Abstract
The transcription factor T-bet was originally described to be important for the differentiation of the CD4+ Th1 subset. More recent investigations implicate T-bet in the lineage commitment of a variety of innate immune cells also. The T-bet appears to have a dual role in the immune system. Under certain conditions T-bet provides a beneficial role, whereas the exaggerated expression of T-bet in certain innate lymphoid cells can be detrimental to the host. Therefore, this transcription factor needs to be carefully regulated. The feedback control and the epigenetic mechanisms involved in the expression of T-bet remain to be fully elucidated.
Keywords: Innate lymphoid cells, T-bet, T helper cell subsets
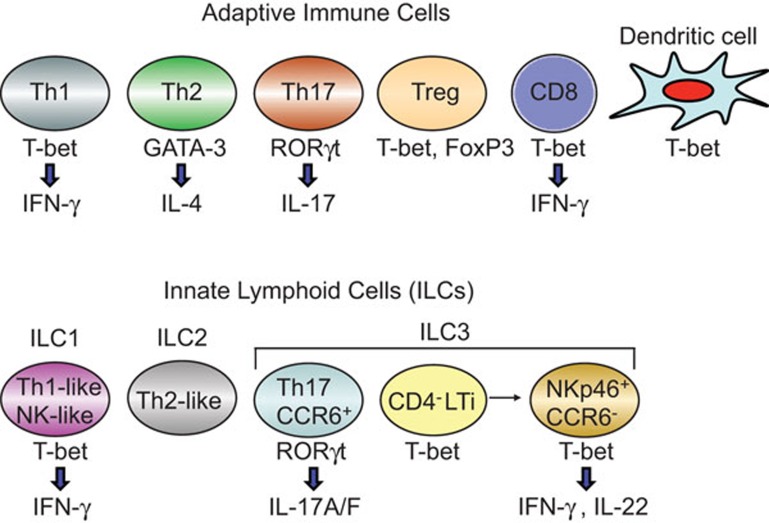
More than a quarter century ago, Mosmann et al.1 reported that a majority of murine CD4+ T-helper (Th) clones could be divided into two functionally distinct subsets, interferon (IFN)-γ-producing Th1 and IL-4-expressing Th2 cells. This dichotomy of Th cells represented a paradigm shift in the understanding of the role of T cells in various immune responses, specifically in pathological conditions. The Th1 cells appear to be relatively more rigid than the Th2 subset. The later identified IL-17-producing Th17 cells remain ‘plastic' and function only after conversion into Th1 type cells in vivo.2,3 The rigidity of the recently described Th9 and follicular Th cells remains to be fully elucidated. Our understanding of how the naive T cells are induced to become functionally distinct Th subsets stems primarily from two seminal observations, which demonstrated the involvement of the ‘master regulators', GATA-3 and T-bet, respectively in Th24 and Th15 lineage commitment (Figure 1). Additional studies implicated RORγt in Th176 and FoxP3 in T-regulatory cell7 development. The T-bet is expressed ubiquitously in many cell types including dendritic cells, natural killer (NK) cells, natural killer T cells, B cells and CD8+ cells, and it also negates the activity of GATA-3 during Th1-cell differentiation.8 It is of great interest to determine how T-bet interacts with other transcription factors to allow the differentiation of distinct cell types involved in the adaptive and innate immune system.
Figure 1.
A model of the expression of various transcription factors and the production of lymphokines in diverse cell types of the adaptive and innate immune system is depicted.
Several recent publications indicate that T-bet is also important in the lineage commitment of innate lymphoid cells (ILCs), similar to the conventional Th subsets (Figure 1). Rankin et al.9 have shown that the transcription factor T-bet (encoded by Tbx21) is essential for the development of ILC3s that are NKp46+CCR6− but not for CD4− lymphoid tissue-inducer (LTi) cells or nuocytes. Lack of NKp46+CCR6− ILC3s in Tbx21−/− mice resulted in increased susceptibility to infection with bacteria in the gut due to diminished IL-22 production by ILCs. These findings are consistent with two other studies showing that mice deficient in T-bet had diminished capacity to differentiate NKp46-expressing RORγt+ ILCs (NK-22 cells) and produce IFN-γ.10,11
To closely examine the role of T-bet in the conversion of LTi cells into NKp46+ ILCs, Rankin et al.9 purified innate lymphocytes from Tbx21-deficient mice and adoptively transferred them into recipient mice lacking all ILCs due to the deletion of recombination-activating gene 2 and the common γ-chain gene. Lack of Tbx21 in mature LTi cells resulted in the failure of conversion into NKp46+ ILCs in vivo. Flow cytometric analysis of purified ILCs revealed an increase in the frequency of CD4− LTi cells in Tbx21−/− mice, indicating that the loss of T-bet may have halted the developmental progression into NKp46+ ILCs. Quantitative PCR analysis revealed lower expression of Notch1 and Notch2 in ILCs sorted from Tbx21−/− mice, indicating that the engagement of the Notch signaling pathway by T-bet is essential for the generation of NKp46+ ILCs. Chromatin immunoprecipitation analysis revealed the T-bet-binding site in Notch2 and not in Notch1 of ILCs derived from Tbx21+/+ mice. Coculture of purified ILC populations from T-bet-deficient mice with Notch ligand DL1-expressing OP9 stromal cells did not result in the generation of NKp46+ ILCs, reiterating the role of Notch signaling in the differentiation of this subset.9 Further understanding of the mechanisms involved in the Notch-dependent differentiation of NKp46+ ILCs will be crucial for devising effective strategies to maximize protection against bacterial infections and to curtail certain autoimmunity in the intestinal tract.
Evidence for the involvement of T-bet in the development of ILC1 subset was recently provided by two groups. Bernink et al.12 identified ILC1 cells, homologous to Th1 cells, in human tonsil and fetal intestinal tissue. These cells expressed T-bet and responded to IL-12 by producing IFN-γ. Importantly, higher numbers of ILC1 cells were found in the inflamed intestine of people with Crohn's disease. Fuchs et al.13 also characterized a similar human ILC1 subset that produces IFN-γ in response to IL-12 and IL-15. In mice, this subset was characterized by CD160 expression and required Tbx21-encoded transcription factor for development. The intraepithelial ILC1 cells were amplified in patients with Crohn's disease and contributed to the anti-CD40 antibody-induced colitis in mice. Taken together, these data underpin the role of T-bet in the development and differentiation of distinct subsets of ILCs and point out that the dysregulation of these cells can have dire consequences.
Although T-bet is the master regulator of Th1 differentiation and IFN-γ production, as expected of transcription factors, it can also bind to DNA regions indiscriminately outside of the Ifng locus. Recent genome-wide analysis by Kanhere et al.14 provided significant insights into how T-bet and GATA-3 are related to each other. In Th2 cells, GATA-3 binds to a unique set of sites containing a GATA motif that are associated with the expression of Th2 genes. Whereas the direct binding of T-bet at Tbx21 resulted in autoactivation in Th1 cells, the occupancy of GATA-3 at sites that are usually bound by T-bet prevented the activation of Th2 genes in Th1 cells. Similarly, it will be interesting to delineate the impact of the binding of T-bet and Rorγt outside of their respective loci on gene expression in distinct T-cell subsets.
Despite the expression of T-bet in a wide range of cell types, surprisingly very little is known about the regulation of this transcription factor by genetic mechanisms including single-nucleotide polymorphisms, copy number variation, as well as gene deletion and duplication, in various patient populations. Although microRNA-mediated regulation of gene expression has been extensively studied in cancer, neurological disorders and certain autoimmune conditions, data on the regulation of T-bet and other T-cell lineage-determining transcription factors by noncoding microRNAs are minimal. Because of the promiscuous nature of the binding of transcription factors to DNA regions, it will be hard to intervene this process for therapeutic purposes. However, altered expression of Tbx21 and other transcription factors under pathological conditions may be modulated using epigenetic mechanisms, including DNA methylation and histone acetylation. In this regard, it is interesting to note that chromatin remodeling mediated by a small molecule inhibitor of histone deacetylases resulted in the upregulation of Tbx21 associated with alleviation of autoimmune diabetes in mice.15 In-depth analysis of epigenetic control of Tbx21 may provide novel information useful for devising treatment strategies to manage a variety of pathological conditions.
References
- Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- Bending D, de la Peña H, Veldhoen M, Phillips JM, Uyttenhove C, Stockinger B, et al. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest. 2009;119:565–572. doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Orozco N, Chung Y, Chang SH, Wang YH, Dong C. Th17 cells promote pancreatic inflammation but only induce diabetes efficiently in lymphopenic hosts after conversion into Th1 cells. Eur J Immunol. 2009;39:216–224. doi: 10.1002/eji.200838475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- Lazarevic V, Glimcher LH. T-bet in disease. Nat Immunol. 2011;12:597–606. doi: 10.1038/ni.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin LC, Groom JR, Chopin M, Herold MJ, Walker JA, Mielke LA, et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol. 2013;14:389–395. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciumé G, Hirahara K, Takahashi H, Laurence A, Villarino AV, Singleton KL, et al. Distinct requirements for T-bet in gut innate lymphoid cells. J Exp Med. 2012;209:2331–2338. doi: 10.1084/jem.20122097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d'Hargues Y, et al. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, et al. Intraepithelial Type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-Responsive IFN-γ-producing cells. Immunitye-pub ahead of print 2013 Feb 26. doi:10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed]
- Kanhere A, Hertweck A, Bhatia U, Gökmen MR, Perucha E, Jackson I, et al. T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nat Commun. 2012;3:1268. doi: 10.1038/ncomms2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T, Patel V, Singh R, Jayaraman S. Chromatin remodeling resets the immune system to protect against autoimmune diabetes in mice. Immunol Cell Biol. 2011;89:640–649. doi: 10.1038/icb.2010.144. [DOI] [PubMed] [Google Scholar]