coculture of human eosinophils and BEAS-2B cells upon NOD1 and NOD2 ligand stimulation. Eosinophils (5×105 cells) and BEAS-2B (8×104 cells) were treated with or without 1% paraformaldehyde for 45 min on ice prior to treatment with or without (a, b) iE-DAP (10 µg/ml) or (c, d) MDP (10 µg/ml) for 24 h. Cell-free culture supernatant was collected, and CCL2 and CXCL8 released into the supernatant were quantified using CBA. Results are expressed as arithmetic mean plus s.d. of three independent experiments. Eos: Eosinophils; Eoŝ: Fixed eosinophils; BE: BEAS-2B cells; BE ^: Fixed BEAS-2B cells. ***P<0.001 when compared between groups denoted by the horizontal lines. #P<0.05 and ###P<0.001 when compared with corresponding unfixed groups. CBA, cytometric bead array; iE-DAP, γ-D-glutamyl-meso-diaminopimelic acid; MDP, muramyl dipeptide; NOD, nucleotide-binding oligomerization domain; s.d., standard deviation.
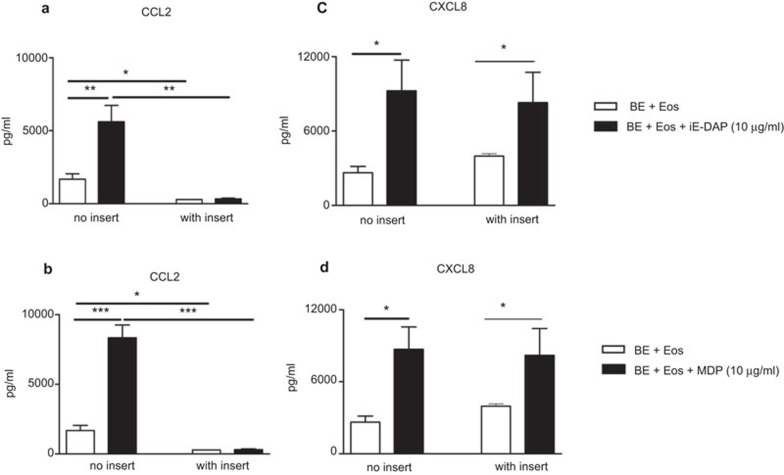
Effects of transwell insert on the induction of CCL2 and CXCL8 release in coculture of human eosinophils and BEAS-2B cells upon NOD1 and NOD2 ligand stimulation
To explore whether the direct interaction was essential for the induction of CCL2 and CXCL8 release in the coculture of eosinophils and BEAS-2B cells upon NOD1 and NOD2 ligands stimulation, transwell insert with pore size of 0.4 µm was used to separate eosinophils and BEAS-2B cells into two compartments in the coculture system. Intercellular communication through soluble mediators, such as cytokines and chemokines, was allowed in this transwell coculture system.
Without any stimulation, induction of CCL2 release in coculture of eosinophils and BEAS-2B cells was totally abolished in the presence of transwell insert, suggesting that CCL2 release in the coculture system may depend on the direct interaction between eosinophils and BEAS-2B cells (Figure 5A and B). Upon iE-DAP or MDP treatment, CCL2 release from the coculture system was also abrogated using the transwell insert (Figure 5A and B). However, the release of CXCL8 in the coculture of eosinophils and BEAS-2B cells with or without iE-DAP or MDP treatment could not be suppressed by the transwell inserts (Figure 5C and D), suggesting that soluble mediators may be used for intercellular communication and induction of CXCL8 release.
Figure 5.
Effects of the transwell insert on the induction of (a, b) CCL2 and (c, d) CXCL8 release in coculture of eosinophils and bronchial epithelial cells upon NOD1 and NOD2 ligand stimulation. Eosinophils (5×105 cells) and bronchial epithelial cells (8×104 cells) were cultured together with (a, c) iE-DAP or (b, dD) MDP (10 µg/ml) for 24 h in the presence or absence of the transwell insert. Cell-free culture supernatant was collected and chemokine released into the supernatant was quantified using CBA. Results are expressed as arithmetic mean plus s.d. of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 when compared between groups denoted by the horizontal lines. BE: BEAS-2B cells; Eos: eoisnophils. CBA, cytometric bead array; iE-DAP, γ-D-glutamyl-meso-diaminopimelic acid; MDP, muramyl dipeptide; NOD, nucleotide-binding oligomerization domain; s.d., standard deviation.
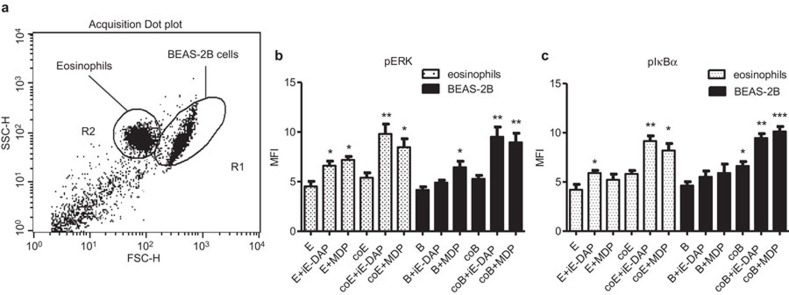
Intracellular signaling pathways involved in the interaction of eosinophils and BEAS-2B cells under NOD1,2 ligand stimulation
To investigate the underlying signaling mechanisms, intracellular staining by quantitative flow cytometry was employed. After fixation and permeabilization, eosinophils and BEAS-2B cells formed discrete populations and were gated on the basis of their forward and side light scatter (Figure 6A). The gated populations in R1 and R2 regions were found to be CCR3-negative and CCR3-positive, respectively, thereby confirming that the R1 and R2 regions were BEAS-2B cells and eosinophils, respectively (data not shown). Figure 6B and C shows that intracellular ERK and IκB-α were differentially phosphorylated in eosinophils and BEAS-2B cells (all P<0.05) upon the stimulation by NOD1 ligand iE-DAP and NOD2 ligand MDP on individual cells alone or coculture.
Figure 6.
Activation of ERK and NF-κB in coculture of eosinophils and BEAS-2B cells upon NOD1 and NOD2 ligand stimulation. Eosinophils (5×105 cells) and confluent BEAS-2B cells (8×104 cells) were cultured either together or separately with or without iE-DAP or MDP (10 µg/ml) stimulation for 10 min. (a) After fixation and permeabilization, eosinophils and BEAS-2B cells formed very discrete populations and were gated based on forward and side light scatter in dot plot using flow cytometry. The intracellular contents of (b) phosphorylated ERK and (c) phosphorylated IκB in permeabilized eosinophils and BEAS-2B cells were measured by intracellular staining using flow cytometry. Results are shown in MFI subtracting corresponding isotypic control and are expressed as the arithmetic mean plus s.d. of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 when compared with medium control of the same cell type. E: eosinophils only; coE: eosinophils in coculture; B: BEAS-2B cells only; coB: BEAS-2B cells in coculture. iE-DAP, γ-D-glutamyl-meso-diaminopimelic acid; MDP, muramyl dipeptide; MFI, mean fluorescence intensity; NOD, nucleotide-binding oligomerization domain; s.d., standard deviation.
In vivo effect of NOD1,2 ligands on IgE and chemokine production in serum and BALF in allergic asthmatic mice
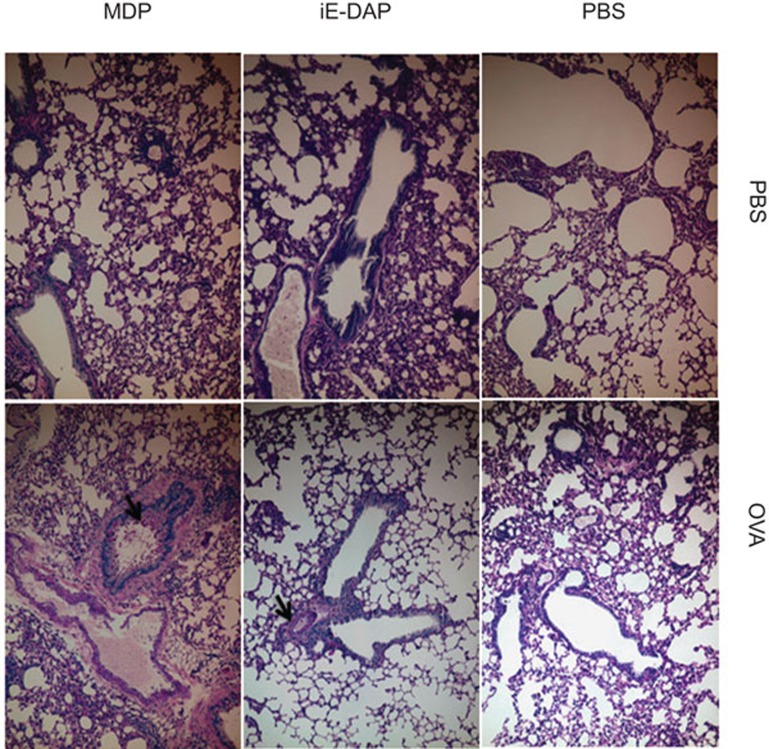
As shown in Figure 7, histological analysis of the lungs revealed that the ovalbumin could initiate the thickness and the fibrotic pattern of the bronchial tissue while iE-DAP and MDP could further increase the number of mucin-secreting goblet cells, the thickness and fibrosis of the bronchial subepithelial tissue (Figure 7). We also observed the presence of eosinophil in BALF in asthmatic mice treated with ovalbumin, iE-DAP and MDP but not in control normal mice. As the number of eosinophils in BALF was too little for cell counting, data were not shown.
Figure 7.
Effect of NOD1 and NOD2 ligands on the morphology of bronchial tissues in allergic asthmatic mice. Fixed lung tissue sections were stained with PAS to evaluate the mucin-producing goblet cells and general morphology of lung tissues from mice with different treatments and representative staining figures were shown. Arrows in digital photomicrographs (×10 magnification) denote mucin-producing goblet cells stained with bright purple. PBS: PBS control; OVA: ovalbumin. NOD, nucleotide-binding oligomerization domain; PAS, periodic acid Schiff; PBS, phosphate-buffered saline.
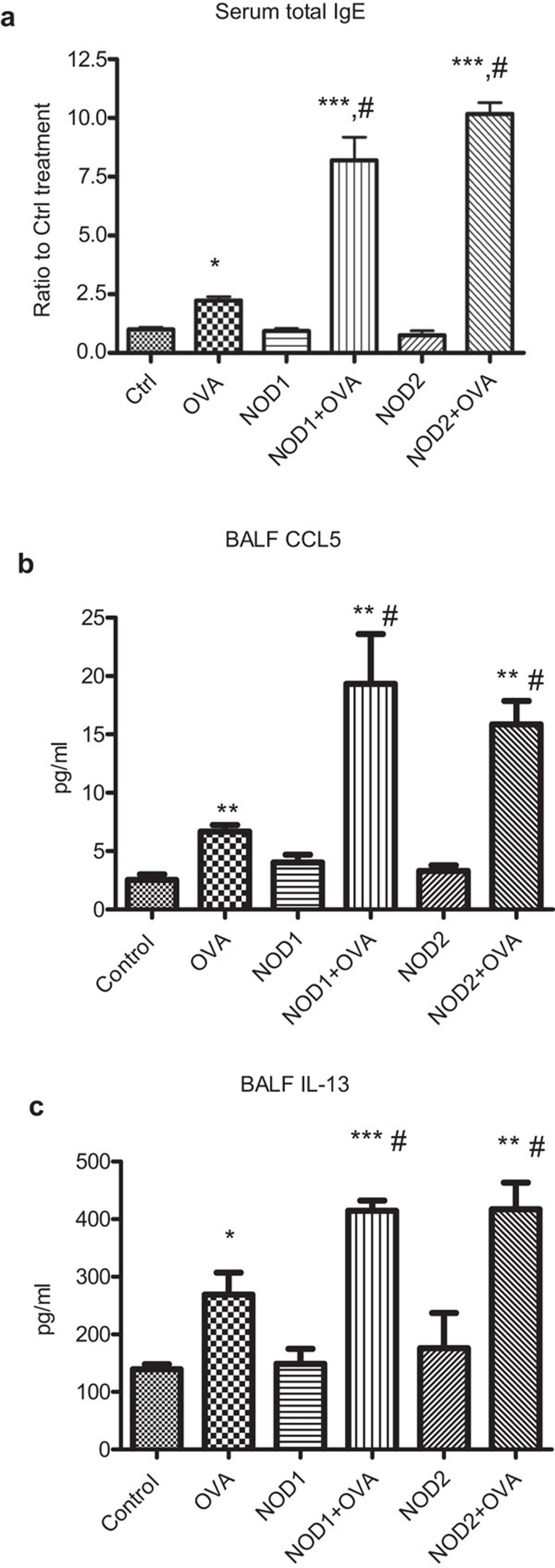
Ovalbumin sensitization could elevate serum total IgE in allergic asthmatic mice (Figure 8A). Although NOD1 and NOD2 ligands alone could not exhibit any effect on IgE level, both NOD1 and NOD2 ligands could further significantly enhance the production of total IgE serum (Figure 8A). Figure 8B and C shows that ovalbumin could significantly induce the production of CCL5 and IL-13 in BALF, concomitantly, NOD1 ligand iE-DAP and NOD2 ligand MDP could further elevate the BALF CCL5 and IL-13 concentration in allergic asthmatic mice (all P<0.05). Nevertheless, we observed that ovalbumin could significantly elevate BALF concentrations of IL-5, tumor-necrosis factor-α and CCL11 but NOD1,2 ligands did not trigger any further enhancement of above cytokines/chemokine in allergic asthmatic mice (data not shown). The discrepancy of cytokines/chemokine production profile between in vitro and in vivo experiments might be due to multiple in vivo immune effector cells participated for the cytokine/chemokine production in mice instead of only eosinophils and bronchial epithelial cells in in vitro coculture study and different sampling times for cytokines and chemokine assay between in vitro and in vivo study.
Figure 8.
Effect of NOD1 and NOD2 ligands on the production of IgE, CCL5 and IL-13 in allergic asthmatic mice. (a) Serum was collected from allergic asthmatic mice with different treatments (n=4) for the determination of the fold increase for the production of serum total IgE using ELISA comparing with control mice. (b, c) Concentrations of CCL5 and IL-13 in BALF from allergic asthmatic mice with different treatments (n=4) were determined using mouse cytokine Bio-Plex assay reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA). *P<0.05, ***P<0.001 when compared with medium control, #P<0.05 when compared with ovalbumin treatment only. Ctrl: PBS control; OVA: ovalbumin; NOD1: NOD1 ligand iE-DAP; NOD2: NOD2 ligand MDP. BALF, bronchoalveolar lavage fluid; iE-DAP, γ-D-glutamyl-meso-diaminopimelic acid; MDP, muramyl dipeptide; NOD, nucleotide-binding oligomerization domain.
Discussion
Activation of NOD1 and NOD2 is involved in Th1 and Th17 cell-mediated diseases, such as Crohn's disease and graft-vs.-host disease.31,32 The polymorphisms of NOD1 and NOD2 have also been shown to be associated with Th2-mediated atopic diseases such as allergic asthma.18,33,34 In the present study, we further confirmed that eosinophils constitutively expressed NOD1 and NOD2. NOD2 ligand MDP exhibits no or weak effects on Th2 cell chemokine CCL2 and neutrophil chemokine CXCL8 release from eosinophils or BEAS-2B cells (Figure 3). NOD1 ligand iE-DAP also showed no effects on eosinophils but significantly activated BEAS-2B cells to secrete chemokine CXCL8. Coculture of eosinophils and BEAS-2B cells could potently augment the secretion of allergic inflammation-related chemokine CXCL8 and CCL2, which could be further enhanced by iE-DAP or MDP treatment (Figure 3). Results of fixation experiments further showed that both eosinophils and BEAS-2B cells were the main source for CXCL8 and CCL2 release in the coculture system upon iE-DAP or MDP stimulation (Figure 4). Moreover, in the presence of transwell insert, the induction of CCL2 release was almost totally abolished while similar amount of CXCL8 was produced upon iE-DAP or MDP treatment in coculture of eosinophils and BEAS-2B cells, thereby suggesting the distinct mechanisms involved in CCL2 and CXCL8 release (Figure 5). The direct intercellular interaction between eosinophils and BEAS-2B cells is therefore partially responsible for CCL2 release and soluble mediators are implicated in CXCL8 release. The above results indicated that bacterial infection could provoke the allergic inflammation via the interaction of eosinophils and bronchial epithelia cells in local inflammatory sites by inducing allergic inflammation-related chemokines. Nevertheless, eosinophils treated with NOD1,2 ligands and cocultured with BEAS-2B cells did not have any effect on the release of eosinophil chemokine CCL11 (all P>0.05) (data not shown). Actually, previous studies only reported that influenza virus A and rhinovirus infection but not bacterial infection could upregulate CCL11 expression in bronchial epithelial cells.35,36
The immunoglobulin gene superfamily member ICAM-1 is constitutively expressed at relatively low levels on airway epithelial cells,37,38 while integrin CD18 is constitutively expressed at moderate levels on eosinophils,39 and the role of ICAM-1 and CD18 in airway inflammation has been well studied.40 Upon iE-DAP or MDP stimulation in the coculture system, the expression of CD18 and ICAM-1 on eosinophils was significantly upregulated, while L-selectin on eosinophils was markedly downregulated (Figure 2). Moreover, ICAM-1 expression on BEAS-2B cells was elevated in coculture system and further augmented upon iE-DAP or MDP stimulation in the coculture system. Cell surface expression of CD18 on eosinophils could be induced by eosinophil chemokine CCL5.41 The direct interaction of ICAM-1 and CD18 are implicated in mediating eosinophil adhesion to bronchial epithelial cells.42 The downregulated L-selectin expression by NOD ligands also contributes to allergic response because L-selectin is responsible for the initial attachment of eosinophils to the endothelial cells before their firm adhesion and diapedesis at sites of inflammation.41,43 Therefore, the modulation of cell surface adhesion molecules upon the interaction between NOD ligand-activated eosinophils and bronchial epithelial cells should play important roles on the bacterial infection-inducing allergic inflammation in asthma. Since previous studies indicated that CCL11 could elicit rapid vesicular transport-mediated release of preformed IL-4 from eosinophils via the intracellular granule-associated IL-4 receptors,44,45 our present result of NOD1-mediated upregulation of IL-4Ra in eosinophils indicated that bacterial infection may provoke the Th2 response in allergic asthma via the activation of eoisnophils (Figure 2E). We found that NOD1 ligand iE-DAP shows less activity to induce the ICAM-1 expression on eosinophils cultured alone or in coculture while comparing with that of NOD2 ligand MDP (Figure 2). In addition, iE-DAP also exhibits less induction of CCL2 in coculture comparing with MDP (Figure 3). This may be due to the less expression of NOD1 than NOD2 in eosinophils (Figure 1C).
NOD1 and NOD2, sharing their structural homology to the apoptosis regulator apoptotic protease activating factor 1, have been reported to regulate apoptosis. Overexpression of NOD1 or NOD2 could promote apoptosis and caspase activation.17,46 On the other hand, NOD1 and NOD2 proteins could also mediate NF-κB and MAPK activation to further induce the expression of antiapoptotic factors, such as A1, c-IAPs and c-FLIP, and proinflammatory factors.47,48,49 Since we could not observe any significant effect of NOD1,2 ligands on the apoptosis of eosinophils or BEAS-2B cells cultured alone (all P>0.05, data not shown), it might imply the in vitro balance between the NOD-mediated antiapoptosis and the NOD-mediated activation of proapoptotic signaling pathways.
Signal transduction mechanisms on the activation of NOD1 and NOD2 have been well characterized in intestinal epithelial cells and macrophages.49,50,51 NOD1 and NOD2-induced activation in eosinophils has recently been shown to be dependent on the NF-κB pathway.20 We further demonstrated that iE-DAP stimulation could activate intracellular ERK and NF-κB pathways in eosinophils, while MDP stimulation could activate ERK in both eosinophils and BEAS-2B cells (Figure 6). The iE-DAP and MDP-activated ERK and NF-κB were further enhanced in coculture (Figure 6). The coculture effects on iE-DAP and MDP-induced activation of signaling pathways showed consistency with that of iE-DAP and MDP-induced release of CCL2 and CXCL8 (at least fivefold increase) from eosinophils and BEAS-2B cells (Figure 3). Results therefore implied that intercellular interaction between eosinophils and BEAS-2B cells plays important role in enhancing the signaling pathway activation, either directly through interaction by adhesion molecules on these two cells or indirectly through mediators released from these two cells, and such enhancement could be the result from intracellular signaling cross-talk.52 iE-DAP and MDP can synergize with IL-32 for the release of IL-1β and IL-6 from PBMC.53 MDP also acted synergistically with TLR4 ligand lipopolysaccharide to stimulate the production of inflammatory cytokines in human monocytes and dendritic cells.54 Thus, intercellular interaction of eosinophils and BEAS-2B cells can therefore synergy with iE-DAP and MDP activation via ERK and NF-κB pathways. We also observed that both ERK inhibitor U0126 and NF-κB inhibitor BAY11-7082 showed significant suppression on the iE-DAP and MDP-induced release of chemokines and counteracted the effects of iE-DAP and MDP on the expression of adhesion molecules on eosinophils and BEAS-2B cells in coculture (all P<0.05). However, there were no significant activations of p38MAPK and JNK upon NOD1,2 ligand stimulation (both P>0.05) (data not shown). The above results therefore complementarily proved the involvement of these ERK and NF-κB signaling pathways upon iE-DAP and MDP stimulation.
Our in vivo animal model using ovalbumin-sensitized allergic asthmatic mice shows that both NOD1 and NOD ligands could increase the number of mucin-secreting goblet cells, bronchial subepithelial fibrosis and the in vivo production of total IgE. It suggests that the bacterial infection may enhance the sensitization to inhaled allergen and the subsequent development of allergic asthma. Intravenous injection of NOD ligands was adopted because it is a more standardized, effective and widely used optimal procedure with known applied amount of NOD ligands.25 In parallel with elevated IgE induction, NOD ligand could further enhance the bronchial subepithelial fibrosis, the production of Th2 cells and eosinophils chemokine CCL5 and asthma Th2 cytokine IL-13, probably at the inflammatory airway. The measured levels of murine CCL5 in BALF (up to 23 pg/ml) and IL-13 (about 100–400 pg/ml) were actually comparable to the previously studies.55,56 In view of the induced serum IgE, BALF CCL5 and IL-13 production, NOD ligand treatment could establish the in vivo Th2-mediated chronic allergic inflammation (Figure 8). In conjunction with the above in vitro coculture results, bacterial infection could therefore exacerbate the allergic inflammation in asthma via NLR-mediated activation and subsequent recruitment of eosinophils, neutrophils, Th2 cells and basophils, and IgE production at inflammatory airway. To further confirm the effects of NOD ligands on other chemokine induction in allergic asthmatic mice, in vitro coculture experiments using other immune effector cells such as basophils, mast cells, Th17 cells and dendritic cells and longitudinal animal studies using eosinophil-deficient mice require further investigations.
In conclusion, this study demonstrated the immunological mechanisms by which NOD1 and NOD2 regulate the interaction between human eosinophils and bronchial epithelial cells in allergic asthma, with both in vitro and in vivo study focusing on the modulation of surface adhesion molecule expression, intracellular IL-4Ra and the stimulation of allergic inflammation-related chemokine and cytokine release. Taken together with previous studies on the potential participation of NOD1 and NOD2 activation in Th2-responses and the polymorphisms of NOD1 and NOD2 with asthma, this mechanistic study provides further evidences of how bacterial infections could be involved in the pathogenesis of asthma. Our results of intracellular signaling mechanisms and in vivo animal study upon NOD ligand stimulation may shed light on the development of novel treatment approaches for eosinophil-associated allergic diseases.
Acknowledgments
This work was supported by the Research Grant Committee General Research Fund, Hong Kong (project reference no.: CUHK 476411, principal investigator: CKW) and direct grant from The Chinese University of Hong Kong (reference no.: 2008.1.018).
The authors declare that they have no competing financial interests.
References
- Hogan SP, Rosenberg HF, Moqbel R, Phipps S, Foster PS, Lacy P, et al. Eosinophils: biological properties and role in health and disease. Clin Exp Allergy. 2008;38:709–750. doi: 10.1111/j.1365-2222.2008.02958.x. [DOI] [PubMed] [Google Scholar]
- Lemière C, Ernst P, Olivenstein R, Yamauchi Y, Govindaraju K, Ludwig MS, et al. Airway inflammation assessed by invasive and noninvasive means in severe asthma: eosinophilic and noneosinophilic phenotypes. J Allergy Clin Immunol. 2006;118:1033–1039. doi: 10.1016/j.jaci.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Takafuji S, Ohtoshi T, Takizawa H, Tadokoro K, Ito K. Eosinophil degranulation in the presence of bronchial epithelial cells. Effect of cytokines and role of adhesion. J Immunol. 1996;156:3980–3985. [PubMed] [Google Scholar]
- Wong CK, Wang CB, Ip WK, Tian YP, Lam CW. Role of p38 MAPK and NF-κB for chemokine release in co-culture of human eosinophils and bronchial epithelial cells. Clin Exp Immunol. 2005;139:90–100. doi: 10.1111/j.1365-2249.2005.02678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CB, Wong CK, Ip WK, Tian YP, Lam CW. Induction of IL-6 in co-culture of bronchial epithelial cells and eosinophils is regulated by p38 MAPK and NF-κB. Allergy. 2005;60:1378–1385. doi: 10.1111/j.1398-9995.2005.00884.x. [DOI] [PubMed] [Google Scholar]
- Cheung PF, Wong CK, Ho AW, Hu S, Chen DP, Lam CW. Activation of human eosinophils and epidermal keratinocytes by Th2 cytokine IL-31: implication for the immunopathogenesis of atopic dermatitis. Int Immunol. 2010;22:453–467. doi: 10.1093/intimm/dxq027. [DOI] [PubMed] [Google Scholar]
- Schröder NW, Crother TR, Naiki Y, Chen S, Wong MH, Yilmaz A. Innate immune responses during respiratory tract infection with a bacterial pathogen induce allergic airway sensitization. J Allergy Clin Immunol. 2008;122:595–602. doi: 10.1016/j.jaci.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crother TR, Schröder NW, Karlin J, Chen S, Shimada K, Slepenkin A, et al. Chlamydia pneumoniae infection induced allergic airway sensitization is controlled by regulatory T-cells and plasmacytoid dendritic cells. PLoS ONE. 2011;6:e20784. doi: 10.1371/journal.pone.0020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos NG, Christodoulou I, Rohde G, Agache I, Almqvist C, Bruno A, et al. Viruses and bacteria in acute asthma exacerbations—a GA2 LEN-DARE systematic review. Allergy. 2011;66:458–468. doi: 10.1111/j.1398-9995.2010.02505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Carneiro LA, Magalhaes JG, Tattoli I, Philpott DJ, Travassos LH. NOD-like proteins in inflammation and disease. J Pathol. 2008;214:136–148. doi: 10.1002/path.2271. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Tsuji Y, Watanabe T, Kudo M, Arai H, Strober W, Chiba T. Sensing of commensal organisms by the intracellular sensor NOD1 mediates experimental pancreatitis. Immunity. 2012;37:326–338. doi: 10.1016/j.immuni.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, et al. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedra JH, Cassel SL, Sutterwala FS. Sensing pathogens and danger signals by the inflammasome. Curr Opin Immunol. 2009;21:10–16. doi: 10.1016/j.coi.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes JG, Fritz JH, le Bourhis L, Sellge G, Travassos LH, Selvanantham T, et al. Nod2-dependent Th2 polarization of antigen-specific immunity. J Immunol. 2008;181:7925–7935. doi: 10.4049/jimmunol.181.11.7925. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frame shift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Hysi P, Kabesch M, Moffatt MF, Schedel M, Carr D, Zhang Y. NOD1 variation, immunoglobulin E and asthma. Hum Mol Genet. 2005;14:935–941. doi: 10.1093/hmg/ddi087. [DOI] [PubMed] [Google Scholar]
- Rosenstiel P, Till A, Schreiber S. NOD-like receptors and human diseases. Microbes Infect. 2007;9:648–657. doi: 10.1016/j.micinf.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Kvarnhammar AM, Petterson T, Cardell LO. NOD-like receptors and RIG-I-like receptors in human eosinophils: activation by NOD1 and NOD2 agonists. Immunology. 2011;134:314–325. doi: 10.1111/j.1365-2567.2011.03492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Cummings R, Usatyuk P, Morris A, Irani K, Natarajan V. Involvement of phospholipases D1 and D2 in sphingosine 1-phosphate-induced ERK (extracellular-signal-regulated kinase) activation and interleukin-8 secretion in human bronchial epithelial cells. Biochem J. 2002;367:751–760. doi: 10.1042/BJ20020586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister F, Henry A, Kreindler JL, Dubin PJ, Ulrich L, Steele C, et al. Role of IL-17A, IL-17F, and the IL-17 receptor in regulating growth-related oncogenealpha and granulocyte colony-stimulating factor in bronchial epithelium: implications for airway inflammation in cystic fibrosis. J Immunol. 2005;175:404–412. doi: 10.4049/jimmunol.175.1.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi M, Kokubu F, Kuga H, Matsukura S, Hoshino H, Ieki K, et al. Modulation of bronchial epithelial cells by IL-17. J Allergy Clin Immunol. 2001;108:804–809. doi: 10.1067/mai.2001.119027. [DOI] [PubMed] [Google Scholar]
- Wong CK, Cao J, Yin YB, Lam CW. Interleukin-17A activation on bronchial epithelium and basophils: a novel inflammatory mechanism. Eur Respir J. 2010;35:883–893. doi: 10.1183/09031936.00088309. [DOI] [PubMed] [Google Scholar]
- Beigelman A, Gunsten S, Mikols CL, Vidavsky I, Cannon CL, Brody SL, et al. Azithromycin attenuates airway inflammation in a noninfectious mouse model of allergic asthma. Chest. 2009;136:498–506. doi: 10.1378/chest.08-3056. [DOI] [PubMed] [Google Scholar]
- Barton JL, Berg T, Didon L, Nord M. The pattern recognition receptor Nod1 activates CCAAT/enhancer binding protein beta signalling in lung epithelial cells. Eur Respir J. 2007;30:214–222. doi: 10.1183/09031936.00143906. [DOI] [PubMed] [Google Scholar]
- Farkas L, Stoelcker B, Jentsch N, Heitzer S, Pfeifer M. Muramyldipeptide modulates CXCL-8 release of BEAS-2B cells via NOD2. Scand J Immunol. 2008;68:315–322. doi: 10.1111/j.1365-3083.2008.02145.x. [DOI] [PubMed] [Google Scholar]
- Bérubé J, Bourdon C, Yao Y, Rousseau S. Distinct intracellular signaling pathways control the synthesis of IL-8 and RANTES in TLR1/TLR2, TLR3 or NOD1 activated human airway epithelial cells. Cell Signal. 2009;21:448–456. doi: 10.1016/j.cellsig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Cheung PF, Wong CK, Ip WK, Lam CW. FAK-mediated activation of ERK for eosinophil migration: a novel mechanism for infection-induced allergic inflammation. Int Immunol. 2008;20:353–363. doi: 10.1093/intimm/dxm146. [DOI] [PubMed] [Google Scholar]
- Lal RB, Edison LJ, Chused TM. Fixation and long-term storage of human lymphocytes for surface marker analysis by flow cytometry. Cytometry. 1988;9:213–219. doi: 10.1002/cyto.990090305. [DOI] [PubMed] [Google Scholar]
- Holler E, Rogler G, Herfarth H, Brenmoehl J, Wild PJ, Hahn J, et al. Both donor and recipient NOD2/CARD15 mutations associate with transplant-related mortality and GvHD following allogeneic stem cell transplantation. Blood. 2004;104:889–894. doi: 10.1182/blood-2003-10-3543. [DOI] [PubMed] [Google Scholar]
- Hugot JP. CARD15/NOD2 mutations in Crohn's disease. Ann N Y Acad Sci. 2006;1072:9–18. doi: 10.1196/annals.1326.011. [DOI] [PubMed] [Google Scholar]
- Reijmerink NE, Bottema RW, Kerkhof M, Gerritsen J, Stelma FF, Thijs C, et al. TLR-related pathway analysis: novel gene-gene interactions in the development of asthma and atopy. Allergy. 2010;65:199–207. doi: 10.1111/j.1398-9995.2009.02111.x. [DOI] [PubMed] [Google Scholar]
- Weidinger S, Klopp N, Rümmler L, Wagenpfeil S, Baurecht HJ, Gauger A, et al. Association of CARD15 polymorphisms with atopy-related traits in a population-based cohort of Caucasian adults. Clin Exp Allergy. 2005;35:866–872. doi: 10.1111/j.1365-2222.2005.02269.x. [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Kokubu F, Kuga H, Tomita T, Matsukura S, Kadokura M, et al. Expression of eotaxin by normal airway epithelial cells after influenza virus A infection. Int Arch Allergy Immunol. 2000;122 Suppl 1:44–49. doi: 10.1159/000053632. [DOI] [PubMed] [Google Scholar]
- Papadopoulos NG, Papi A, Meyer J, Stanciu LA, Salvi S, Holgate ST, et al. Rhinovirus infection up-regulates eotaxin and eotaxin-2 expression in bronchial epithelial cells. Clin Exp Allergy. 2001;31:1060–1066. doi: 10.1046/j.1365-2222.2001.01112.x. [DOI] [PubMed] [Google Scholar]
- Bloemen PG, van den Tweel MC, Henricks PA, Engels F, Wagenaar SS, Rutten AA, et al. Expression and modulation of adhesion molecules on human bronchial epithelial cells. Am J Respir Cell Mol Biol. 1993;9:586–593. doi: 10.1165/ajrcmb/9.6.586. [DOI] [PubMed] [Google Scholar]
- Look DC, Rapp SR, Keller BT, Holtzman MJ. Selective induction of intercellular adhesion molecule-1 by interferon- in human airway epithelial cells. Am J Physiol. 1992;263:L79–87. doi: 10.1152/ajplung.1992.263.1.L79. [DOI] [PubMed] [Google Scholar]
- Cheung PF, Wong CK, Ip WK, Lam CW. IL-25 regulates the expression of adhesion molecules on eosinophils: mechanism of eosinophilia in allergic inflammation. Allergy. 2006;61:878–885. doi: 10.1111/j.1398-9995.2006.01102.x. [DOI] [PubMed] [Google Scholar]
- Proud D, Leigh R. Epithelial cells and airway diseases. Immunol Rev. 2011;242:186–204. doi: 10.1111/j.1600-065X.2011.01033.x. [DOI] [PubMed] [Google Scholar]
- Kitayama J, Fuhlbrigge RC, Puri KD, Springer TA. P-selectin, L-selectin, and alpha 4 integrin have distinct roles in eosinophil tethering and arrest on vascular endothelial cells under physiological flow conditions. J Immunol. 1997;159:3929–3939. [PubMed] [Google Scholar]
- Burke-Gaffney A, Hellewell PG. A CD18/ICAM-1-dependent pathway mediates eosinophil adhesion to human bronchial epithelial cells. Am J Respir Cell Mol Biol. 1998;19:408–418. doi: 10.1165/ajrcmb.19.3.3179. [DOI] [PubMed] [Google Scholar]
- Michail S, Mezoff E, Abernathy F. Role of selectins in the intestinal epithelial migration of eosinophils. Pediatr Res. 2005;58:644–647. doi: 10.1203/01.PDR.0000180572.65751.F4. [DOI] [PubMed] [Google Scholar]
- Spencer LA, Melo RC, Perez SA, Bafford SP, Dvorak AM, Weller PF. Cytokine receptor-mediated trafficking of preformed IL-4 in eosinophils identifies an innate immune mechanism of cytokine secretion. Proc Natl Acad Sci USA. 2006;103:3333–3338. doi: 10.1073/pnas.0508946103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandeira-Melo C, Sugiyama K, Woods LJ, Weller PF. Cutting edge: eotaxin elicits rapid vesicular transport-mediated release of preformed IL-4 from human eosinophils. J Immunol. 2001;166:4813–4817. doi: 10.4049/jimmunol.166.8.4813. [DOI] [PubMed] [Google Scholar]
- Geddes BJ, Wang L, Huang WJ, Lavellee M, Manji GA. Human CARD12 is a novel CED4/Apaf-1 family member that induces apoptosis. Biochem Biophys Res Commun. 2001;284:77–82. doi: 10.1006/bbrc.2001.4928. [DOI] [PubMed] [Google Scholar]
- Inohara M, Chamaillard M, McDonald C, Nuñez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–360. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J. NF-kappaB signals induce the expression of c-FLIP. Mol Cell Biol. 2001;21:5299–5305. doi: 10.1128/MCB.21.16.5299-5305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauleau AL, Murray PJ. Role of Nod2 in the response of macrophages to toll-like receptor agonists. Mol Cell Biol. 2003;23:7531–7539. doi: 10.1128/MCB.23.21.7531-7539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nuñez G, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- Park JH, Kim YG, Shaw M, Kanneganti TD, Fujimoto Y, Fukase K, et al. Nod1/RICK and TLR signaling regulate chemokine and antimicrobial innate immune responses in mesothelial cells. J Immunol. 2007;179:514–521. doi: 10.4049/jimmunol.179.1.514. [DOI] [PubMed] [Google Scholar]
- Cheung PF, Wong CK, Lam CW. Molecular mechanisms of cytokine and chemokine release from eosinophils activated by IL-17A, IL-17F, and IL-23: implication for Th17 lymphocytes-mediated allergic inflammation. J Immunol. 2008;180:5625–5635. doi: 10.4049/jimmunol.180.8.5625. [DOI] [PubMed] [Google Scholar]
- Netea MG, Azam T, Ferwerda G, Girardin SE, Walsh M, Park JS, et al. IL-32 synergizes with nucleotide oligomerization domain (NOD) 1 and NOD2 ligands for IL-1beta and IL-6 production through a caspase 1-dependent mechanism. Proc Natl Acad Sci USA. 2005;102:16309–16314. doi: 10.1073/pnas.0508237102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JH, Girardin SE, Fitting C, Werts C, Mengin-Lecreulx D, Caroff M, et al. Synergistic stimulation of human monocytes and dendritic cells by Toll-like receptor 4 and NOD1- and NOD2-activating agonists. Eur J Immunol. 2005;35:2459–2470. doi: 10.1002/eji.200526286. [DOI] [PubMed] [Google Scholar]
- Weigt SS, Elashoff RM, Keane MP, Strieter RM, Gomperts BN, Xue YY, et al. Altered levels of CC chemokines during pulmonary CMV predict BOS and mortality post-lung transplantation. Am J Transplant. 2008;8:1512–1522. doi: 10.1111/j.1600-6143.2008.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueders MM, Paulissen G, Crahay C, Quesada-Calvo F, Hacha J, van Hove C, et al. Mouse models of asthma: a comparison between C57BL/6 and BALB/c strains regarding bronchial responsiveness, inflammation, and cytokine production. Inflamm Res. 2009;58:845–854. doi: 10.1007/s00011-009-0054-2. [DOI] [PubMed] [Google Scholar]