Preface
We have extended our understanding of the molecular biology underlying adult glioblastoma over many years. In contrast, high-grade gliomas in children and adolescents have remained a relatively under-investigated disease. The latest large-scale genomic and epigenomic profiling studies have yielded an unprecedented abundance of novel data and revealed deeper insights into gliomagenesis across all age groups, highlighting key distinctions, but also some commonalities. As we are on the verge of dissecting glioblastomas into meaningful biological subgroups, this Review summarizes the hallmark genetic alterations associated with distinct epigenetic features and patient characteristics in both paediatric and adult disease, and examines the complex interplay between the glioblastoma genome and epigenome.
Introduction
Glioblastoma (GBM; synonymous with the formerly used term ‘glioblastoma multiforme’) is the most frequent and most aggressive malignant primary brain tumour1, 2, classified as grade IV in the World Health Organization (WHO) classification of tumours of the central nervous system (CNS)1. GBM can develop from lower-grade diffuse astrocytoma (WHO grade II) and anaplastic astrocytoma (WHO grade III), and is then termed ‘secondary’ GBM. ‘Primary’ (de novo) GBMs are more common, and typically manifest rapidly after a short clinical history and without recognizable signs of a preceding precursor lesion3. Diagnostic criteria for GBM, including high mitotic activity, microvascular proliferation and/or necrotic areas, are fulfilled by both primary and secondary GBM, making them indistinguishable by histology alone1.
Overall, primary GBM accounts for 16% of primary brain tumours2. The incidence increases with age, affecting an average of 7.2 per 100,000 adults (> 19 years of age) every year, with a peak age at diagnosis between 75 and 84 years (annual incidence rate 14.6 per 100,000)2. Due to the lower incidence in children, studies on paediatric high-grade gliomas (HGG) have traditionally combined GBM, anaplastic astrocytomas and diffuse intrinsic pontine gliomas (DIPG). Although most DIPG histologically present as high-grade tumours and share a universally fatal outcome, they comprise tumours with varying histological grade and divergent differentiation4–7. Together, up to 0.8 per 100,000 children (age < 19) are estimated to develop high-grade gliomas each year, making them the most common group of malignant CNS neoplasms in this age group alongside embryonal tumours2. The current treatment strategy for GBM patients consists of surgery, radiotherapy and chemotherapy. Complete surgical resection of these infiltrative tumours is virtually impossible and surgery is typically not attempted for DIPG patients due to their location. Concurrent adjuvant radiotherapy in combination with temozolomide (TMZ) represents the standard of care for newly diagnosed GBM, but still less than 5% of patients survive longer than five years post diagnosis2, 8–10. Since no chemotherapeutic drugs have proven to be effective in the treatment of DIPG, radiation therapy alone represents the current standard regimen11, 12. Prognosis here is even worse, with less than 10% of DIPG patients surviving more than two years after diagnosis2, 13.
Over recent years, large-scale research efforts spearheaded by The Cancer Genome Atlas (TCGA) network and the paediatric neurooncology community have been directed at studying the molecular biology underlying GBM, and have generated detailed catalogues of genomic and epigenomic alterations. The emerging insights into gliomagenesis have triggered an avalanche of novel perceptions on the epigenomic and genomic landscape, biological subgroups and putative cells of origin of GBM, fuelling hopes for more effective treatment strategies in the near future. In this Review, we discuss the major recent advances in GBM molecular research, and we aim to convey important details, as well as a general overview of GBM biology across all ages.
Molecular subtypes of glioblastoma
Gene expression profiling of glioblastoma
Genome-wide profiling studies using gene expression microarrays were successfully used to compare gene expression patterns in primary vs. secondary GBM14–17 or adult vs. paediatric GBM18, and to identify differentially expressed genes that could be used to distinguish between different groups. In parallel, early reports showed the utility of expression arrays in diagnostic assignment and prognostication within larger glioma cohorts of different grades19–21 and within GBM cohorts only22. Phillips and colleagues23 described three subclasses of high-grade glioma (termed proneural, mesenchymal and proliferative, based on the functional annotation of signature genes) as being associated with different outcomes, namely prolonged survival of the proneural subclass. Similar subclasses of GBM were also detected in a large cohort of mixed gliomas and data from this cohort also revealed a distinct gene expression cluster enriched for secondary GBM24. Partially in line with these previous findings, unsupervised clustering of gene expression data from 200 adult GBM samples from TCGA network25, 26 identified four different molecular subtypes: proneural, neural, classical and mesenchymal. The proneural subtype was largely characterized by abnormalities in platelet derived growth factor receptor α (PDGFRA) or isocitrate dehydrogenase 1 (IDH1), whereas mutation of the epidermal growth factor receptor (EGFR) was found in the classical subgroup and mutations in neurofibromin (NF1) were common in mesenchymal tumours. This classification system has been refined by subdividing proneural GBM into glioma-CpG island methylator phenotype (G-CIMP)-positive and G-CIMP-negative GBM subsets based on characteristic DNA methylation patterns that strongly correspond with IDH1 mutation status27 (Figure 1; discussed below). Comparing the transcriptional classification schemes of Phillips et al.23 with those of Verhaak et al.25 suggests that the proneural and mesenchymal signatures are the most robust and account for the greatest transcriptional variance, while the exact number and composition of other signatures may depend on the sample cohort compositions and experimental designs28, 29. The transcription factors signal transducer and activator of transcription 3 (STAT3), C/EBPβ and TAZ have been identified as epigenetic master regulators of the mesenchymal signature and might predict poor clinical outcome30, 31. Moreover, CTNND2 (encoding cyclin D2) and RHPN2 have recently emerged as negative and positive genetic drivers of mesenchymal transformation, respectively32, 33. There is also evidence of plasticity between the proneural and mesenchymal subtypes in GBM23 that might be driven by the tumour microenvironment, perhaps through microglia and the NF-κB pathway34. The role that the proneural and mesenchymal signatures and other signatures might have as predictors of clinical outcome remains to be clarified. The initially reported better prognosis of the proneural subclass23, 25 was recently shown to only prove true for the G-CIMP subset, while non-G-CIMP proneural and mesenchymal GBM tend to show less favourable outcomes in the first twelve months post diagnosis compared with other GBM subtypes26.
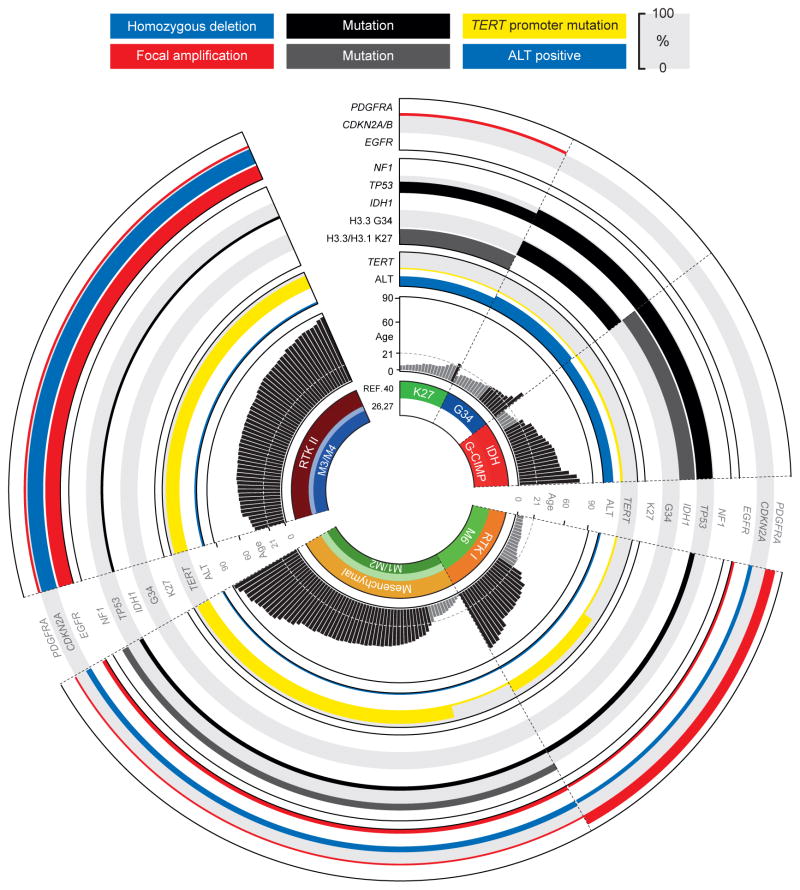
Figure 1. Age-based genomic and epigenomic features of biological glioblastoma subgroups.
Simplified schematic overview of glioblastoma subgroups depicting recurrent associations between genomic and epigenomic features: (from inside to outside) DNA methylation subclass affiliation (TCGA methylation [REFS26, 27], and dkfz methylation [REF.40]), patient age (years), telomere maintenance mechanisms, mutational status (H3.3 and H3.1 K27, H3.3 G34, IDH1, TP53 and NF1), and copy-number aberrations, grouped by biological subgroup and sorted by patient age. Copy-number states, presence of mutations and ALT positivity are represented by different colours as indicated. Height of bars represents an estimated percentage of cases positive for a specific feature. ALT, alternative lengthening of telomeres; CDKN2A and B, cyclin-dependent kinase inhibitor 2A and 2B; EGFR, epidermal growth factor receptor; G-CIMP, glioma-CpG island methylator phenotype; IDH1, isocitrate dehydrogenase 1; NF1, neurofibromin 1; PDGFRA, platelet-derived growth factor receptor type alpha; RTK, receptor tyrosine kinase; TERT, telomerase reverse transcriptase.
Initial gene expression studies of paediatric HGG highlighted significant differences compared to tumours from adult patients, and suggested the existence of molecularly diverse subsets within paediatric cohorts18, 35–37. Activation of a PDGFRA-driven gene expression signature has been described in a large proportion of paediatric HGG4, 36, correlating with the higher frequency of PDGFRA amplifications observed in this age group36–40. Interestingly, one of these studies further revealed shared gene expression patterns between DIPG and a proportion of midline/thalamic GBM, pointing towards the closely related pathogenesis of these tumours, which was later confirmed by genome sequencing studies40–42. Indeed, comparing midline GBM and DIPG that harboured a K27M amino acid change owing to a mutation in H3F3A, one of the histone H3.3 genes, with hemispheric GBM with H3F3A mutations that result in amino acid changes at G34 revealed specific gene expression profiles that are distinct from each other and from tumours with wild-type H3F3A status40, 41, 43, 44.
DNA methylation profiling of glioblastoma
A number of studies have reported promoter-associated hypermethylation of specific loci in GBM, frequently affecting the expression of genes with known tumour suppressor function such as cyclin dependent kinase inhibitor 2A (CDKN2A), RB1, PTEN and TP5345–48, as well as previously unrecognised regulatory genes involved in cell proliferation (epithelial membrane protein 3 (EMP3) and PDGFB)49, 50, invasion (protocadherin γ subfamily A, member 11 (PCDHGA11))51, or radiation sensitivity (suppressor of cytokine signalling 1 (SOCS1))52, 53. Most prominently, promoter methylation of MGMT (encoding O6-methylguanine methyltransferase) occurs in ~ 45% of GBM in adult patients26, 54–57, leading to gene silencing and consequently to a reduced proficiency for repairing DNA damage induced by alkylating agent chemotherapy58–61. Thus, methylation at this locus has been established as a biomarker for predicting the benefit of TMZ chemotherapy, particularly in elderly GBM patients56, 57, 61, 62, while both the frequency (16–50%)63–66 and prognostic significance of MGMT silencing in childhood HGG remain controversial63, 66.
DNA microarray techniques have successfully been applied to study the GBM methylome in a genome-wide manner, and have led to the discovery of clearly defined GBM subgroups based on their global DNA methylation patterns (Figure 1). The first large study, with a purely adult patient cohort, reported three DNA methylation subgroups. One group was tightly associated with IDH1 mutations and displayed concerted hypermethylation at a large number of loci, and was therefore termed G-CIMP-positive27. A later study comparing DNA methylation patterns across both paediatric and adult GBM patients found a similar clustering in tumours from adult patients, and further identified three more distinct clusters composed predominantly of children and adolescents40. Two of these corresponded strictly with recurrent age-specific (K27 or G34) mutations in H3F3A, and another group of tumours was enriched for PDGFRA alterations (and termed receptor tyrosine kinase 1 (RTK I) and consisted of patients from a more widespread age range40. Aberrations of PDGFRA are present in approximately half of G-CIMP-negative GBM of the proneural subtype in children and adults25, 40, 67. Epigenetic silencing by promoter hypermethylation of the neural lineage markers OLIG1 and OLIG2 was observed as a novel and exclusive characteristic in GBM from the G34 cluster and points towards a different pathogenesis of this subgroup. These tumours also showed a considerable decrease in overall DNA methylation on a global scale40. This has been previously reported as a common epigenetic feature in primary GBM and was linked to genomic instability of affected regions, deficiency of the methyl group metabolism gene MTHFR and increased proliferative activity68. A distinct G-CIMP-positive IDH cluster was also identified and, in line with previous reports, tumours from this group were found among younger adult patients, showed proneural gene expression patterns, lacked typical copy-number aberrations (gain of chromosome 7, loss of chromosome 10, EGFR amplification, CDKN2A and CDKN2B deletion), and were associated with a favourable prognosis compared to all other GBM subgroups27, 40, explaining the increased survival observed in the proneural GBM subtype23, 25. The remaining G-CIMP-negative DNA methylation clusters comprised GBM from elderly patients, displaying predominantly classical gene expression, combined gain of chromosome 7 and loss of chromosome 10, and EGFR, CDKN2A and CDKN2B alterations (termed cluster #2 and RTK II)27, 40, and GBM enriched for mesenchymal gene expression patterns and mutations in NF1 and PTEN (termed cluster #3 and mesenchymal subtype)27, 40.
Although mutations in H3F3A and IDH1 seem to map one-to-one with three distinct DNA methylation subclasses, recent in-depth analysis of genome-wide DNA methylation and sequencing data by TCGA has found no similarly striking pathognomonic mutations for the additional non-G-CIMP subclasses of adult GBM26. This study further confirmed the notion that, despite considerable overlap, established gene expression subclasses do not directly correspond to those defined by DNA methylation profiling26, unlike DNA methylation profiling in medulloblastoma69. Whether DNA methylation profiling provides a more robust and clinically useful platform for GBM subgrouping remains to be tested, but the logistics involved are probably less error-prone than for any RNA-based analyses.
Structural variations in glioblastoma
DNA copy-number aberrations (CNAs) are commonly observed in GBM, and can affect a significant fraction of the tumour genome67, 70. Assessing DNA copy-number using high-resolution techniques in large collections of tumour samples has allowed the precise characterization of focal CNA target regions, and genome-wide sequencing studies are beginning to unravel more complex structural rearrangements and hitherto unknown fusion events.
Chromosomal aberrations and genomic rearrangements
Genomic gains of chromosome 7 and losses of chromosome 10 (most often occurring concomitantly; 7+/10−) represent by far the most common gross chromosomal abnormalities in GBM, being detected in 83–85% of GBM in adults26, 67 (Table 1). Consistently throughout reported studies, 7+/10− is more frequent in older (age ≥ 70 years) compared to younger (age ≤ 40 years) patients71, and is therefore highly enriched (> 95 %) in GBM from the classical (or RTK II) subclass, but less common in GBM of the proneural gene expression subtype23, 25, and virtually absent in H3F3A- or IDH-mutant G-CIMP-positive tumours27, 40 (Figure 1). Further broad genomic copy-number changes seen at high frequency (> 20%) in adult GBM patients include gains of chromosomes 19 and 20 (35–40%; enriched in the classical (or RTK II) subtype), and losses affecting chromosomes 9p (38%), 22q (33%), 13q (33%), 14q (27%) and 6q (22%)26, 67 (Table 1). The number of chromosomal imbalances is generally lower in childhood HGG and a proportion of these tumours (~ 15%) lack any detectable copy-number abnormalities36, 38, 40, 72. Paediatric tumours also differ from their adult counterparts in displaying frequent gain of chromosome 1q (20%; enriched in H3F3A G34-mutated GBM), while only rarely harbouring aberrations of chromosomes 7 and 1036, 38.
Table 1.
Frequent chromosomal copy-number alterations in glioblastoma
| Genomic region | Copy-number change | Frequency (%)adult GBM* | Frequency (%)childhood HGG¶ | Subtype enrichment | References |
|---|---|---|---|---|---|
| Chromosome 1q | Single copy gain | 9 | 19–29 | G34 | 26, 36, 38 |
|
|
|||||
| Chromosome 6q | Single copy loss | 22 | 4–5 | non-H3/non-G-CIMP | 26, 36, 38 |
|
|
|||||
| Chromosome 7 | Single copy gain | 74–83 | 13–19 | non-H3/non-G-CIMP | 26, 36, 38, 40 |
|
|
|||||
| Chromosome 9p | Single copy loss | 33–38 | 11–18 | non-K27 | 26, 36, 38 |
|
|
|||||
| Chromosome 10 | Single copy loss | 80–86 | 16–38 | non-H3/non-G-CIMP | 26, 36, 38, 40 |
|
|
|||||
| Chromosome 13q | Single copy loss | 31–33 | 24–34 | G34, RTK I | 26, 36, 38 |
|
|
|||||
| Chromosome 14q | Single copy loss | 26–27 | 16–29 | RTK I | 26, 36, 38 |
|
|
|||||
| Chromosome 19 | Single copy gain | 35–40 | 1–13 | Classical, RTK II | 26, 36, 38 |
|
|
|||||
| Chromosome 20 | Single copy gain | 39 | 3–6 | Classical, RTK II | 26, 36, 38 |
|
|
|||||
| Chromosome 22q | Single copy loss | 33 | 1–16 | G-CIMP, MES, RTK II | 26, 36, 38 |
≥ 21 years;
< 21 years, diffuse intrinsic pontine glioma (DIPG), anaplastic astrocytoma WHO grade III, GBM; G-CIMP, glioma-CpG island methylator phenotype; RTK, receptor tyrosine kinase.
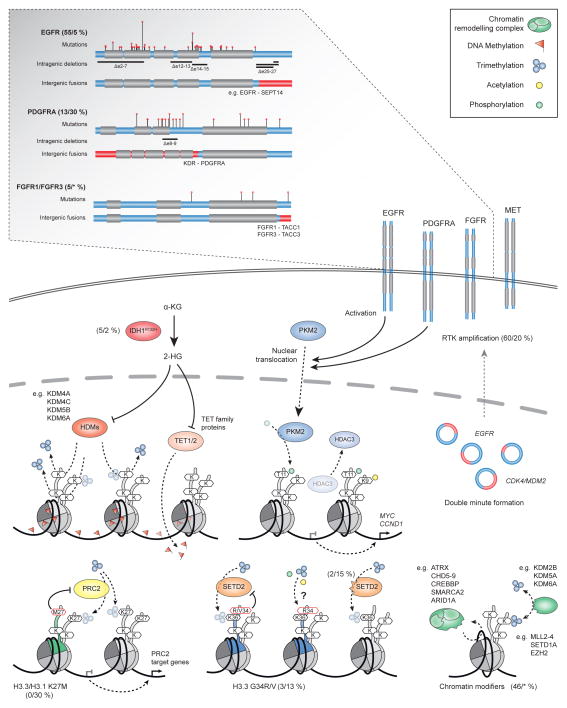
Recent data indicate that the incidence of complex chromosomal rearrangements in the context of a single catastrophic event (chromothripsis) is significantly higher in GBM (> 30%) relative to most other tumour types (9%), including breast, colon and lung cancer70. Interchromosomal, intrachromosomal (intergenic) and intragenic rearrangements can be detected in the majority (69%) of GBM samples, and these frequently (56%) co-occur with intragenic copy-number differences at the breakpoints73. The most prominent intragenic deletions in GBM target parts of the gene encoding either the extracellular domain of EGFR (exons 2–7 to form EGFRvIII) or the carboxy terminus73–76, and are almost always associated with amplification and co-expression of the wild-type EGFR allele26, 76, 77 (Figure 2 and Table 2). Similarly, a fraction of PDGFRA-amplified GBM from both adults and children harbour age-specific intragenic deletion rearrangements of this kinase with constitutively increased activity78, 79. Further intragenic deletions disrupt the function of the tumour suppressor candidate gene Fas-associated factor 1 (FAF1) as a result of focal deletion of the adjacent gene, CDKN2C73.
Figure 2. Interplay between the glioblastoma genome and epigenome.
Frequent genomic alterations in GBM impacting the epigenomic machinery. Receptor tyrosine kinases (RTKs) are commonly activated by somatic mutations or structural variations (see magnification), leading to PKM2 nuclear translocation and expression of MYC and CCND1. Recurrent somatic mutations in IDH1, IDH2, histone proteins, SETD2 and various other chromatin modifying proteins result in the disruption of multiple epigenetic regulatory processes by affecting histone modification, DNA methylation and chromatin remodelling. Numbers in brackets represent estimated frequencies of alterations observed in GBM of adults and childhood HGG, respectively (see Table 2 for exact numbers; *unknown). 2-HG, 2-hydroxyglutarate; α-KG, α-ketoglutarate; ARID1A, AT-rich interactive domain-containing protein 1A; ATRX, alpha thalassemia/mental retardation syndrome X-linked; CDK4, cyclin-dependent kinase 4; CHD, chromodomain helicase DNA binding protein; CREBBP, CREB-binding protein; EGFR, epidermal growth factor receptor; EZH2, enhancer of zeste homolog 2; FGFR, fibroblast growth factor receptor; HDM, histone demethylase; IDH1, isocitrate dehydrogenase 1; KDM, lysine (K)-specific demethylase; KDR, kinase insert domain receptor; MDM2, mouse double minute 2 homolog; MLL, mixed-lineage leukemia; PDGFRA, platelet-derived growth factor receptor type alpha; PKM2, pyruvate kinase muscle isozyme; PRC2, polycomb repression complex 2; RTK, receptor tyrosine kinase; SEPT14, septin 14; SETD1A, SET domain containing 1A; SETD2, SET domain containing 2; SMARCA2, SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2; TACC, transforming acidic coiled-coil.
Table 2.
Recurrent somatic alterations in glioblastoma
| Target gene(s) | Alteration type | Frequency (%)adult GBM* | Frequency (%)childhood HGG¶ | Subtype enrichment | References |
|---|---|---|---|---|---|
| EGFR | Focal amplification | 43 | 1–11 | Classical, Neural, RTK II | 25, 26, 36, 38, 40, 67, 74 |
|
| |||||
| Point mutation | 18–26 | 4 | Classical | 26, 27, 41 | |
|
| |||||
| Intragenic deletion (Δe2-7; EGFRvIII) | 10–64 | 17 | Classical | 26, 74, 76 | |
|
| |||||
| Intragenic deletion (Δe25-27) | 3–9 | NA | Classical | 26 | |
|
| |||||
| Intragenic deletion (Δe12-13/Δe14-15) | 29/3 | NA | Classical | 26 | |
|
| |||||
| Fusion | 8 | NA | Classical, Mesenchymal | 26, 32 | |
|
| |||||
| PDGFRA | Focal amplification | 11–26 | 8–39 | Proneural non-G-CIMP, RTK I | 25, 26, 36, 38, 40, 84, 217 |
|
| |||||
| Mutation | 3–4 | 4–9 | 4, 26, 41, 79 | ||
|
| |||||
| Intragenic deletion | 7–18 | 4 | 26, 78, 79 | ||
|
| |||||
| PTEN | Focal deletion | 8–10 | 0–1 | 26, 36, 38 | |
|
| |||||
| Mutation | 23–31 | 1–12 | non-G-CIMP | 26, 41, 218, 219 | |
|
| |||||
| NF1 | Focal deletion | 1 | 0–1 | Mesenchymal | 25, 26, 36, 38 |
|
| |||||
| Mutation | 9–10 | 24 | Mesenchymal | 25, 26, 41 | |
|
| |||||
| RB1 | Focal deletion | 3–4 | 0–3 | 26, 36, 38 | |
|
| |||||
| Mutation | 7–9 | 10 | Mesenchymal | 26, 41 | |
|
| |||||
| CDKN2A and CDKN2B | Focal deletion | 62 | 10–19 | non-H3/non-G-CIMP | 25, 26, 36, 38, 217 |
|
|
|||||
| TERT | Promoter mutation | 55–83 | 3–11 | non-H3/non-G-CIMP | 26, 91–93, 95 |
|
|
|||||
| TP53 | Mutation | 20–29 | 34–37 | K27, G34, G-CIMP | 25–27, 72, 220 |
|
|
|||||
| CDK4 | Focal amplification | 13–18 | 3–4 | Proneural non-G-CIMP | 26, 36, 38 |
|
|
|||||
| FGFR1 or FGFR3 | Fusion | 3–8 | NA | 82, 83 | |
|
|
|||||
| IDH1 | Mutation | 5–12 | 0–16 | G-CIMP | 25–27, 36, 40, 41, 86, 113, 221 |
|
|
|||||
| PIK3CA | Mutation | 7–21 | 8–20 | 26, 41, 222,§ | |
|
|
|||||
| PIK3R1 | Mutation | 6–11 | 10–12 | 26, 41,§ | |
|
|
|||||
| H3F3A G34R/V | Mutation | 0–3 | 12–14 | G34 | 26, 40, 41, 101 |
|
|
|||||
| H3F3A K27M | Mutation | 0–1 | 23–43 | K27 | 26, 40, 41, 101,§,‡ |
|
|
|||||
| HIST1H3B K27M or HIST1H3C K27M | Mutation | 0 | 12–31¥ | 26, 67, 101,§,‡ | |
|
|
|||||
| ATRX | Mutation | 6 | 14–29 | G34, G-CIMP | 26, 41, 97 |
|
|
|||||
| SETD2 | Mutation | 1–2 | 15 | 26, 110 | |
|
|
|||||
| ACVR1 | Mutation | 0 | 20–22¥ | 26,§,‡ | |
|
|
|||||
| MYCN | Focal amplification | 3 | 3–5 | 26, 36, 38, 217 | |
|
|
|||||
| MYC | Focal amplification | 2 | 3–4 | G-CIMP | 26, 36, 38 |
|
|
|||||
| BRAFV600E | Mutation | 2 | 10–25 | 26, 41, 223, 224 | |
≥ 21 years;
< 21 years, diffuse intrinsic pontine glioma (DIPG), anaplastic astrocytoma WHO grade III, GBM; ACVR1, activin a receptor type I; ATRX, alpha thalassemia/mental retardation syndrome X-linked; CDK4, cyclin-dependent kinase 4; CDKN2A and B, cyclin-dependent kinase inhibitor 2A and 2B; EGFR, epidermal growth factor receptor; FGFR, fibroblast growth factor receptor; G-CIMP, glioma-CpG island methylator phenotype; IDH1, isocitrate dehydrogenase 1; NF1, neurofibromin 1; PDGFRA, platelet-derived growth factor receptor type alpha; PIK3CA, phosphatidylinositol 3-kinase catalytic subunit alpha; PIK3R1, phosphatidylinositol 3-kinase regulatory subunit alpha; PTEN, phosphatase and tensin homolog; RB1, retinoblastoma 1; RTK, receptor tyrosine kinase; SETD2, SET domain containing 2; TERT, telomerase reverse transcriptase;
DIPG only;
C.J., J.G., O.B., C.H., J.M., N.J., unpublished observations;
Suzanne J. Baker, personal communication.
A breakpoint-enriched region on chromosome 12q (12q14-15), identified in ~ 5% of GBM, was recently shown to harbour copy-number equal co-amplification of the two known GBM oncogenes CDK4 and MDM2, assembled into double minute chromosomes73, 80. Additional fusion transcripts between genes included in the two amplicon segments were also observed73. This phenomenon was found to be mutually exclusive with CDKN2A and CDKN2B deletions and TP53 mutations, and associated with adverse patient survival73. Similarly, a number of gliomas were previously found to harbour double minute chromosomes bearing amplified copies of the EGFR gene81 (Figure 2). More recently, EGFR was shown to be further activated by recurrent translocations in 7% of GBM samples, most frequently being fused in-frame to septin 14 (SEPT14) or phosphoserine phosphatase (PSPH) as the 3′ gene segment, and almost always occurring within amplified regions of the fusion partner genes26, 32. In a smaller fraction of GBM (~ 3%), local inversion and in-frame fusion of the tyrosine kinase coding region of fibroblast growth factor receptor 1 (FGFR1) to the transforming acidic coiled-coil (TACC) coding domain of TACC1 (or alternatively fusion of FGFR3 to TACC3) results in constitutive kinase activity82, 83. Therefore, recurrent fusion events involving RTK-encoding genes may represent a promising therapeutic target and provide a strong rationale for the inclusion of these patients in future clinical trials using RTK inhibitors.
Focal copy-number aberrations
The spectrum of high-amplitude focal copy-number alterations in adult GBM highlights a key role of EGFR amplifications (43% of cases) and CDKN2A and CDKN2B deletions (53% of cases)26, although these events are rarely detected in paediatric HGG (< 5% and ~ 20% of cases, respectively)38, 40, 72 (Figure 1 and Table 2). Both events are enriched in the classical (or RTK II) and neural molecular subtypes25 and co-occur with EGFR intragenic deletions and/or point mutations25, 26, 76. Known genes less frequently targeted by homozygous deletion in adult GBM include PTEN (10%), RB1 (3%), CDKN2C and FAF1 (3%), QKI (1.6%), NF1 (1.3%), NPAS3 (1.3%) and TP53 (1%)26. High-level amplifications of CDK4 (13%), PDGFRA (11%), MDM2 (8%), MDM4 (7%), MET (2%) and CDK6 (1.5%) often co-occur in a variety of combinations26, with common co-amplification of RTK-encoding genes in a mosaic-like pattern (Box 1). Amplifications of CCND2, CCNE1, SOX2, MYC and MYCN represent recurrent but less frequent events in adult GBM (< 3%)26. Although to some extent most of these gains are also observed in paediatric HGG, focal amplifications of PDGFRA (14%) and MYC or MYCN (8%) are more frequent in the paediatric population38, 40, 72, 84. Recurrent focal amplification of PDGFRA has been suggested as a key oncogenic event in DIPG in a number of studies4, 37, 85. With increasing molecular data on treatment-naïve DIPG, however, the occurrence of this feature seems to be slightly lower than previously reported (< 40%), and enriched in DIPG harbouring histone H3 mutations40 (C.J., J.G., O.B., C.H. J.M. and N.J., unpublished observations). As PDGFRA amplification is somewhat more commonly observed in radiation-induced gliomas36, one might hypothesize a possible link between PDGFRA amplifications and prior radiotherapy, a caveat when studying post mortem DIPG (and GBM) tissue obtained from autopsy cases.
Box 1. Tumour heterogeneity in glioblastoma.
Intratumoural heterogeneity, referring to the presence of multiple, epigenetically and genetically different cell sub-populations within a single tumour, might contribute to tumour growth, progression and treatment failure180. In glioblastoma (GBM), the previously used term ‘multiforme’ describes the histopathologically observed co-existence of morphologically heterogeneous areas1. On a molecular level, this phenomenon is well reflected by different area-specific chromosome aberrations181–183, mutations184 and gene expression patterns22, 185, 186. Multiple receptor tyrosine kinase (RTK) genes (epidermal growth factor receptor (EGFR), platelet derived growth factor receptor α (PDGFRA) and MET) can be amplified and activated in a mutually exclusive manner in adjacent intermingled tumour cells178, 179, 187, and even multiple EGFR mutant alleles can commonly be expressed in a single tumour26. Arising from a single precursor and following an early driver event (such as loss of cyclin dependent kinase inhibitor 2A (CDKN2A) and CDKN2B)186, 187, activation of different RTKs in equally proliferating sub-populations may have distinct effects on downstream signalling pathways not only within but also between and among heterogeneous sub-populations via cell-cell interactions187. Indeed, a paracrine mechanism has been identified by which cells expressing EGFRvIII recruit wild-type EGFR-expressing cells into accelerated proliferation through upregulation of interleukin 6 (IL-6), leukaemia inhibitor factor (LIF) and GP13, thereby potently driving growth of the entire tumour mass and maintaining tumour cell heterogeneity188. These observations have important implications regarding the effects of RTK-targeted therapies, as tumours that have mosaic driver RTK amplifications will probably require the simultaneous use of more than one targeted agent178, 189. Furthermore, the impact of sampling bias over space and time is an important diagnostic consideration, particularly in the era of targeted therapy186. Tumour heterogeneity in GBM may also arise from clonal evolution, i.e. through the expansion and acquisition of mutations during tumour progression, promoting genetic variability190. This concept may be linked with the hierarchical cancer stem cell (CSC) model, which postulates a small sub-population of stem-like cells that give rise to a tumour and are maintained through their capability to self-renew and to produce diverse daughter cells that populate the tumour bulk191.
Mutational spectrum of glioblastoma
Advances in sequencing technology have led to the identification of increasing numbers of recurrent somatic mutations in the GBM genome of adult patients (Table 2). The number of coding mutations per tumour sample is highly variable (median: ~ 53; range: 3–179)26. Frequent mutations in genes such as PTEN (29%), TP53 (29%), EGFR (20%), NF1 (9%), RB1 (8%), phosphatidylinositol 3 catalytic α (PIK3CA; 7%), PIK3R1 (6%) and IDH1 (5%) have been reported as significant GBM signature events in earlier studies67, 86. The increasing number of samples with genome-wide sequencing data has now facilitated the detection of lower-frequency events in both cancer-related as well as previously un-associated genes such as spectrin α (SPTA1; 9%), ATRX (6%), KEL (5%), gaba-aminobyteric acid α6 (GABRA6; 4%), leucine zipper-like transcriptional regulator 1 (LZTR1; 3%), BCOR (2%), BRAF (2%), cateninδ2 (CTNND2 (2%)) and QKI (2%)26. Some of those genes (such as LZTR1, CTNND2, QKI, BRAF and BCOR) are recurrently targeted by both copy-number changes and point mutations, thus underscoring their role as new driver genes32. Of note, there is a non-random pattern of mutual exclusivities (such as PIK3R1 and PIK3CA)26, 67 (C.J., J.G., J.M. and N.J., unpublished observations) and co-occurrences (such as IDH1, TP53 and ATRX)26, 87, 88, and multiple but overall mutually exclusive mutations in genes implicated in regulation of chromatin modification26, a phenomenon recently observed in various other cancer types89, 90. Based on genome-wide sequencing data from 284 GBM samples generated by TCGA26, 46% of cases were found to have at least one somatic mutation in a gene associated with DNA methylation (IDH1), histone modifications (SET domain containing protein 2 (SETD2) and lysine-specific demethylase 6A (KDM6A), mixed lineage leukaemia 2 (MLL2), MLL3, MLL4, EZH2 and histone deacetylase 2 (HDAC2)) or chromatin remodelling (ATRX, CREB binding protein (CREBBP), chromodomain helicase DNA binding protein 5 (CHD5), CHD6, CHD7, CHD8, CHD9 and SWI/SNF-related matrix-associated, actin-dependent regulator of chromatin A2 (SMARCA2)) (Figure 2).
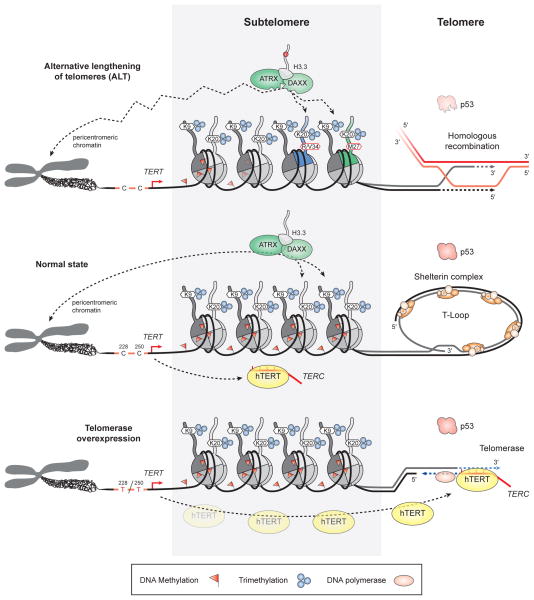
Importantly, 58–84% of primary GBM in adults have hotspot mutations in the TERT gene promoter. These mutations are tightly coupled with EGFR amplifications and inversely correlated with IDH1, IDH2, TP53 and ATRX mutations26, 91–95, and are associated with increased telomerase expression and activity26, 92, 94, 96 (Figure 3). TERT promoter mutations are found at a much lower rate (3–11%) in childhood GBM91, 95, which instead frequently display loss of ATRX and an alternative lengthening of telomeres (ALT) phenotype to maintain or increase telomere length41, 97.
Figure 3. Telomere maintenance mechanisms in glioblastoma.
Maintenance of telomere length is accomplished by different mechanisms in GBM. In normal cells (middle panel), chromosome ends are protected from undergoing non-homologous end joining or homologous recombination by forming a T-Loop structure. Alternative lengthening of telomeres (ALT), thought to be mediated by homologous recombination, is more prevalent in the paediatric setting (upper panel). While mutations in ATRX or DAXX (mediating incorporation of histone H3.3 into pericentromeric and subtelomeric regions) are known to promote ALT, the contribution of p53 loss, H3.3 mutations and/or subtelomeric DNA hypomethylation (such as in GBM harbouring H3.3 G34R/V mutation) needs further elucidation. Telomere length in adult GBM (lower panel) in the presence of functional p53 is mainly maintained by upregulated telomerase (hTERT) expression as a consequence of TERT promoter hotspot mutations C228T or C250T, for example. ATRX, alpha thalassemia/mental retardation syndrome X-linked; DAXX, death-domain associated protein; T-Loop, telomere loop; TERC, telomerase RNA component; TERT, telomerase reverse transcriptase.
Moreover, recent work has identified novel oncogenic mutations in PDGFRA in a proportion of paediatric HGG, often in combination with amplification of the PDGFRA locus4, 79. Of further note and clinical relevance, inactivating mutations in DNA mismatch repair (MMR) genes (such as MSH6) can be induced during TMZ treatment and evoke a GBM hypermutator phenotype, which is causally related to TMZ resistance67, 98–100.
Histone mutations in childhood high-grade gliomas
The first recurrent histone mutations in human cancer were uncovered by sequencing studies on a large number of paediatric GBM and DIPG patient samples41, 101. Heterozygous mutations in H3F3A result in amino acid substitutions at positions K27 or G34, and K27 was also found to be mutated in the H3.1 histone genes HIST1H3B and HIST1H3C41, 101 (C.J., J.G., O.B., C.H., J.M. and N.J., unpublished observations). These mutations directly or indirectly target important sites on the histone tail for posttranslational modifications (Figure 2). Occurring exclusively in ~ 38% of childhood and young adult (≤ 30 years) HGG41, 102 (Table 2) and in a mutually exclusive fashion with each other and with mutations in IDH1, each histone mutation shows a strong association with distinct molecular features as well as clinical patient characteristics5, 40, 42 (Figure 1). Mutations in H3F3A that result in amino acid changes at G34 occur in adolescents around the age of 20 years that have disease exclusively located in hemispheric regions. A significant number of these tumours have mutations in TP53, ATRX and (less frequently) DAXX40, 41, and commonly display a global pattern of DNA hypomethylation and an ALT phenotype40. Mutations in H3F3A that result in amino acid changes at K27 occur in 70–80% of midline GBM and DIPG in younger children5, 40, 101. The same substitutions in HIST1H3B or HIST1H3C seem to be less common and specific to DIPG (11–31%) and to coincide with novel activating mutations in ACVR1 (also known as ALK2) identified in ~ 20% of DIPG cases (C.J., J.G., O.B., C.H., J.M. and N.J., unpublished observations; Suzanne J. Baker, personal communication), providing a potentially druggable target in this subset. Interestingly, recent studies also detected H3F3A K27M mutations in a small fraction (< 2%) of midline pilocytic astrocytomas (WHO grade I)103, 104, conceivably indicating some overlap between the genetics of paediatric low-grade and high-grade gliomas or, alternatively, a remaining degree of diagnostic uncertainty. GBM harbouring mutations in histone H3 genes are mostly devoid of hallmark cytogenetic aberrations, such as EGFR alterations and CDKN2A and CDKN2B deletions observed in adult GBM patients40 (Figure 1). Interestingly, mutations at K27 seem to confer a dismal prognosis5, 40, while G34 mutations might be associated with slightly prolonged overall survival40. However, these initial observations will have to be tested in larger, prospective patient cohorts.
A gain-of-function K27M mutation in H3.3 or H3.1 leads to global downregulation of the repressive histone mark H3K27me3 through inhibition of the Polycomb repressive complex 2 (PRC2)44, 105–107. K27M-mutant H3.3 was shown to aberrantly bind PRC2 and interfere with the enzymatic activity of EZH2 – the catalytic subunit of PRC2 that establishes the H3K27me3 mark – possibly due to methionine mimicking the structure of monomethyl lysine106. Consequent loss of repressive PRC2 activity resulting from a combination of reduced H3K27me3 and DNA hypomethylation has therefore been proposed as the main driver of gliomagenesis in K27M-mutant HGG44, 106 (Figure 2).
In a similar vein, G34R and G34V-mutant H3.3 can interfere with the regulatory H3K36me3 modification43, 106. Genome-wide mapping of H3K36me3 in a G34V-mutant GBM cell line identified MYCN among the genes most strongly enriched for H3K36me3 marks, with increased RNA polymerase II binding and transcriptional upregulation of this locus in G34V-mutant cells43. Therefore, in addition to MYCN focal amplification, H3.3 G34 mutations may represent an alternative mechanism to overexpress MYCN, which was recently shown to drive glioma formation in neural stem cells (NSCs) in vivo108, and could potentially be targeted by bromodomain inhibition109. H3K36me3 is further disrupted by mutations in the H3K36 trimethyltransferase SETD2 in children and young adults, which are mutually exclusive with H3F3A G34 mutations110 (Figure 2). Besides its role in transcriptional elongation106, H3K36me3 was found to be important as a recruitment platform for MMR proteins111. Depletion of SETD2 and therefore H3K36me3 levels in HeLa cells leads to an increased spontaneous mutation frequency and chromosomal instability111. In line with this, altered histone modifications in subtelomeric regions have been suggested to contribute to deregulated telomere length, also resulting in chromosomal instability. This is reflected by a particularly high number of paediatric GBM displaying ALT41 (Figure 3). Although genetic hits in ATRX and DAXX (both essential components for H3.3 incorporation at pericentromeric heterochromatin and at telomeres) are known to promote the ALT phenotype, the contribution of mutant p53112, histone modifications and subtelomeric DNA hypomethylation to GBM development needs further investigation.
IDH mutations in glioblastomas of young adults
In the early stages of the next-generation sequencing era, the first genome-wide exon sequencing effort in glioma identified heterozygous hotspot mutations at codon 132 (most commonly R132H) in IDH1 in 12% of GBM86. Today, mutations in IDH1 (and to a lesser extent in IDH2) are commonly established as a hallmark molecular feature of secondary GBM in young adults (age 25–45 years; being present in ~ 70% compared to < 5% in primary GBM)113–116 with predominant localisation in the frontal and temporal lobes117. The small number of IDH-mutant primary GBM may therefore have rapidly progressed from clinically undiagnosed lower-grade astrocytomas. Mutations in the IDH genes are thought to be causative of G-CIMP within the proneural GBM subgroup23, 25, 27, 40, 118 (Figure 1). The role of mutations in IDH genes in gliomas with respect to molecular pathogenesis as well as clinical implications has recently been reviewed in detail119, 120. IDH mutations seem to require cooperating mutations in TP53 and ATRX87, 88, and possibly SETD2110, while being less frequently detected in GBM with classical RTK pathway alterations40, 67, 121. Besides being a powerful diagnostic marker122, IDH-mutant GBM have a favourable clinical outcome compared to their non-mutated counterparts26, 123. The main mechanism by which mutant IDH contributes to the pathogenesis of GBM is ascribed to the neomorphic enzymatic activity of mutant IDH proteins, which produce high amounts of the R enantiomer of 2-hydroxyglutarate (2-HG) (a putative oncometabolite) from 2-oxoglutarate (2-OG; also known as α-ketoglutarate)124, 125. 2-HG has been shown to inhibit a variety of 2-oxoglutarate-dependent dioxygenases, including key epigenetic regulators such as the DNA demethylating enzymes of the TET126 or Jumonji domain families127, 128 (Figure 2). Hence, the methylome of IDH-mutant GBM is dominated by widespread DNA hypermethylation at gene promoters (G-CIMP)118, irrespective of patient age40. The interaction of TET2 with particular transcription factors, such as EBF1, might result in specific rather than random gene promoter hypermethylation129. Although there is a strong association of IDH mutation with G-CIMP, early passage neurosphere cultures from IDH wild-type GBM can show a G-CIMP-like phenotype, suggesting intratumoural heterogeneity and/or the involvement of other, as yet unknown, factors34.
The cells of origin of glioblastoma
Although multiple genetic and epigenetic alterations are known to initiate or promote gliomagenesis, answers as to the cell of origin that acquires the first tumour-promoting mutation remain elusive. Several cell types including NSCs130, 131, more differentiated progeny such as oligodendrocyte precursors132, 133, or cells of the astrocytic lineage are discussed as putative glioma-initiating cells. Indeed, all these cell types can give rise to glioma if genetically manipulated (Box 2). Interestingly, malignant transformation seems not to be limited to undifferentiated cell types – transformation of mature neurons or astrocytes by depletion of Nf1 and Trp53 has also been shown to generate malignant gliomas in vivo134. Various pieces of evidence, including differences in tumour location, patient age, mutational spectrum or expression of neuronal lineage markers have led to growing speculation that in addition to acquiring distinct genetic events, at least some of the GBM subgroups described above also have unique cellular origins. This hypothesis might especially prove true for the tight correlations between molecular and clinical characteristics observed in DIPG and midline GBM, which possibly originate from recently identified pontine precursor-like cells135.
Box 2. In vivo tumour models of glioblastoma.
In vivo tumour models provide an indispensable tool to study the contribution of cancer-related genes or develop and evaluate novel treatment options. Strategies to reproduce glioblastoma (GBM) tumours in animals include xenograft transplantation models, germline modification of mouse strains and/or somatic gene transfer192, 193. Orthotopic injection of primary human GBM cells into immunocompromised mice gives rise to tumours partially retaining histological features and key molecular characteristics of human GBM, but with the disadvantages of altered tumour immunology and absence of a natural microenvironment194–197. To recapitulate the diversity of human GBM subgroups23, 25, 40, genetically engineered mouse models allowing for spontaneous tumour growth have been generated by adopting key glioma signature mutations in receptor tyrosine kinase (RTK) genes, TP53, PTEN, RB, NF1 and in cell-cycle pathways192, 193, 198, 199. While overexpression of epidermal growth factor receptor (EGFR) alone or EGFRvIII alone is insufficient in effectively promoting tumour formation from neuroglial precursors200 or astrocytic cells200, 201, concomitant overexpression of EGFRvIII (and less efficiently wild-type EGFR) with loss of the cyclin dependent kinase inhibitor 2A (Cdkn2a) and/or Pten and Trp53 loci is capable of inducing GBM202–205 that might resemble human tumours of the classical transcriptome subtype25, 40. Similarly, PDGF-driven gliomagenesis recapitulates GBM formation in mice when combined with germline loss of Cdkn2a, Pten, or Trp53206–208. This model has been successfully used to study paediatric and adult GBM biology192, 206, 209, 210, and is significantly enriched in proneural and RTK I GBM subtypes23, 25, 40. The mesenchymal subtype of GBM might be best investigated using established models based on loss-of-function of Nf1, which cooperates with early Trp53 mutations (and Pten haploinsufficiency) to induce astrocytomas of different grades131, 211–215. Although molecular profiling of genetically engineered GBM mouse models suggests significant similarities with the adult human disease199, they may not reflect intratumoural heterogeneity to the same degree observed in primary tumours, and many novel findings of recent genome-wide studies have not yet been successfully transferred into in vivo models, including recurrent mutations in isocitrate dehydrogenase 1 (IDH1)216, H3F3A, HIST1H3B106, ATRX and others. These are now being assessed in the context of reasonable co-oncogenic alterations in order to complement the currently available repertoire of GBM in vivo models.
It is widely thought that IDH1-mutant GBM might represent a distinct disease entity that probably arises from a different cellular origin compared with GBM in which IDH is not mutated136. Based on a current model, mutations of IDH1 represent an early event in gliomagenesis, priming quiescent neural progenitor cells for additional tumour-promoting genetic hits such as TP53 mutations or 1p/19q co-deletions, leading to the manifestation of WHO grade II/III gliomas. Subsequent malignant transformation into grade IV GBM is mediated by additional key genomic alterations (such as RTK pathway activation or PTEN mutations)136. This stepwise progression of secondary, IDH1-mutant GBM, with preservation of a proneural expression pattern, substantially differs from de novo GBM, which seem to originate as the result of genetic hits in a distinct cell population more closely resembling a NSC136.
Lineage tracing experiments have indicated that oligodendrocyte precursor cells (OPCs) can be considered as cells of origin for Nf1 and Trp53-mutant GBM, although identical genetic hits can also give rise to glioma when introduced into NSCs132. In a premalignant state, in vivo proliferation of Nf1;Trp53-mutant cells was found to be dramatically increased only in the OPC lineage and not NSCs or progeny lineages of astrocytes, neurons or oligodendrocytes. Fascinatingly, direct introduction of Nf1 and Trp53 mutations into OPCs led to the generation of gliomas that molecularly and histologically resembled tumours initiated from NSCs, with transcriptomes of both models corresponding well with the proneural subtype132.
Assuming that tumours partially retain the gene expression signature of their cellular origin, transcriptional profiles of H3F3A K27- and G34-mutant GBM were found to resemble those of distinct anatomical regions at different stages in the developing human brain40, 137. Expression of OLIG1 and OLIG2 was originally thought to be a common feature of human gliomas138–140 and OLIG2 was considered essential for promoting gliomagenesis in vivo141, 142. However, H3F3A G34-mutant GBM presumably arise from NSCs or undifferentiated progenitor cells (not of oligodendroglial lineage) which epigenetically repress OLIG1 and OLIG2 expression40 in order to preserve multipotency, unlike other glioma subtypes initiated from OLIG2-positive progenitor-like cells of the subventricular zone143.
Novel strategies in glioblastoma therapy
Current postoperative standard treatment for GBM patients is mainly based on unselective induction of DNA damage via radiotherapy and alkylating agents such as TMZ8. The majority of drugs specifically targeting key signalling pathways and mechanisms of gliomagenesis, such as RTK signalling (erlotinib144–147, gefitinib147, 148, cetuximab149 and imatinib150–153) or angiogenesis (bevacizumab154, 155 and cediranib156) do not provide a significant survival benefit when tested alone or in combination with other therapies (reviewed in157). Comparable results were obtained when combining these drugs with pharmacological histone deacetylase (HDAC) inhibitors (such as panobinostat158 and vorinostat158, 159), although the use of an HDAC inhibitor such as valproic acid in addition to radiotherapy was associated with a better outcome in patients with GBM160, 161. It should be noted, however, that efforts to identify molecularly defined patient subgroups for targeted therapies have generally been lacking, and it is possible that within the context of a negative clinical trial, subgroups of patients could be defined that would benefit from such a specific therapeutic approach. Indeed, the identification of several different molecular subgroups25, 27, 40 would argue against the use of one standardised therapy for GBM and the increasing knowledge about the genetic and/or epigenetic events driving these subgroups offers the opportunity to design innovative patient-tailored treatment protocols.
Given the high prevalence of IDH mutations in GBM and other malignancies such as acute myeloid leukaemia (AML), several studies have sought to therapeutically target the neomorphic enzymatic activity of mutant IDH1 and IDH2 using specific inhibitors162–164. Recently, the small molecule AGI-5198 was shown to inhibit R132H-mutant IDH1, leading to reduced levels of 2-HG and substantial growth reduction of glioma cells in vitro and human glioma xenografts in vivo162. Interestingly, pharmacological blockade of mutant IDH1 did not reverse the G-CIMP in glioma xenografts, but induced expression of genes associated with astrocytic and oligodendrocytic differentiation162. Although not tested in GBM yet, a recent study has reported on the successful inhibition of mutant IDH2 by a small molecule (AGI-6780) in AML164. This encouraging data demonstrates that these new drugs might represent an effective treatment option for patients with an IDH-mutant GBM.
In contrast, tumour growth and disease progression of GBM lacking mutations of IDH1 or IDH2 have recently been shown to utilize alternative energy resources such as branched-chain amino acids (BCAA) to compensate for increased metabolic demand. Expression of branched-chain amino acid transaminase 1 (BCAT1) was found to be highly dependent on intracellular levels of 2-OG, which is mainly produced by (wild-type) IDH1165. Impaired BCAA metabolism via knockdown of BCAT1 or overexpression of mutant IDH1 resulted in reduced cell proliferation165, rendering the reprogramming of energy metabolism an attractive target for innovative therapeutic approaches in GBM. Interestingly, the metabolic enzyme pyruvate kinase M2 (PKM2) was shown to have a non-metabolic function as a key regulator of histone phosphorylation and acetylation. Activation of EGFR (and also of PDGFR) results in activation of ERK2, which was found to phosphorylate PKM2 leading to nuclear translocation and promotion of the Warburg effect166. Nuclear PKM2 was found to phosphorylate histone H3 at threonine 11, causing dissociation of HDAC3 from the CCND1 and MYC gene promoters167 (Figure 2). Subsequent expression of cyclin D1 and MYC due to increased acetylation of histone H3 at lysine 9 was found to induce cell proliferation and gliomagenesis166.
The identification of histone mutations in a large proportion of HGG from paediatric and adolescent patients41, 101 has raised expectations for targeted treatment approaches that tackle these tumours40, 43. Although the precise mechanism of K27M-mediated PRC2 inhibition is not fully understood, pharmacological intervention targeting K27M-mutant H3.3 or downstream consequences of this change might represent the ultimate therapeutic goal. Care must be taken here, however, since the role of the PRC2 complex in oncogenesis seems to be highly context-dependent, and unspecific pharmacological H3K27me3 upregulation in K27M-mutant tumours could in fact promote disease progression, as has been shown for other malignancies harbouring EZH2 activating mutations168–170. The existence of ALT as the predominant telomere maintenance mechanism in the majority of H3-mutant HGG (especially those with G34R and G34V alterations)41 might also represent a promising biological target for the development of more powerful drugs in the future, as we begin to understand the underlying molecular details. In contrast, pharmacological inhibition of telomerase activity might be more effective in GBM in adult patients, given their dependence on high levels of telomerase expression171 (Figure 3). Despite the manifold differences between GBM subtypes, the evolving overall theme of epigenetic deregulation holds promise for epigenetic modulators to be active across subsets of epigenetically distinct tumours, as has recently been shown for a new class of agents targeting the BET domain proteins172.
Despite the identification of these multiple new targets in certain patient subgroups, bridging the gap between identification of recurrent genomic drivers, such as EGFR, and successful targeted therapies remains a considerable challenge, as indicated by the above-mentioned failures of many of the agents tested to date. The development of novel drugs requires accurate assessment of pharmacodynamic and pharmacokinetic parameters of drug activity in preclinical and clinical trials, and penetrance of the blood brain barrier is an additional challenge (although the integrity of this barrier in GBM is debatable). Although some drugs show detectable activity both in vitro and in vivo, downstream signal transducers may remain unaffected173, and even for drugs able to induce a clinical response in GBM patients, treated tumours ultimately progress. One reason for the failure of early clinical trials may be a lack of patient selection for a specific therapy, implying that molecular determinants of the response to a chosen intervention are needed174. Another could be pre-existing175 or emerging tumour resistance mechanisms in response to treatment, for example through activation of alternative RTKs as a result of EGFR inhibition176.
Due to the inter- and intratumoural molecular heterogeneity of GBM, it is likely that only a small proportion of patients will benefit from therapeutic interventions aimed at any one target, but synergistic combination approaches may prove more effective177–179. This likelihood raises the need for prospective tissue collection in order to maximize information gained from clinical trials. Molecular pathological stratification criteria are needed in this and other cancers to optimize enrolment in the therapeutic arm most adapted to tumour biology, and efficacy of a specific agent should be considered in the context of known and clinically relevant biological subsets of glioma.
Summary
Recent large-scale profiling studies have revealed a number of novel findings that have changed the way we look at the genomic and epigenomic landscape of GBM. The community is slowly gaining insight into the complex interactions between genetic alterations and changes in DNA methylation, histone modifications, chromatin remodelling and gene expression, and how deregulation on different levels might contribute to GBM pathogenesis. IDH and H3-mutated GBM are outstanding examples whereby disrupted epigenetic mechanisms due to genetic mutations result in the establishment of mutation-specific epigenetic and transcriptional programs. These and other recurrent genetic aberrations occur in a specific context of cellular origin, co-oncogenic hits and epigenetic phenomena, and are present in distinct patient populations. The biological discrimination of GBM subgroups should therefore guide the design of future clinical trials. In addition, the complex interplay of ‘hard-wired’ genetic events and potentially reversible alterations of the GBM epigenome offers novel opportunities for the development of molecularly targeted therapies, which might represent a promising strategy to tackle this deadly brain tumour.
Key points.
Glioblastoma is the most frequent and most aggressive malignant primary brain tumour and remains almost universally incurable in both children and adults.
Comprehensive molecular profiling studies have greatly broadened our knowledge of the underlying genomic and epigenomic aberrations associated with glioblastoma initiation and progression.
Genetic lesions result in disrupted epigenetic control mechanisms by altering histone modifications, DNA methylation and gene expression patterns in a large proportion of glioblastomas.
Based on recurrent combinations of genomic and/or epigenomic features with distinct patient characteristics, glioblastomas across all ages are being dissected into meaningful biological subgroups, which are likely to guide future clinical trial design.
The complex interplay between the glioblastoma genome and epigenome opens the avenue for the development of novel innovative therapeutic strategies that are urgently needed to tackle this deadly brain tumour.
Glossary
- WHO Classification of tumours of the central nervous system
Classification system where histological grading is applied as a means of predicting the biological behaviour of a tumour. It ranges from benign tumours (grade I) to highly aggressive, rapidly progressing tumours with frequently fatal outcome (grade IV)
- Glioma
Glioma refers to tumours that have histologic features similar to normal glial cells, i.e. astrocytes (astrocytoma), oligodendrocytes (oligodendroglioma), or ependymal cells (ependymoma), but is often used to imply only astrocytic or oligodendroglial tumours
- Diffuse intrinsic pontine glioma (DIPG)
Highly infiltrative glial tumour arising in the pons. Occurs almost exclusively in children, with a peak age at diagnosis of between 5 and 9 years
- Temozolomide (TMZ)
Alkylating chemotherapeutic agent used for the treatment of GBM. Triggers tumour cell death through extensive DNA damage
- CpG Island Methylator Phenotype (CIMP)
DNA methylation pattern of widespread CpG island promoter methylation. CIMP is frequently reported to be associated with distinct tumour subgroups, patient prognosis and response to treatment
- Chromothripsis
Clustered chromosomal rearrangements in one or a few chromosomes during cancer development, thought to occur through a one-step catastrophic genomic event
- High amplitude focal copy-number alteration
Small fragment (typically ~ 3 Mb or smaller in size) of amplified or homozygously deleted DNA, often resulting in numerous copies of oncogenes or deletion of both copies of tumour suppressor genes
- Double minute chromosomes
Small circular fragment of extrachromosomal DNA frequently harbouring one or more oncogenes
- Alternative lengthening of telomeres (ALT)
One or more mechanism(s) by which 5–10% of human cancers maintain or increase the overall length of their telomeres without the need of increased telomerase activity. The exact molecular mechanism(s) of ALT remain elusive, but may rely on recombination-mediated elongation
- Warburg effect
Predominant production of energy by a high rate of glycolysis followed by lactic acid fermentation in the cytosol observed in most cancer cells in the presence of oxygen
- Polycomb repressive complex 2 (PRC2)
One of two classes of polycomb-group proteins. PRC2 has methyltransferase activity and primarily trimethylates histone H3 on lysine 27 (i.e. H3K27me3), a mark of transcriptionally silent chromatin
Biographies
Dominik Sturm
Dominik Sturm is a physician scientist in the Division of Pediatric Neurooncology at the German Cancer Research Center (DKFZ), Heidelberg, Germany. His recent research activities focused on elucidating the molecular biology underlying childhood high-grade gliomas and diffuse intrinsic pontine gliomas, and applying novel molecular diagnostic tools to improve childhood brain tumour classification. Having received his MD in 2011, he is currently in clinical training as a paediatrician at the Department of Pediatric Oncology and Hematology at the Heidelberg University Medical Center for Children and Adolescents.
Sebastian Bender
Sebastian Bender is a postdoctoral research fellow in S.M.P.’s laboratory at the German Cancer Research Center (DKFZ), Heidelberg, Germany. He received his PhD from the University of Heidelberg, Germany, in 2013 for studying the molecular consequences of histone H3.3 mutations in paediatric high-grade gliomas. His main research focus is the characterization of molecular mechanisms driving tumourigenesis in childhood brain tumours.
David T.W. Jones
David Jones is a senior postdoctoral scientist at the DKFZ, where he has played a major part in the International Cancer Genome Consortium (ICGC) PedBrain Tumour sequencing project. He received his PhD from the University of Cambridge in 2009, working in the group of V. Peter Collins. In 2008, he was the first to describe a highly recurrent BRAF fusion gene occurring in two-thirds of the most common childhood brain tumour (pilocytic astrocytoma). His primary research focus is the application of cutting-edge genomics techniques to identify new diagnostic, prognostic and therapeutic targets in the field of neurooncology.
Peter Lichter
Peter Lichter pioneered the development of technologies to delineate almost any chromosomal region by fluorescence in situ hybridization (FISH) and to detect DNA copy-number alterations via high resolution comparative genomic hybridization (arrayCGH). Applying these, as well as next-generation DNA sequencing approaches, he made major contributions to decipher the higher order genome organization and to elucidate pathomechanisms of tumour aetiology and progression, including the description of novel prognostic or predictive gene signatures.
Jacques Grill
Jacques Grill is the Head of the Pediatric Brain Tumour Program at Gustave Roussy, Villejuif, France, and Principal Investigator of Several International Trials including HERBY, a randomized phase II study for paediatric non-brainstem high-grade gliomas and BIOMEDE, a biology-driven trial for diffuse intrinsic pontine glioma. He is the group leader of a research team of the CNRS Unit 8203 dedicated to the discovery of new biomarkers and therapeutics targets in glial tumours.
Oren Becher
Oren Becher is an assistant professor in the Departments of Pediatrics and Pathology at Duke University. His laboratory is focused on the development of genetically engineered Diffuse Intrinsic Pontine Glioma (DIPG) mouse models, dissection of the function of novel genetic alterations in DIPG, and the evaluation of the novel therapeutics.
Cynthia Hawkins
Cynthia Hawkins is a paediatric neuropathologist and scientist at The Hospital for Sick Children (SickKids) in Toronto, Canada. Her clinical practice includes diagnosis of paediatric brain tumours and she serves as a central pathology reviewer for several national and international paediatric brain tumour clinical trials. The Hawkins laboratory is focused on the study of paediatric high grade glioma, in particular diffuse intrinsic pontine glioma. Her lab uses cancer genetic techniques in combination with in vitro and in vivo models to better understand what cellular processes underlie paediatric high grade glioma and how we can use this information to improve our ability to diagnose and treat children with brain tumours.
Jacek Majewski
Dr. Jacek Majewski obtained his BS in Physics and MS in Electrical Engineering from Stanford, and PhD in Biology from Wesleyan University. Having vowed never to live in New York, he nevertheless followed his fiancée to the Rockefeller University as a post-doctoral fellow in statistical genetics. He is currently a Canada Research Chair and Associate Professor at the Department of Human Genetics at McGill University. His research involves genomics and analysis of high throughput data aimed at understanding human genetic disorders. The approaches include RNA sequencing, exome, and whole genome sequencing, with specific applications to monogenic disorders and cancer.
Chris Jones
Chris Jones is a research team leader at in the Divisions of Molecular Pathology and Cancer Therapeutics at The Institute of Cancer Research (ICR) in London, UK. He completed his first degree in toxicology and pharmacology and later a PhD in molecular biology at the School of Pharmacy, University of London, UK. He moved to the ICR in 2001 and took up his post focusing on the genomics of childhood cancer in 2003. He is a Fellow of the Royal College of Pathologists by published works and is active in international paediatric brain tumour collaborative groups and clinical trials.
Joseph F. Costello
Dr. Costello is a Professor of Neurosurgery at UCSF, holds the Karen Osney Brownstein Endowed Chair in Neuro-oncology and serves as the Director of the UCSF-based NIH Roadmap Epigenome Mapping Center. Dr. Costello’s laboratory investigates how genetic and epigenetic mechanisms interact in the control of gene expression. His research includes epigenetic technology development and its application to human stem cells and tissues from different developmental stages. His translational research investigates tumour genome and epigenome evolution by integrating treatment, advanced imaging and histologic analyses with patterns of mutations and epimutations.
Antonio Iavarone
Antonio Iavarone received his MD at the Catholic University of Rome, where he also received the Residency degree in Paediatrics and performed a fellowship training in Paediatric Oncology. He worked at UCSF, Memorial Sloan Kettering Cancer Center and Albert Einstein College of Medicine in New York. He is Professor of Neurology and Pathology at Columbia University. One of the first discoveries made by his group is the now widely accepted notion that the most aggressive forms of brain tumours are invariably associated with loss of differentiated features. In more recent years his laboratory identified the master regulators and key genomic drivers (gene fusions and mutations) responsible for initiation, progression and maintenance of malignant glioma.
Kenneth Aldape
Kenneth Aldape is a neuropathologist with an interest in basic and translational research in brain tumours. He has a particular interest in molecular classification of glioma. His research has encompassed expression profiling and use of other genome-wide platforms to identify subclasses of these tumours. He is particularly interested in clinically relevant molecular alterations that can inform treatment and potentially lead to precision medicine for patients with primary and metastatic brain tumours.
Cameron W. Brennan
Cameron Brennan is an oncologic neurosurgeon and a laboratory investigator focused on tumour molecular profiling and brain tumour drug responses. He received his MD and neurosurgery training at New York Hospital – Cornell Medical Center, followed by postdoctoral work in Ronald DePinho’s laboratory at the Dana-Farber Cancer Institute. He joined Memorial Sloan-Kettering Cancer Center in 2004 and opened a laboratory in 2009 within the Human Oncology and Pathogenesis Program. He has been a part of The Cancer Genome Atlas from its inception, initially developing methods to identify DNA structural rearrangements and recently as external co-chair of the glioblastoma initiative.
Nada Jabado
Nada Jabado is an Associate Professor with tenure in the Department of Paediatrics and an associate member of the departments of Human Genetics, Oncology, and Experimental Medicine at McGill University. She obtained her MD with a specialization in Paediatrics and a PhD in Immunology from the Université de Paris VI in France. Her work aims to identify molecular alterations underlying paediatric gliomas. She has authored/co-authored 99 peer-reviewed papers in her career and received several awards recognizing her contribution to the field of cancer including the William E. Rawl young investigator award from the National Canadian Cancer Research Council in 2012.
Stefan M. Pfister
Stefan M. Pfister was appointed Head of the Division of Pediatric Neurooncology at the DKFZ in 2012. He is a paediatrician by training, and received his MD from Tübingen University, Germany, and his clinical education at Mannheim and Heidelberg University Hospitals. As a physician scientist, he completed postdoctoral fellowships with Christopher Rudd at the Dana-Faber Cancer Institute/Harvard Medical School, Boston, USA, and with Peter Lichter at the DKFZ, Division of Molecular Genetics, Heidelberg, Germany. His research is focused on the genetic characterization of childhood brain tumours through the application of next-generation profiling methods and subsequently translating novel findings into a clinical context.
Footnotes
Copyright information
None of the figures or other display items have been modified or reproduced from any other publication (either in print or online).
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO Classification of tumors of the central nervous system. IARC; Lyon: 2007. [Google Scholar]
- 2.Dolecek T, Propp J, Stroup N, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro-oncology. 2012;14 (Suppl 5):49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puget S, et al. Mesenchymal transition and PDGFRA amplification/mutation are key distinct oncogenic events in pediatric diffuse intrinsic pontine gliomas. PLoS One. 2012;7:e30313. doi: 10.1371/journal.pone.0030313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khuong-Quang DA, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124:439–47. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren K. Diffuse intrinsic pontine glioma: poised for progress. Front Oncol. 2012;2:205. doi: 10.3389/fonc.2012.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cage T, et al. Feasibility, safety, and indications for surgical biopsy of intrinsic brainstem tumors in children. Child Nerv Syst. 2013;29:1313–9. doi: 10.1007/s00381-013-2101-0. [DOI] [PubMed] [Google Scholar]
- 8.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. This was the first study to show that the addition of temozolomide to radiotherapy results in a statistically significant survival benefit for newly diagnosed glioblastoma patients. [DOI] [PubMed] [Google Scholar]
- 9.Stupp R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 10.Cohen KJ, et al. Temozolomide in the treatment of high-grade gliomas in children: a report from the Children’s Oncology Group. Neuro-Oncology. 2011;13:317–23. doi: 10.1093/neuonc/noq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hargrave D, Bartels U, Bouffet E. Diffuse brainstem glioma in children: critical review of clinical trials. Lancet Oncol. 2006;7:241–248. doi: 10.1016/S1470-2045(06)70615-5. [DOI] [PubMed] [Google Scholar]
- 12.Janssens G, et al. Hypofractionation vs conventional radiation therapy for newly diagnosed diffuse intrinsic pontine glioma: a matched-cohort analysis. Int J Radiat Oncol. 2013;85:315–320. doi: 10.1016/j.ijrobp.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Jansen MH, van Vuurden DG, Vandertop WP, Kaspers GJ. Diffuse intrinsic pontine gliomas: a systematic update on clinical trials and biology. Cancer Treat Rev. 2012;38:27–35. doi: 10.1016/j.ctrv.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Godard S, et al. Classification of human astrocytic gliomas on the basis of gene expression: a correlated group of genes with angiogenic activity emerges as a strong predictor of subtypes. Cancer Res. 2003;63:6613–6625. [PubMed] [Google Scholar]
- 15.Shai R, et al. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene. 2003;22:4918–4923. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- 16.Maher E, et al. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res. 2006;66:11502–11513. doi: 10.1158/0008-5472.CAN-06-2072. [DOI] [PubMed] [Google Scholar]
- 17.Tso CL, et al. Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res. 2006;66:159–167. doi: 10.1158/0008-5472.CAN-05-0077. [DOI] [PubMed] [Google Scholar]
- 18.Faury D, et al. Molecular profiling identifies prognostic subgroups of pediatric glioblastoma and shows increased YB-1 expression in tumors. J Clin Oncol. 2007;25:1196–208. doi: 10.1200/JCO.2006.07.8626. [DOI] [PubMed] [Google Scholar]
- 19.Nutt C, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- 20.Freije W, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 21.Shirahata M, et al. Gene expression-based molecular diagnostic system for malignant gliomas is superior to histological diagnosis. Clin Cancer Res. 2007;13:7341–7356. doi: 10.1158/1078-0432.CCR-06-2789. [DOI] [PubMed] [Google Scholar]
- 22.Liang Y, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. P Natl Acad Sci USA. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips H, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. One of the first studies to classify glioblastoma into three distinct molecular subclasses based on gene expression profiling signatures associated with differences in patient survival. [DOI] [PubMed] [Google Scholar]
- 24.Gravendeel L, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 25.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. This TCGA study discovered four distinct subtypes of glioblastoma distinguished by gene expression patterns associated with distinct genetic aberrations and clinical characteristics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. This most recent TCGA study describes the landscape of somatic genomic alterations based on multi-dimensional and comprehensive characterization of more than 500 glioblastoma tumors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. This was the first TCGA study to apply genome-wide DNA methylation profiling and identified a Glioma-CpG Island Methylator Phenotype (G-CIMP) to be linked to less severe outcome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huse J, Phillips H, Brennan C. Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia. 2011;59:1190–1199. doi: 10.1002/glia.21165. [DOI] [PubMed] [Google Scholar]
- 29.Zheng S, Chheda MG, Verhaak RG. Studying a complex tumor: potential and pitfalls. Cancer J. 2012;18:107–14. doi: 10.1097/PPO.0b013e3182431c57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–25. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhat K, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–2609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frattini V, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–9. doi: 10.1038/ng.2734. This manuscript reports the genomic landscape of driver genes targeted by both mutations and copy-number aberrations through an integrated computational and experimental pipeline. It also reports the landscape of gene fusions in GBM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danussi C, et al. RHPN2 Drives Mesenchymal Transformation in Malignant Glioma by Triggering RhoA Activation. Cancer Res. 2013;73:5140–5150. doi: 10.1158/0008-5472.CAN-13-1168-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhat KP, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–46. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haque T, et al. Gene expression profiling from formalin-fixed paraffin-embedded tumors of pediatric glioblastoma. Clin Cancer Res. 2007;13:6284–6292. doi: 10.1158/1078-0432.CCR-07-0525. [DOI] [PubMed] [Google Scholar]
- 36.Paugh BS, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28:3061–8. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paugh BS, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29:3999–4006. doi: 10.1200/JCO.2011.35.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bax DA, et al. A distinct spectrum of copy number aberrations in paediatric high grade gliomas. Clin Cancer Res. 2010;16:3368–3377. doi: 10.1158/1078-0432.CCR-10-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qu HQ, et al. Genome-wide profiling using single-nucleotide polymorphism arrays identifies novel chromosomal imbalances in pediatric glioblastomas. Neuro-Oncology. 2010;12:153–63. doi: 10.1093/neuonc/nop001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturm D, et al. Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell. 2012;22:425–37. doi: 10.1016/j.ccr.2012.08.024. This study applied genome-wide DNA methylation profiling to a combined cohort of paediatric and adult patients and described biological subgroups of glioblastomas associated with distinct molecular aberrations and clinical characteristics. [DOI] [PubMed] [Google Scholar]
- 41.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. Together with Wu et al. 2012, these were the first studies to identify recurrent H3F3A/HIST1H3B mutations in paediatric high-grade gliomas and the first reports on somatic histone mutations in human cancer. [DOI] [PubMed] [Google Scholar]
- 42.Fontebasso A, Liu XY, Sturm D, Jabado N. Chromatin remodeling defects in pediatric and young adult glioblastoma: a tale of a variant histone 3 tail. Brain Pathol. 2013;23:210–216. doi: 10.1111/bpa.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bjerke L, et al. Histone H3.3 Mutations Drive Pediatric Glioblastoma through Upregulation of MYCN. Cancer Discov. 2013;3 (5):512–519. doi: 10.1158/2159-8290.CD-12-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bender S, et al. Reduced H3K27me3 and DNA Hypomethylation Are Major Drivers of Gene Expression in K27M Mutant Pediatric High-Grade Gliomas. Cancer cell. 2013;24:660–72. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 45.Costello J, Berger M, Huang H, Cavenee W. Silencing of p16/CDKN2 expression in human gliomas by methylation and chromatin condensation. Cancer Res. 1996;56:2405–2410. [PubMed] [Google Scholar]
- 46.Baeza N, Weller M, Yonekawa Y, Kleihues P, Ohgaki H. PTEN methylation and expression in glioblastomas. Acta Neuropathol. 2003;106:479–485. doi: 10.1007/s00401-003-0748-4. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura M, Yonekawa Y, Kleihues P, Ohgaki H. Promoter hypermethylation of the RB1 gene in glioblastomas. Lab Invest. 2001;81:77–82. doi: 10.1038/labinvest.3780213. [DOI] [PubMed] [Google Scholar]
- 48.Amatya V, Naumann U, Weller M, Ohgaki H. TP53 promoter methylation in human gliomas. Acta Neuropathol. 2005;110:178–184. doi: 10.1007/s00401-005-1041-5. [DOI] [PubMed] [Google Scholar]
- 49.Alaminos M, et al. EMP3, a myelin-related gene located in the critical 19q13.3 region, is epigenetically silenced and exhibits features of a candidate tumor suppressor in glioma and neuroblastoma. Cancer Res. 2005;65:2565–2571. doi: 10.1158/0008-5472.CAN-04-4283. [DOI] [PubMed] [Google Scholar]
- 50.Bruna A, et al. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer cell. 2007;11:147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 51.Waha A, et al. Epigenetic silencing of the protocadherin family member PCDH-gamma-A11 in astrocytomas. Neoplasia. 2005;7:193–199. doi: 10.1593/neo.04490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou H, et al. Reciprocal regulation of SOCS 1 and SOCS3 enhances resistance to ionizing radiation in glioblastoma multiforme. Clin Cancer Res. 2007;13:2344–2353. doi: 10.1158/1078-0432.CCR-06-2303. [DOI] [PubMed] [Google Scholar]
- 53.Zardo G, et al. Integrated genomic and epigenomic analyses pinpoint biallelic gene inactivation in tumors. Nat Genet. 2002;32:453–458. doi: 10.1038/ng1007. [DOI] [PubMed] [Google Scholar]
- 54.Esteller M, Hamilton S, Burger P, Baylin S, Herman J. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. First study to show inactivation by promoter hypermethylation of the MGMT gene in glioblastoma patient samples. [PubMed] [Google Scholar]
- 55.Felsberg J, et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer. 2011;129:659–670. doi: 10.1002/ijc.26083. [DOI] [PubMed] [Google Scholar]
- 56.Wick W, et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13:707–715. doi: 10.1016/S1470-2045(12)70164-X. [DOI] [PubMed] [Google Scholar]
- 57.Malmström A, et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13:916–926. doi: 10.1016/S1470-2045(12)70265-6. References 56 and 57 demonstrated MGMT promoter methylation to be a predictive biomarker for the benefit from alkylating agent chemotherapy particularly in elderly glioblastoma patients. [DOI] [PubMed] [Google Scholar]
- 58.Costello J, Futscher B, Tano K, Graunke D, Pieper R. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem. 1994;269:17228–17237. First study to report on hypermethylation of the MGMT gene in human glioma cells. [PubMed] [Google Scholar]
- 59.Costello J, Futscher B, Kroes R, Pieper R. Methylation-related chromatin structure is associated with exclusion of transcription factors from and suppressed expression of the O-6-methylguanine DNA methyltransferase gene in human glioma cell lines. Mol Cell Biol. 1994;14:6515–6521. doi: 10.1128/mcb.14.10.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Esteller M, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 61.Hegi ME, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. This study demonstrated that the presence of an inactivated MGMT gene confers benefit from alkylating agent chemotherapy using Temozolomide. [DOI] [PubMed] [Google Scholar]
- 62.Olson R, Brastianos P, Palma D. Prognostic and predictive value of epigenetic silencing of MGMT in patients with high grade gliomas: a systematic review and meta-analysis. J Neuro-Oncol. 2011;105:325–335. doi: 10.1007/s11060-011-0594-5. [DOI] [PubMed] [Google Scholar]
- 63.Donson A, Addo-Yobo S, Handler M, Gore L, Foreman N. MGMT promoter methylation correlates with survival benefit and sensitivity to temozolomide in pediatric glioblastoma. Pediatr Blood Cancer. 2007;48:403–407. doi: 10.1002/pbc.20803. [DOI] [PubMed] [Google Scholar]
- 64.Buttarelli F, et al. Evaluation status and prognostic significance of O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation in pediatric high grade gliomas. Child Nerv Syst. 2010;26:1051–1056. doi: 10.1007/s00381-010-1191-1. [DOI] [PubMed] [Google Scholar]
- 65.Srivastava A, et al. MGMT gene promoter methylation in pediatric glioblastomas. Child Nerv Syst. 2010;26:1613–1618. doi: 10.1007/s00381-010-1214-y. [DOI] [PubMed] [Google Scholar]
- 66.Lee J, et al. MGMT promoter gene methylation in pediatric glioblastoma: analysis using MS-MLPA. Child Nerv Syst. 2011;27:1877–1883. doi: 10.1007/s00381-011-1525-7. [DOI] [PubMed] [Google Scholar]
- 67.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. This first comprehensive TCGA study on 206 glioblastoma samples reported three core signalling pathways (RTK/RAS/PI-3K, P53, and RB) implicated in glioblastoma development and treatment resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cadieux B, Ching TT, VandenBerg S, Costello J. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 69.Hovestadt V, et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta neuropathologica. 2013;125:913–916. doi: 10.1007/s00401-013-1126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malhotra A, et al. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 2013;23:762–776. doi: 10.1101/gr.143677.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bozdag S, et al. Age-specific signatures of glioblastoma at the genomic, genetic, and epigenetic levels. PloS One. 2013;8:e62982. doi: 10.1371/journal.pone.0062982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones C, Perryman L, Hargrave D. Paediatric and adult malignant glioma: close relatives or distant cousins? Nat Rev Clin Oncol. 2012;9:400–413. doi: 10.1038/nrclinonc.2012.87. [DOI] [PubMed] [Google Scholar]
- 73.Zheng S, et al. A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival. Genes Dev. 2013;27:1462–1472. doi: 10.1101/gad.213686.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bax D, et al. EGFRvIII deletion mutations in pediatric high-grade glioma and response to targeted therapy in pediatric glioma cell lines. Clin Cancer Res. 2009;15:5753–5761. doi: 10.1158/1078-0432.CCR-08-3210. [DOI] [PubMed] [Google Scholar]
- 75.Cho J, et al. Glioblastoma-derived epidermal growth factor receptor carboxyl-terminal deletion mutants are transforming and are sensitive to EGFR-directed therapies. Cancer Res. 2011;71:7587–7596. doi: 10.1158/0008-5472.CAN-11-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gan H, Cvrljevic A, Johns T. The epidermal growth factor receptor variant III (EGFRvIII): where wild things are altered. FEBS J. 2013;280:5350–70. doi: 10.1111/febs.12393. [DOI] [PubMed] [Google Scholar]
- 77.Biernat W, Huang H, Yokoo H, Kleihues P, Ohgaki H. Predominant expression of mutant EGFR (EGFRvIII) is rare in primary glioblastomas. Brain Pathol. 2004;14:131–136. doi: 10.1111/j.1750-3639.2004.tb00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ozawa T, et al. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24:2205–2218. doi: 10.1101/gad.1972310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paugh B, et al. Novel Oncogenic PDGFRA Mutations in Pediatric High-Grade Gliomas. Cancer Res. 2013;73:6219–29. doi: 10.1158/0008-5472.CAN-13-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanborn J, et al. Double minute chromosomes in glioblastoma multiforme are revealed by precise reconstruction of oncogenic amplicons. Cancer Res. 2013;73:6036–45. doi: 10.1158/0008-5472.CAN-13-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vogt N, et al. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. P Natl Acad Sci USA. 2004;101:11368–73. doi: 10.1073/pnas.0402979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh D, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–5. doi: 10.1126/science.1220834. The first report of a recurrent gene fusion (FGFR-TACC) in GBM. This study has pawed the way to a personalized tailored therapeutic approach with FGFR inhibitors for GBM harbouring FGFR-TACC fusions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parker B, et al. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest. 2013;123:855–865. doi: 10.1172/JCI67144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Phillips J, et al. PDGFRA Amplification is Common in Pediatric and Adult High-Grade Astrocytomas and Identifies a Poor Prognostic Group in IDH1 Mutant Glioblastoma. Brain Pathol. 2013;23:565–73. doi: 10.1111/bpa.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zarghooni M, et al. Whole-Genome Profiling of Pediatric Diffuse Intrinsic Pontine Gliomas Highlights Platelet-Derived Growth Factor Receptor {alpha} and Poly (ADP-ribose) Polymerase As Potential Therapeutic Targets. J Clin Oncol. 2010;28:1337–44. doi: 10.1200/JCO.2009.25.5463. [DOI] [PubMed] [Google Scholar]
- 86.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. This comprehensive analysis discovered of a variety of genes that were not known to be altered in glioblastoma, most notably recurrent hotspot mutations in IDH1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu XY, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124:615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 88.Jiao Y, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dawson M, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 90.Shen H, Laird P. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Killela P, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. P Natl Acad Sci USA. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. This study was the first to report recurrent somatic mutations in the TERT gene promoter as a likely mechanism of telomerase activation in a large proportion of glioblastomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arita H, et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013;126:267–276. doi: 10.1007/s00401-013-1141-6. [DOI] [PubMed] [Google Scholar]
- 93.Nonoguchi N, et al. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013;126:931–7. doi: 10.1007/s00401-013-1163-0. [DOI] [PubMed] [Google Scholar]
- 94.Vinagre J, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 95.Koelsche C, et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013;126:907–15. doi: 10.1007/s00401-013-1195-5. [DOI] [PubMed] [Google Scholar]
- 96.Boldrini L, et al. Telomerase activity and hTERT mRNA expression in glial tumors. Int J Oncol. 2006;28:1555–1560. doi: 10.3892/ijo.28.6.1555. [DOI] [PubMed] [Google Scholar]
- 97.Heaphy C, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333(425) doi: 10.1126/science.1207313. This was the first study to report on the presence of an alternative lengthening of telomeres (ALT) phenotype ocurring in tumours with mutation in ATRX or DAXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hunter C, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cahill D, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yip S, et al. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15:4622–4629. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–3. doi: 10.1038/ng.1102. Together with Schwartzentruber et al. 2012, these were the first studies to identify recurrent H3F3A/HIST1H3B mutations in paediatric high-grade gliomas and the first reports on somatic histone mutations in human cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Downing J, et al. The Pediatric Cancer Genome Project. Nat Genet. 2012;44:619–622. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang J, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jones DTW, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Venneti S, et al. Evaluation of Histone 3 Lysine 27 Trimethylation (H3K27me3) and Enhancer of Zest 2 (EZH2) in Pediatric Glial and Glioneuronal Tumors Shows Decreased H3K27me3 in H3F3A K27M Mutant Glioblastomas. Brain Pathol. 2013;23:558–564. doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lewis P, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. This was the first study to show the functional inhibition of PRC2 by a gain-of-function mutation at position K27 of histone variant H3.3, leading to globally reduced levels of H3K27me3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chan KM, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Swartling FJ, et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell. 2012;21:601–13. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Puissant A, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer discovery. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fontebasso A, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 2013;125:659–669. doi: 10.1007/s00401-013-1095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li F, et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen YJ, et al. Association of mutant TP53 with alternative lengthening of telomeres and favorable prognosis in glioma. Cancer Res. 2006;66:6473–6476. doi: 10.1158/0008-5472.CAN-06-0910. [DOI] [PubMed] [Google Scholar]
- 113.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. This study identified IDH1/2 mutations in the majority of lower grade gliomas and secondary glioblastomas associated with better outcome than those with wild-type IDH genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ichimura K, et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro-Oncology. 2009;11:341–7. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang C, Moore L, Li X, Yung W, Zhang W. IDH1/2 mutations target a key hallmark of cancer by deregulating cellular metabolism in glioma. Neuro-oncology. 2013;15:1114–26. doi: 10.1093/neuonc/not087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19:764–772. doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- 117.Larjavaara S, et al. Incidence of gliomas by anatomic location. Neuro-oncology. 2007;9:319–325. doi: 10.1215/15228517-2007-016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. This study was the first to show a causal link between mutations in IDH1 and the Glioma-CpG Island Methylator Phenotype (G-CIMP) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim W, Liau L. IDH mutations in human glioma. Neurosurg Clin N Am. 2012;23:471–480. doi: 10.1016/j.nec.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ichimura K. Molecular pathogenesis of IDH mutations in gliomas. Brain Tumor Pathol. 2012;29:131–139. doi: 10.1007/s10014-012-0090-4. [DOI] [PubMed] [Google Scholar]
- 121.Lass U, et al. Clonal analysis in recurrent astrocytic, oligoastrocytic and oligodendroglial tumors implicates IDH1- mutation as common tumor initiating event. PloS One. 2012;7:e41298. doi: 10.1371/journal.pone.0041298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hartmann C, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120:707–718. doi: 10.1007/s00401-010-0781-z. [DOI] [PubMed] [Google Scholar]
- 123.Hartmann C, et al. Longterm survival in primary glioblastoma with versus without isocitrate dehydrogenase mutations. Clin Cancer Res. 2013;19:5146–57. doi: 10.1158/1078-0432.CCR-13-0017. [DOI] [PubMed] [Google Scholar]
- 124.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Figueroa M, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chowdhury R, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–9. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guilhamon P, et al. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat Commun. 2013;4:2166. doi: 10.1038/ncomms3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Koso H, et al. Transposon mutagenesis identifies genes that transform neural stem cells into glioma-initiating cells. P Natl Acad Sci USA. 2012;109:E2998–3007. doi: 10.1073/pnas.1215899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Alcantara Llaguno S, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu C, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–21. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sugiarto S, et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer cell. 2011;20:328–340. doi: 10.1016/j.ccr.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Friedmann-Morvinski D, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–4. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Monje M, et al. Hedgehog-responsive candidate cell of origin for diffuse intrinsic pontine glioma. P Natl Acad Sci USA. 2011;108:4453–4458. doi: 10.1073/pnas.1101657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lai A, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29:4482–90. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kang H, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ohnishi A, et al. Expression of the oligodendroglial lineage-associated markers Olig1 and Olig2 in different types of human gliomas. J Neuropathol Exp Neurol. 2003;62:1052–9. doi: 10.1093/jnen/62.10.1052. [DOI] [PubMed] [Google Scholar]
- 139.Lu QR, et al. Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. P Natl Acad Sci USA. 2001;98:10851–6. doi: 10.1073/pnas.181340798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Marie Y, et al. OLIG2 as a specific marker of oligodendroglial tumour cells. Lancet. 2001;358:298–300. doi: 10.1016/S0140-6736(01)05499-X. [DOI] [PubMed] [Google Scholar]
- 141.Barrett LE, et al. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer Cell. 2012;21:11–24. doi: 10.1016/j.ccr.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 142.Appolloni I, et al. Antagonistic modulation of gliomagenesis by Pax6 and Olig2 in PDGF-induced oligodendroglioma. Int J Cancer. 2012;131:E1078–87. doi: 10.1002/ijc.27606. [DOI] [PubMed] [Google Scholar]
- 143.Wang Y, et al. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer cell. 2009;15:514–526. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Prados MD, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27:579–84. doi: 10.1200/JCO.2008.18.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Brown PD, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North Central Cancer Treatment Group Study N0177. J Clin Oncol. 2008;26:5603–9. doi: 10.1200/JCO.2008.18.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.van den Bent MJ, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol. 2009;27:1268–74. doi: 10.1200/JCO.2008.17.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lassman AB, et al. Molecular study of malignant gliomas treated with epidermal growth factor receptor inhibitors: tissue analysis from North American Brain Tumor Consortium Trials 01-03 and 00-01. Clin Cancer Res. 2005;11:7841–50. doi: 10.1158/1078-0432.CCR-05-0421. [DOI] [PubMed] [Google Scholar]
- 148.Franceschi E, et al. Gefitinib in patients with progressive high-grade gliomas: a multicentre phase II study by Gruppo Italiano Cooperativo di Neuro-Oncologia (GICNO) Br J Cancer. 2007;96:1047–51. doi: 10.1038/sj.bjc.6603669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Neyns B, et al. Stratified phase II trial of cetuximab in patients with recurrent high-grade glioma. Ann Oncol. 2009;20:1596–603. doi: 10.1093/annonc/mdp032. [DOI] [PubMed] [Google Scholar]
- 150.Raymond E, et al. Phase II study of imatinib in patients with recurrent gliomas of various histologies: a European Organisation for Research and Treatment of Cancer Brain Tumor Group Study. J Clin Oncol. 2008;26:4659–65. doi: 10.1200/JCO.2008.16.9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wen PY, et al. Phase I/II study of imatinib mesylate for recurrent malignant gliomas: North American Brain Tumor Consortium Study 99-08. Clin Cancer Res. 2006;12:4899–907. doi: 10.1158/1078-0432.CCR-06-0773. [DOI] [PubMed] [Google Scholar]
- 152.Reardon DA, et al. Multicentre phase II studies evaluating imatinib plus hydroxyurea in patients with progressive glioblastoma. Br J Cancer. 2009;101:1995–2004. doi: 10.1038/sj.bjc.6605411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dresemann G, et al. Imatinib in combination with hydroxyurea versus hydroxyurea alone as oral therapy in patients with progressive pretreated glioblastoma resistant to standard dose temozolomide. J Neurooncol. 2010;96:393–402. doi: 10.1007/s11060-009-9976-3. [DOI] [PubMed] [Google Scholar]
- 154.Friedman HS, et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol. 2009;27:4733–40. doi: 10.1200/JCO.2008.19.8721. [DOI] [PubMed] [Google Scholar]
- 155.Lai A, et al. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. J Clin Oncol. 2011;29:142–8. doi: 10.1200/JCO.2010.30.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Batchelor TT, et al. Phase III Randomized Trial Comparing the Efficacy of Cediranib As Monotherapy, and in Combination With Lomustine, Versus Lomustine Alone in Patients With Recurrent Glioblastoma. J Clin Oncol. 2013;31:3212–8. doi: 10.1200/JCO.2012.47.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tanaka S, Louis DN, Curry WT, Batchelor TT, Dietrich J. Diagnostic and therapeutic avenues for glioblastoma: no longer a dead end? Nat Rev Clin Oncol. 2013;10:14–26. doi: 10.1038/nrclinonc.2012.204. [DOI] [PubMed] [Google Scholar]
- 158.Drappatz J, et al. Phase I study of panobinostat in combination with bevacizumab for recurrent high-grade glioma. J Neurooncol. 2012;107:133–8. doi: 10.1007/s11060-011-0717-z. [DOI] [PubMed] [Google Scholar]
- 159.Friday BB, et al. Phase II trial of vorinostat in combination with bortezomib in recurrent glioblastoma: a north central cancer treatment group study. Neuro-Oncology. 2012;14:215–21. doi: 10.1093/neuonc/nor198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Barker CA, Bishop AJ, Chang M, Beal K, Chan TA. Valproic acid use during radiation therapy for glioblastoma associated with improved survival. Int J Radiat Oncol. 2013;86:504–9. doi: 10.1016/j.ijrobp.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Weller M, et al. Prolonged survival with valproic acid use in the EORTC/NCIC temozolomide trial for glioblastoma. Neurology. 2011;77:1156–64. doi: 10.1212/WNL.0b013e31822f02e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rohle D, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–30. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Davis M, et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2010. [PubMed] [Google Scholar]
- 164.Wang F, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–6. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 165.Tonjes M, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19:901–8. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yang W, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang W, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.McCabe MT, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) P Natl Acad Sci USA. 2012;109:2989–94. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ryan RJ, et al. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PloS One. 2011;6:e28585. doi: 10.1371/journal.pone.0028585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sneeringer CJ, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. P Natl Acad Sci USA. 2010;107:20980–5. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Marian C, et al. The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin Cancer Res. 2010;16:154–163. doi: 10.1158/1078-0432.CCR-09-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cheng Z, et al. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hegi M, et al. Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib--a phase II trial. Mol Cancer Ther. 2011;10:1102–1112. doi: 10.1158/1535-7163.MCT-11-0048. [DOI] [PubMed] [Google Scholar]
- 174.Mellinghoff I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 175.Fenton T, et al. Resistance to EGF receptor inhibitors in glioblastoma mediated by phosphorylation of the PTEN tumor suppressor at tyrosine 240. P Natl Acad Sci USA. 2012;109:14164–14169. doi: 10.1073/pnas.1211962109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jun H, Bronson R, Charest A. Inhibition of EGFR induces a c-MET driven stem cell population in Glioblastoma. Stem cells 2013. 2013 Oct 1; doi: 10.1002/stem.1554. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bielen A, et al. Enhanced efficacy of IGF1R inhibition in pediatric glioblastoma by combinatorial targeting of PDGFRα/β. Mol Cancer Ther. 2011;10:1407–1418. doi: 10.1158/1535-7163.MCT-11-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Szerlip N, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. P Natl Acad Sci USA. 2012;109:3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Little S, et al. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res. 2012;72:1614–1620. doi: 10.1158/0008-5472.CAN-11-4069. [DOI] [PubMed] [Google Scholar]
- 180.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochimica et biophysica acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Harada K, et al. Intratumoral cytogenetic heterogeneity detected by comparative genomic hybridization and laser scanning cytometry in human gliomas. Cancer Res. 1998;58:4694–4700. [PubMed] [Google Scholar]
- 182.Jung V, et al. Evidence of focal genetic microheterogeneity in glioblastoma multiforme by area-specific CGH on microdissected tumor cells. J Neuropath Exp Neur. 1999;58:993–999. doi: 10.1097/00005072-199909000-00009. [DOI] [PubMed] [Google Scholar]
- 183.Nobusawa S, et al. Intratumoral patterns of genomic imbalance in glioblastomas. Brain Pathol. 2010;20:936–944. doi: 10.1111/j.1750-3639.2010.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ren ZP, et al. Molecular genetic analysis of p53 intratumoral heterogeneity in human astrocytic brain tumors. J Neuropath Exp Neur. 2007;66:944–954. doi: 10.1097/nen.0b013e318156bc05. [DOI] [PubMed] [Google Scholar]
- 185.Hegi M, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 186.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. P Natl Acad Sci USA. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Snuderl M, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 188.Inda M-d-M, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Stommel J, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 190.Greaves M, Maley C. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Jordan C, Guzman M, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 192.Hambardzumyan D, Parada L, Holland E, Charest A. Genetic modeling of gliomas in mice: new tools to tackle old problems. Glia. 2011;59:1155–1168. doi: 10.1002/glia.21142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chen L, Zhang Y, Yang J, Hagan J, Li M. Vertebrate animal models of glioma: Understanding the mechanisms and developing new therapies. Biochimica et biophysica acta. 2013;1836:158–165. doi: 10.1016/j.bbcan.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 195.Radaelli E, et al. Immunohistopathological and neuroimaging characterization of murine orthotopic xenograft models of glioblastoma multiforme recapitulating the most salient features of human disease. Histol Histopathol. 2009;24:879–891. doi: 10.14670/HH-24.879. [DOI] [PubMed] [Google Scholar]
- 196.Zhao Y, et al. An extensive invasive intracranial human glioblastoma xenograft model: role of high level matrix metalloproteinase 9. Am J Pathol. 2010;176:3032–3049. doi: 10.2353/ajpath.2010.090571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Luchman H, et al. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro-oncology. 2012;14:184–191. doi: 10.1093/neuonc/nor207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jacques T, et al. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J. 2010;29:222–235. doi: 10.1038/emboj.2009.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chow L, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer cell. 2011;19:305–316. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Holland E, Hively W, DePinho R, Varmus H. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ding H, et al. Oligodendrogliomas result from the expression of an activated mutant epidermal growth factor receptor in a RAS transgenic mouse astrocytoma model. Cancer Res. 2003;63:1106–1113. [PubMed] [Google Scholar]
- 202.Holland E. A mouse model for glioma: biology, pathology, and therapeutic opportunities. Toxicol Pathol. 2000;28:171–177. doi: 10.1177/019262330002800122. [DOI] [PubMed] [Google Scholar]
- 203.Weiss W, et al. Genetic determinants of malignancy in a mouse model for oligodendroglioma. Cancer Res. 2003;63:1589–1595. [PubMed] [Google Scholar]
- 204.Wei Q, et al. High-grade glioma formation results from postnatal pten loss or mutant epidermal growth factor receptor expression in a transgenic mouse glioma model. Cancer Res. 2006;66:7429–7437. doi: 10.1158/0008-5472.CAN-06-0712. [DOI] [PubMed] [Google Scholar]
- 205.Zhu H, et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. P Natl Acad Sci USA. 2009;106:2712–2716. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hambardzumyan D, Amankulor NM, Helmy KY, Becher OJ, Holland EC. Modeling Adult Gliomas Using RCAS/t-va Technology. Transl Oncol. 2009;2:89–95. doi: 10.1593/tlo.09100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hede SM, et al. GFAP promoter driven transgenic expression of PDGFB in the mouse brain leads to glioblastoma in a Trp53 null background. Glia. 2009;57:1143–1153. doi: 10.1002/glia.20837. [DOI] [PubMed] [Google Scholar]
- 208.Lei L, et al. Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS One. 2011;6:e20041. doi: 10.1371/journal.pone.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Becher O, Holland E. Genetically engineered models have advantages over xenografts for preclinical studies. Cancer Res. 2006;66:3355. doi: 10.1158/0008-5472.CAN-05-3827. [DOI] [PubMed] [Google Scholar]
- 210.Becher O, et al. Preclinical evaluation of radiation and perifosine in a genetically and histologically accurate model of brainstem glioma. Cancer Res. 2010;70:2548–2557. doi: 10.1158/0008-5472.CAN-09-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Reilly K, Loisel D, Bronson R, McLaughlin M, Jacks T. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet. 2000;26:109–113. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
- 212.Bajenaru M, et al. Astrocyte-specific inactivation of the neurofibromatosis 1 gene (NF1) is insufficient for astrocytoma formation. Mol Cell Biol. 2002;22:5100–5113. doi: 10.1128/MCB.22.14.5100-5113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Reilly K, et al. Susceptibility to astrocytoma in mice mutant for Nf1 and Trp53 is linked to chromosome 11 and subject to epigenetic effects. P Natl Acad Sci USA. 2004;101:13008–13013. doi: 10.1073/pnas.0401236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhu Y, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kwon CH, et al. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 2008;68:3286–3294. doi: 10.1158/0008-5472.CAN-07-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sasaki M, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26:2038–2049. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Barrow J, et al. Homozygous loss of ADAM3A revealed by genome-wide analysis of pediatric high-grade glioma and diffuse intrinsic pontine gliomas. Neuro-oncology. 2011;13:212–222. doi: 10.1093/neuonc/noq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Raffel C, et al. Analysis of oncogene and tumor suppressor gene alterations in pediatric malignant astrocytomas reveals reduced survival for patients with PTEN mutations. Clin Cancer Res. 1999;5:4085–4090. [PubMed] [Google Scholar]
- 219.Pollack IF, et al. Rarity of PTEN deletions and EGFR amplification in malignant gliomas of childhood: results from the Children’s Cancer Group 945 cohort. J Neurosurg. 2006;105:418–24. doi: 10.3171/ped.2006.105.5.418. [DOI] [PubMed] [Google Scholar]
- 220.Pollack I, et al. Age and TP53 mutation frequency in childhood malignant gliomas: results in a multi-institutional cohort. Cancer Res. 2001;61:7404–7407. [PubMed] [Google Scholar]
- 221.Pollack I, et al. IDH1 mutations are common in malignant gliomas arising in adolescents: a report from the Children’s Oncology Group. Child Nerv Syst. 2011;27:87–94. doi: 10.1007/s00381-010-1264-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Gallia G, et al. PIK3CA gene mutations in pediatric and adult glioblastoma multiforme. Mol Cancer Res. 2006;4:709–714. doi: 10.1158/1541-7786.MCR-06-0172. [DOI] [PubMed] [Google Scholar]
- 223.Schiffman J, et al. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010;70:512–519. doi: 10.1158/0008-5472.CAN-09-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Nicolaides T, et al. Targeted therapy for BRAFV600E malignant astrocytoma. Clin Cancer Res. 2011;17:7595–7604. doi: 10.1158/1078-0432.CCR-11-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]