Abstract
The restriction of immunoglobulin (Ig) expression to B lymphocytes is well established. However, several reports have confirmed that the Ig gene can be expressed in many non-B cancer cells and/or some normal cells. Our aim is to determine whether the Ig gene promoter can be activated in non-B cancer cells and to identify the regulatory mechanism for Ig gene expression. Our results show that the Ig promoter of VH4-59 was activated in several non-B cancer cell lines. Moreover, two novel positive regulatory elements, an enhancer-like element at −800 to −610 bp and a copromoter-like element at −610 to −300 bp, were identified in two epithelial cancer cell lines, HeLa S3 and HT-29. The octamer element (5′-ATGCAAAT-3′) located in the Ig promoter, a crucial element for B-cell-derived Ig gene transcription, was also very important for non-B-cell-derived Ig gene transcription. More importantly, we confirmed that octamer-related protein-1 (Oct-1), but not Oct-2, was a crucial transcriptional factor for Ig gene transcription due to its ability to bind to the octamer element of the Ig promoter in epithelial cancer cells. These results suggested the presence of a distinct regulatory mechanism for Ig gene expression in non-B cancer cells.
Keywords: Ig gene transcription, Oct-1, promoter, transcription regulation, VH4-59
Introduction
It was thought that immunoglobulin (Ig) genes are selectively expressed in B lymphocytes, and the promoters for the variable region are normally inactivated in non-B cells.1 However, in 1989, we unexpectedly found immunoglobulin G (IgG)-like immunostaining in breast cancer cells. Furthermore, we have found that both IgG and rearranged Ig gene transcripts exist in some epithelial cancer cells.2, 3 Since then, we have demonstrated that many non-B human cancer cells and some normal cells can produce Ig.4, 5, 6, 7, 8, 9 These findings have been confirmed by other laboratories as well.10, 11, 12, 13, 14, 15, 16, 17
IgH and Igκ loci are sequentially activated during B-lymphocyte maturation by multiple lymphocyte-specific transcription factors, including the E box protein E2A, early B-cell factor (EBF), paired box 5 (Pax5), octamer-related protein-1 (Oct-1), Oct-2 and Bob-1. Generation of the earliest B-cell progenitors depends on E2A and EBF, which coordinately activate B-cell gene expression and IgH gene rearrangement at the onset of B-cell lymphopoiesis. In E2A- or EBF-deficient mice, Ig gene rearrangement is lacking.18, 19, 20 The Pax5 transcription factor is specifically expressed by B cells and is essential for Ig rearrangement, Ig class switching and pre-B-cell-receptor signaling.21, 22 Oct-1, Oct-2 and Bob-1 coordinately drive Ig gene transcription by binding to the Ig promoters and enhancers. Oct-2 and Bob-1 are mature B-cell-specific transcription factors, whereas Oct-1 is expressed during the early development of B cells and other types of cell.23, 24 The aforementioned specific transcription factors regulate orderly Ig gene rearrangement and transcription in B cells through site-specific Ig gene regulators.
Key cis-regulators of B-cell-specific expression of IgH genes are VH promoters, Ig region promoters located upstream of each constant region gene, and two sets of enhancers (the intronic enhancer [Eμ] located in the JH-Cμ intron and a complex regulatory region that lies 3′ of the IgH gene locus [3′ RR]).25 Although the Ig heavy- and light-chain enhancers contain binding sites for numerous different classes of transcription factors, Ig gene variable region promoters seem to be less complex and predominantly contain a motif termed the octamer element (5′-ATGCAAAT-3′), which is located 10–25 nucleotides upstream of the TATA box. The canonical octamer or its reverse complement is conserved in all Ig heavy- and light-chain variable region promoters.26 The highly conserved octamer element plays a crucial role in promoting Ig gene transcription in B cells. The importance of this element in mediating Ig transcription has been demonstrated with the use of transgenic mice. A point mutation in the octamer region reduced expression of an Ig transgene to 5% of its normal level.27 The activity of Ig gene promoters is directly regulated by two related transcription factors, Oct-1 and Oct-2, which bind to the same sequence within the octamer element. Oct-1 and Oct-2 are differentially expressed in vivo. Oct-2 is specifically expressed in B lymphocytes and has been implicated in the control of mature B-lymphocyte-specific Ig gene expression. By contrast, Oct-1 may be a critical regulator of Ig gene transcription during the early development of B cells. Oct-1 can also bind and regulate the promoter of Igα, a component of the B-cell receptor on the B-cell surface. However, Oct-1 is a ubiquitous transcription factor that has been implicated in the control of non-immune-associated gene expression by all cell types, such as the histone H2B gene and the genes for snRNA, U2 and U6.28, 29, 30, 31
Although the regulatory mechanism of Ig transcription in B cells is well known, it is not yet clear how the non-B-cell-derived Ig transcription is controlled and whether the regulatory mechanism is equivalent to that of B-cell-derived Ig. In this study, we first tested whether the transcription factors associated with B-lymphocyte-specific Ig gene expression were expressed in several non-B cancer cell lines. We then explored whether the Ig VH promoter could indeed drive Ig expression in these non-B cancer cell lines and whether the regulatory mechanism of the VH promoter for non-B-cell-derived Ig gene transcription is similar to that of B cells. Our results clearly showed that the genes involved in variable, diversity and joining (VDJ) recombination and transcription, E2A, EBF, Oct-1 and Oct-2, but not Pax5, were expressed in the non-B cancer cells. We further cloned the 5′-upstream fragment of VH4-59 containing IgH promoter and found the promoter activity of VH4-59 in 6 of the 10 cancer cell lines. Using a series of 5′-truncated reporter constructs, we found two novel positive regulatory elements in epithelial cancer cell lines, but not in the Daudi cell line used as a positive control. Additionally, we confirmed that Oct-1, but not Oct-2, is a crucial transcriptional factor for Ig gene transcription in non-B cells because of its ability to bind to the octamer element on the Ig gene promoter.
Results
Transcription factors related to Ig gene expression are detected in non-B cancer cell lines
To elucidate whether the Ig gene promoter can be expressed in non-B cancer cells, we tested whether Ig gene expression-related transcription factors were expressed in several non-B cancer cell lines. Our results showed that the transcription factors E2A and Oct-1 were widely expressed in all of the cancer cell lines assessed. EBF and Oct-2, the B-cell-specific transcription factors, were expressed in a few of the non-B cell lines assessed. However, Pax5 was not detected in any of the non-B cancer cell lines assessed (Figure 1).
Figure 1.
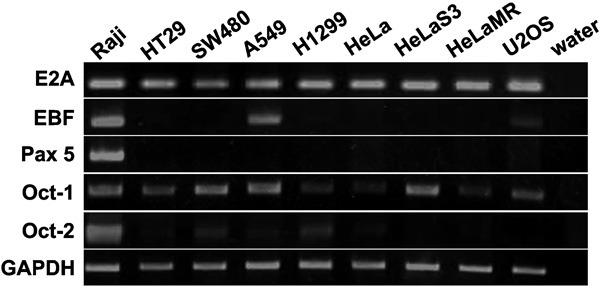
Ig-related transcription factors are expressed by non-B cancer cells. The expression of transcription factors (E2A, EBF, Pax5, Oct-1 and Oct-2) at the mRNA level was analyzed by RT-PCR in several non-B tumor cell lines. The B-cell-derived Daudi cell line was used as a positive control. GAPDH was used as an internal control. EBF, early B-cell factor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Oct-1, octamer-related protein-1; Oct-2, octamer-related protein-2; Pax5, paired box 5; RT-PCR, reverse transcriptase-polymerase chain reaction.
The 5′-flanking sequence of VH4-59 has promoter activity in non-B cancer cell lines
We amplified and identified a 1.2-kb 5′-flanking sequence of the Ig VH4-59 gene from HeLa S3 cells and cloned it into a firefly luciferase expression vector, pGL3-basic. We then tested the promoter activity of the 1.2-kb 5′-flanking sequence of Ig VH4-59 by a transient transfection assay. The 5′-flanking sequence of VH4-59 yielded higher promoter activity in six cell lines (HeLa, HeLa MR, HeLa S3, HT-29, U2OS and Daudi) than that of the pGL3 control and even higher activity in the above six cell lines than that of the Simian Virus 40 (SV40) promoter, which has a high activity in the mammalian cell lines (Figure 2a). No promoter activity was detected in A549, HepG2, SW480 or Jurkat cell lines (Figure 2b).
Figure 2.
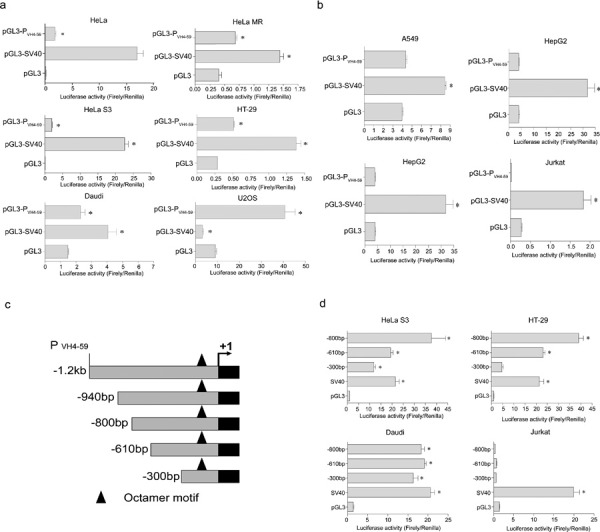
5′-flanking sequence of VH4-59 has promoter activity in non-B cancer cells. (a) The 1.2-kb 5′-flanking sequence of VH4-59 (PVH4-59) showed promoter activity after being transfected into HeLa, HeLa MR, HeLa S3, HT-29, Daudi or U2OS cell lines. (b) PVH4-59 had no promoter activity after being transfected into A549, HepG2, SW480 or Jurkat cell lines. (c) 5′-deletion truncations of the 1.2-kb pGL3 promoter luciferase construct. (d) The three 5′-deletion truncations of 1.2-kb pGL3 were transfected into HeLa S3, HT-29, Daudi or Jurkat cells. Luciferase activity was measured using a dual luciferase reporter system. The results are representative of three independent experiments after normalization to renilla luciferase activity (internal controls). N=3, *P<0.001. Each bar represents mean±SD.
To further define the regulatory elements in the VH4-59 promoter, a series of deletions of the 1.2-kb fragment containing the VH4-59 promoter were generated by PCR amplification (Figure 2c). The promoter activities of the various deletions were tested as described above. Among all of the deletion constructs containing the complete promoter region, the strongest promoter activity was observed with both the −800- and −940-bp fragments. This promoter activity was stronger than that of the −610-bp fragment (1.8 times stronger in the HeLa S3 cell line and 1.7 times stronger in the HT-29 cell line) or that of the −300-bp fragment (3.1 times stronger in the HeLa S3 cell line and 7.3 times stronger in the HT-29 cell line) (Figure 2d). In the Daudi cell line, all of the deletions showed the same level of promoter activity, whereas in the Jurkat cell line, no promoter activity was detected (Figure 2d). Thus, the two novel regulatory elements (−800 to −610 bp and −610 to −300 bp) on the 59-flanking sequence of Ig VH4-5959 are utilized in cancer cells.
The octamer element is crucial for non-B-cell-derived Ig gene transcription in HeLa S3 and HT-29 cell lines
To identify the role of the octamer element located in the promoter region of the Ig VH4-59 gene in cancer cells, the deletion constructs above were mutated by deleting the four base pairs in the middle of the octamer motif (Figure 3a). Our results show that the −300-bp mutant lacking the intact octamer site had no activity in HeLa S3, HT-29 or Daudi cells. Of interest, the −610- and −800-bp mutants lacking the intact octamer motif showed promoter activity in HeLa S3 and HT-29 cells, although the promoter-like activity was much lower than that with the wild-type octamer motif. In contrast, neither the −610-bp nor the −800-bp mutant showed any activity in Daudi cells (Figure 3b). These results suggested that, similar to B cells, the octamer motif is an important transcriptional element for Ig gene transcription in non-B cancer cells. Furthermore, we found another novel promoter-like element with lower promoter activity at −610 to −300 bp in the two cancer cell lines assessed, but not in B-lymphoma cells.
Figure 3.
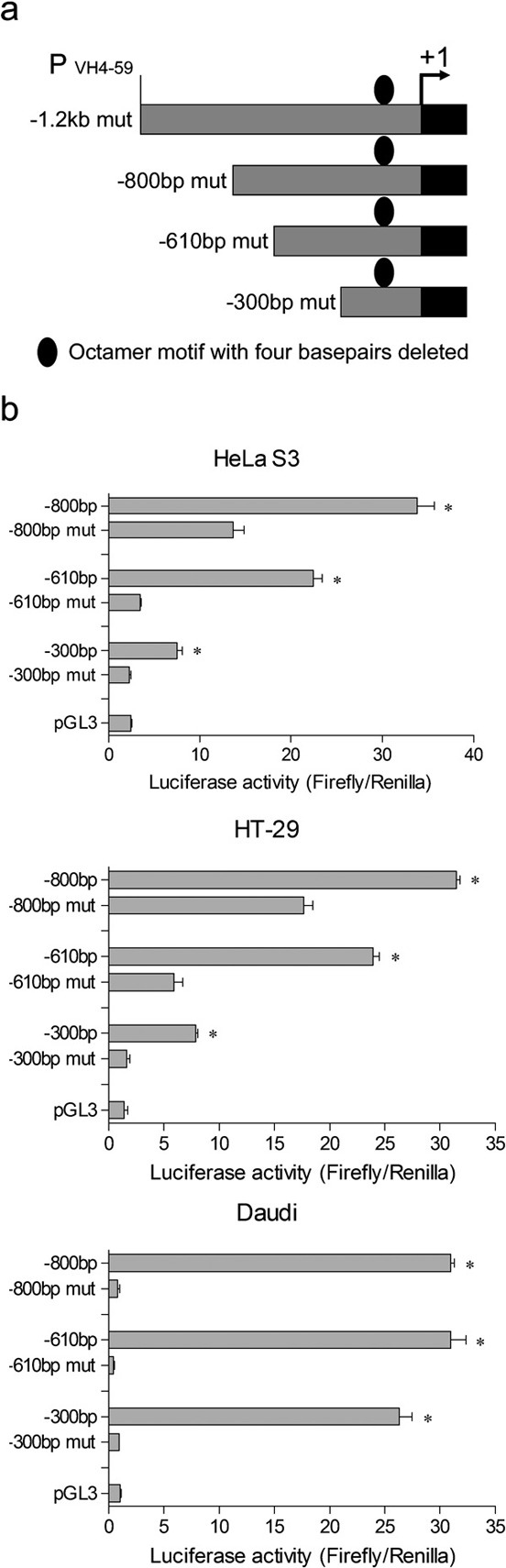
The octamer element is a crucial element for non-B-cell-derived Ig gene transcription in HeLa S3 and HT-29. (a) 5′-deletion truncations of the 1.2-kb pGL3 with a 4-bp deletion in the octamer motif (ATGCAAAT). (b) The intact and mutated 5′-deletion truncations of 1.2-kb pGL3 were transfected into HeLa S3, HT-29 or Daudi cell lines. Luciferase activity was measured using a dual luciferase reporter system. The results are representative of three independent experiments after normalization to renilla luciferase activity (internal controls). N=3, *P<0.001. Each bar represents mean±SD.
Oct-1, but not Oct-2, regulates Ig VH4-59 promoter activity and induces IgG gene expression
Oct-1, but not Oct-2, can bind the octamer element on electrophoretic mobility shift assay (EMSA) and gel supershift assay
We performed an EMSA to evaluate the binding of transcriptional factors to the octamer element of the Ig VH4-59 gene in non-B cancer cells. Double-stranded, 41-bp oligonucleotides containing the octamer motif and corresponding to nucleotides −130 to −89 interacted with nuclear protein fractions from HeLa S3 cells (Figure 4a). Binding of the 41-bp probe with HeLa S3 nuclear protein fractions was inhibited significantly by an excess (125-fold) of unlabeled 41-bp probe and to a lesser extent by the double-stranded, 33-bp oligonucleotides without the octamer motif, which were used as a negative control. Antibodies directed against Oct-1 strongly supershifted the binding complex of HeLa S3 nuclear proteins, whereas anti-Oct-2 antibody did not significantly impair the binding interaction (Figure 4b). These results indicate that Oct-1 interacts with the Ig VH4-59 promoter via the octamer element in HeLa S3 cells.
Figure 4.
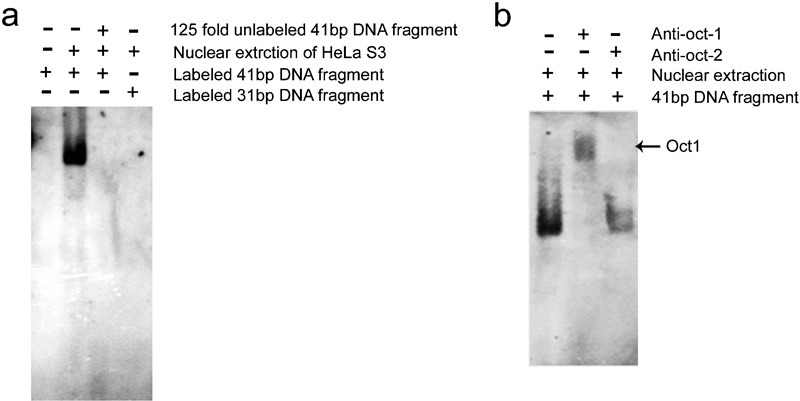
Oct-1, not Oct-2, binds to the octamer element by electrophoretic mobility shift assay (EMSA and gel supershift). (a) The EMSA assay for octamer motif binding factor located in the promoter region of the VH4-59 gene in HeLa S3 cells. 41-bp DNA fragment: upstream VH4-59 gene with the octamer motif; 33-bp DNA fragment: 41-bp DNA fragment with an 8-bp deletion in the octamer motif; 33-bp fragment: negative control. (b) Supershift assay for octamer motif binding factor with the addition of anti-Oct-1 or anti-Oct-2 antibody in the binding reaction system. The results are representative of three independent experiments. EMSA, electrophoretic mobility shift assay; Oct-1, octamer-related protein-1; Oct-2, octamer-related protein-2.
Oct-1 binds the VH4-59 promoter in vivo as assessed by chromatin immunoprecipitation (ChIP)
To confirm the binding of Oct-1 to the Ig VH4-59 promoter, soluble chromatin from HeLa S3 cells was prepared and incubated with or without polyclonal anti-Oct-1 antibody. The antibody-bound DNA was analyzed by PCR using primers specific for the human Ig VH4-59 promoter sequence containing the octamer motif to produce a 233-bp product. As shown in Figure 5, anti-Oct-1 antibody was able to immunoprecipitate the Ig VH4-59 promoter region; the region could not be pulled down without the antibody. Collectively, these data confirm that Oct-1 protein binds in vivo to the octamer motif located within the promoter region of the Ig VH gene in the HeLa S3 epithelial cancer cell line.
Figure 5.
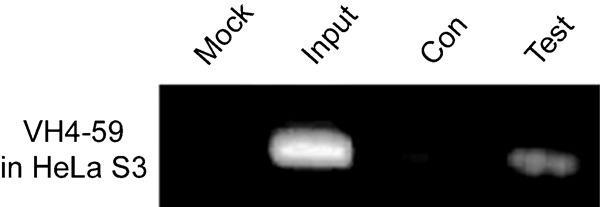
The Oct-1 binding DNA fragment of VH4-59 promoter was amplified by Chip-related PCR. Mock: no template in the PCR reaction system; Input DNA (positive control): the sonicated chromatin fragments of the cells were used as the PCR template; Con: no antibody added to the IP system; Test: dilutions of IP were used as templates for PCR to amplify the Oct-1 binding DNA sequence. Results are representative of three independent experiments.
Oct-1 significantly increases Ig gene transcription and expression
We constructed expression plasmids (−800-bp pGL3 luciferase reporter plasmids) containing Oct-1 or Oct-2 and found that VH4-59 promoter activity was significantly higher with the plasmids containing Oct-1 than with the plasmids containing Oct-2 in HeLa S3 and HT-29 cells (Figure 6a). Similarly, overexpression of Oct-1, but not Oct-2, significantly increased IgG expression (Figure 6b). In addition, we synthesized antisense oligonucleotides for Oct-1 and, using flow cytometry, found that when Oct-1 expression was blocked by antisense oligonucleotide, IgG expression was also reduced in HeLa S3 (Figure 6c). These results suggested that Oct-1 is the crucial transcriptional factor for Ig gene transcription in non-B cancer cells.
Figure 6.
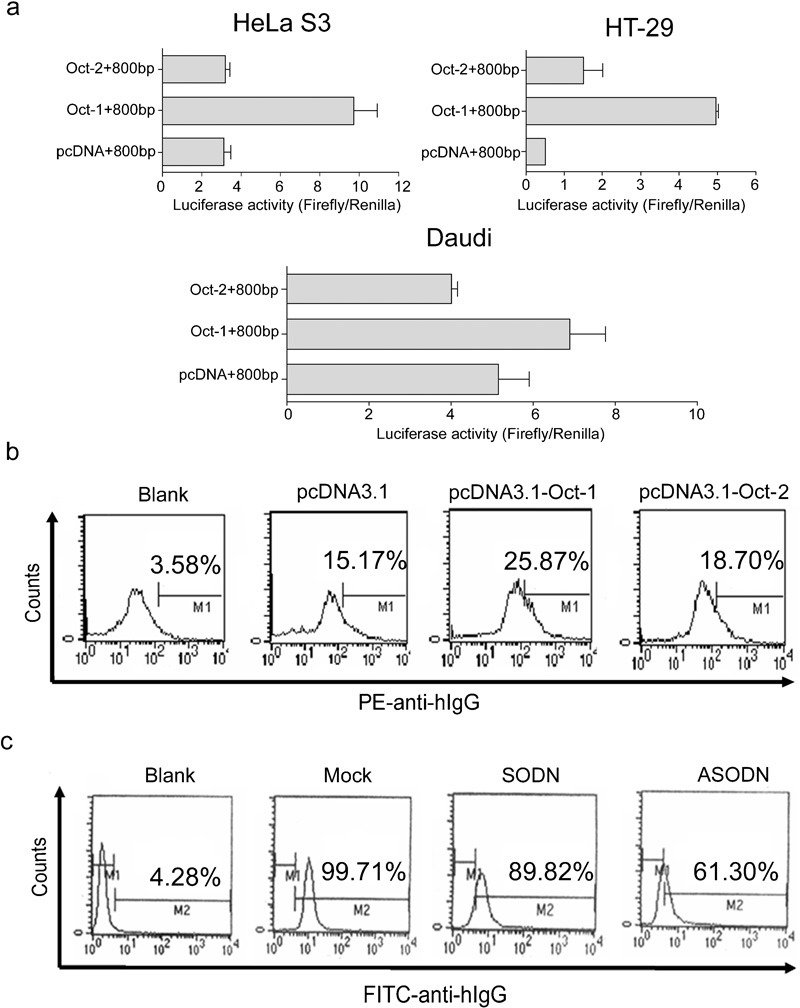
Oct-1 significantly increases Ig gene transcription and expression. (a) pcDNA3.1(−), pcDNA-Oct-1 or pcDNA-Oct-2, with the −800-bp pGL3 luciferase reporter plasmid, were transfected into HeLa S3, HT-29 or Daudi cell lines. Luciferase activity was measured using a dual luciferase reporter system. The results are representative of three independent experiments after normalization to renilla luciferase activity (internal control). Each bar represents mean±SD. (b) Overexpression of Oct-1 by pcDNA-Oct-1 increased IgG protein expression in HeLa S3 as detected by flow cytometry. (c) Antisense oligonucleotide for Oct-1 reduced IgG expression in HeLa S3 as detected by flow cytometry. The results are representative of three independent experiments. ASODN, antisense oligodeoxynucleotides; IgG, immunoglobulin G; Oct-1, octamer-related protein-1; SODN, oligodeoxynucleotides.
Discussion
Ig gene rearrangement and transcription occur in non-B cancer cells
It was thought that transcription of the Ig gene is silenced in non-B lymphocytes.1 However, in 1996, we first reported the detection of IgG-like molecules in epithelial cancer cells in breast and colon carcinoma biopsy tissues (Qiu and Yang 1996).3 We then confirmed that the IgG-like molecules were indeed IgG, and IgG was widely expressed in almost all epithelial carcinomas and epithelial cancer cell lines as well as some normal epithelial cells, neurons, and germ cells from patients without cancer and healthy mice (Qiu, Zhu et al. 2003; Zhu, Li et al. 2008; Huang, Sun et al. 2008, Huang, Zhang et al. 2009; Zhang, Mao et al. 2009).4, 6, 7, 8, 9 Moreover, to determine the functionally rearranged Ig gene repertoires in primary cancer cells, we evaluated the Ig gene transcripts and repertoires after laser capture microdissection of cells from 10 carcinomas of the colon, breast, oral cavity or lung, and found that the cancer-derived Ig genes had a distinct repertoire.9 These results have been subsequently confirmed by other groups (Li, Feng et al. 2004; Babbage, Ottensmeier et al. 2006; Chen and Gu 2007; Liu, Zheng et al. 2007; Lee 2009).11, 12, 13, 14, 17 In this study, we used a mouse model to investigate the regulatory mechanism of Ig gene transcription in epithelial cancer cells and further confirmed that some genes involved in VDJ recombination and Ig gene transcription (E2A, EBF, Oct-1 and Oct-2, but not Pax5) were also expressed in non-B cancer cells. In addition, a much higher level of Ig VH promoter activity was found in some non-B cancer cell lines. More importantly, we proved that a distinct regulatory mechanism for Ig gene transcription was present in the non-B cancer cells.
Sun and Kitchingman had found that VH6-1 promoter was very active in HeLa cells, but they presumed that VH6-1 promoter activity in cancer cells was non-tissue-specific for two reasons: first, no Ig gene transcripts were detected in HeLa cells; and second, they believed that Ig genes, including the VH6-1 gene, were located in the closed chromatin region lacking DNase I hypersensitive sites and would not be transcribed in HeLa cells.32 However, our data and those of others showed that functionally rearranged Ig gene transcripts could be detected in cancer cell lines and primary cancer cells, such as breast and colon cancer cells.9 In this study, we cloned the 5′-flanking sequence of VH4-59 containing the conserved octamer element (5′-ATGCAAAT-3′) and used it as a model to detect the activity of Ig heavy chain promoter. We detected high Ig promoter activity in several cancer cell lines. Furthermore, results from the ChIP assay proved that Oct-1 could bind to the octamer sequences located in the VH4-59 promoter regions in HeLa S3 cells, which indicated that these Ig VH genes were located in the open chromatin DNA region. These results suggested that the non-B-derived cancer cells could drive Ig gene transcription.
Although Ig VH4-59-driven expression was mainly shown in HeLa S3 and HT-29 cells, but not in other cancer cell lines (including Daudi cells) used in this study (data not shown), the Ig VH4-59 promoter contained the critical motif, termed the octamer element (5′-ATGCAAAT-3′), for all Ig gene transcription. Therefore, we proposed that the Ig VH4-59 promoter could be driven by a variety of transcription factors expressed in cancer cells, although these transcription factors may preferentially bind and drive other VH family promoters rather than VH4-59 promoter. In this study, we used the Ig VH4-59 promoter as a model to detect the activity of Ig heavy chain promoter. We transiently transfected the firefly luciferase expression vector pGL3-basic with the 5′-flanking sequence of Ig VH4-59 as the promoter into several types of cancer cell lines: lung cancer cell line A549; cervical cancer cell lines HeLa, HeLa MR and HeLa S3; hepatocellular carcinoma cell line HepG2; colon cancer cell lines HT-29 and SW480; osteosarcoma cell line U2OS; B-lymphoma cell line Daudi (as positive control); and T-leukemia cell line Jurkat (as negative control). We found that a high level of VH4-59 promoter activity not only showed in two VH4-59 family-expressed cancer cell lines, HT-29 and HeLa S3, but also in these no VH4-59 family-expressed cancer cell lines, HeLa, HeLa MR, U2OS and Daudi cells. Suggest that although the Ig gene expression-related transcription factors in these no VH4-59 family-expressed cancer cell lines preferentially bind and drive other VH family promoters, but not the VH4-59, in natural state, but in this study, these transcription factors also cross-driven the transiently transfected VH4-59 promoter via binding the critical octamer element. However, no promoter activity was exhibited in A549, HepG2 and SW480 cells, suggesting that the transcription factors that drive Ig gene transcription in these cancer cell lines cannot bind and drive expression from the VH4-59 promoter containing the canonical octamer element. These results imply that although the octamer element is a critical motif for all Ig gene variable region promoters, the unique sequence in each Ig VH promoter is also necessary for transcriptional factors to preferentially select and drive expression from the Ig VH promoter.
A specific regulatory mechanism for Ig gene transcription exists in non-B cancer cells
It is well known that the Ig gene promoter is necessary for driving Ig transcription in B lymphocytes and that no other activation element is located within 1 kb of the upstream sequence of the VH gene promoter. It has also been found that all of the functional enhancers for Ig VH gene transcription are located downstream of the gene, between the JH, μ region, and the 3′-end of the IgH gene.33, 34, 35 However, in this study, we used a series of 5′-truncated reporter constructs and found new enhancer-like activation elements upstream of the VH4-59 promoter region at −610 to −800 bp in HeLa S3 and HT-29 cells, but not in Daudi cells. In addition, we found that the octamer element, 5′-ATGCAAAT-3′, was an absolutely necessary Ig variable region promoter, but we did not find any other activation elements in the 5′-flanking region of the Ig variable region promoter in B cells.
In this study, although we confirmed that the octamer element was very important for non-B-cell-derived Ig gene transcription, we found another novel promoter-like element with lower activity at −610 to −300 bp in two cancer cell lines (HeLa S3 and HT-29), but not in Daudi cells. These findings suggest that, unlike in B lymphocytes, some unknown cell type-specific transcription factors may exist in cancer cells that can interact with the potential activation elements. However, the mechanism of these novel regulatory elements for Ig gene expression in non-lymphocytic cancer cells remains unclear.
Oct-1 and Oct-2 are POU (pronounced pow) homeodomain proteins that recognize the same octamer motif (ATGCAAAT) that regulates the activity of the Ig gene promoter. Oct-2 is a B lineage-restricted factor that has been shown to control B-lymphocyte-specific expression of genes such as the Ig VH gene.36 Our results showed that although Oct-2 was also expressed in some non-B cancer cell lines, its expression level was much lower than that of Oct-1. We conclude that the transcription factor Oct-1, but not Oct-2, can preferentially bind to the octamer sequences at the open chromatin region and promote Ig VH gene expression in these non-B cancer cells. Furthermore, it has been shown that the coactivator Bob-1 can interact efficiently with the POU (pronounced pow) domains of Oct-1 and Oct-2 as well as the octamer element and is necessary for Ig promoter activation in B lymphocytes.24, 26, 33, 34, 35, 37 However, we did not detect Bob-1 expression in HeLa S3 or HT-29 cells (data not shown). Our findings of the overexpression of Oct-1, but not of Oct-2, indicate that Oct-1 has a major role in promoting Ig expression in non-B cancer cells.
Finally, in addition to Oct-1, we confirmed that E2A, which is involved in VDJ recombination, was also expressed in non-B cancer cells. However, another important transcription factor, EBF, which is also involved in VDJ recombination, was found in only a few non-B cancer cell lines. Pax5, a well-known Ig gene expression-related transcription factor that is essential for Ig rearrangement, Ig class switching, and pre-B-cell-receptor signaling, was not found in any of the non-B cancer cells assessed. Our overall findings suggest that a distinct regulatory mechanism for Ig gene transcription is present in non-lymphocytic cancer cells.
Experimental procedures
Cell lines
The following cell lines were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA): human lung cancer cell line A549; human cervical cancer cell lines HeLa, HeLa MR and HeLa S3; human hepatocellular carcinoma cell line HepG2; human colon cancer cell lines HT-29 and SW480; human osteosarcoma cell line U2OS; B-lymphoma cell line Daudi; and T leukemia cell line Jurkat. HeLa S3 and HT-29 were cultured in DMEM (Gibco/Invitrogen, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (Hyclone/Fisher Scientific, Pittsburgh, PA, USA), 2 mM glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin. Daudi and Jurkat were cultured in RPMI 1640 medium (Gibco) supplemented as above. All of the cells were maintained in an incubator with 5% CO2 at 37 °C.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was extracted from cells with the use of TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RT-PCR was performed with the ThermoSCRIPT RT-PCR system (Invitrogen) according to the manufacturer's instructions. We used l µg of total RNA for first-strand cDNA synthesis using oligo (dT) 15 primer. The primer sequences and PCR conditions for the VDJ recombination-associated transcription factors are shown in Table 1.
Table 1. PCR conditions and primers.
| Genes | Primer sequences | Annealing temperature* | Size (bp) |
|---|---|---|---|
| EBF | F: CCGCCTTGATCTTCTAAGTGC | 57 °C (5)–55 °C (5)–53 °C (31) | 521 |
| R: ATCCTGCTCCGTCCTTATCC | |||
| E2A | F: ATGAACCAGCCGCAGAGGATG | 64 °C (5)–62 °C (5)–60 °C (5)–58 °C (25) | 295 |
| R: CCGAGTCCCGGTCCCAGGAAT | |||
| Pax5 | F: TTGCTCATCAAGGTGTCAGG | 57 °C (5)–55 °C (5)–53 °C (31) | 214 |
| R: CTGATCTCCCAGGCAAACAT | |||
| Oct-1 | F: GCAACACAGGCACACAAACC | 58 °C (5)–56 °C (5)–54 °C (5)–52 °C (25) | 649 |
| R: TTGGCTTTGCTGAGGTAGTT | |||
| Oct-2 | F: CCTGCTCAGTTCCTGCTACC | 60 °C (5)–58 °C (5)–56 °C (31) | 497 |
| R: GATGCTGGTCCTCTTCTTGC |
Abbreviations: EBF, early B-cell factor; F, forward; Oct-1, octamer-related protein-1; Oct-2, octamer-related protein-2; Pax5, paired box 5; PCR, polymerase chain reaction; R, reverse.
Touch down PCR was performed. The amplifying cycles at each temperature are shown in parentheses.
Plasmid construction
The 1.2-kb 5′-flanking sequence of the VH4-59 gene containing the conserved octamer motif was amplified from HeLa S3 cells by PCR using the forward primer 5′-GGGGTACCAGTTGTTCAGGTCCTAAGAAGAAAGC-3′ and the reverse primer 5′-TCCCCCGGGTTTCATGTTCTTGCACAGGAGGTCCA-3′. The PCR products were cloned into the pGEM-T easy vector (Promega, Madison, WI, USA) and sequenced using an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, CA, USA). The inserts were cut with the restriction enzymes KpnΙ and SmaΙ, and were subcloned into the pGL-3 vector (Promega) to construct the 1.2-kb pGL3 luciferase reporter plasmid. The four 5′-truncated reporter constructs, −940-bp pGL3, −800-bp pGL3, −610-bp pGL3 and −300-bp pGL3, were generated from the 1.2-kb pGL3 by PCR amplification. These amplified sequences were also cloned into the pGL3 vector.
Oct-1 and Oct-2 cDNA, kindly provided by Dr W. Herr, were excised from the pCGN-Oct-1 and pCGN-Oct-2 constructs, respectively, using the restriction enzymes XbaI and BamHI, and were subcloned using the same restriction enzymes into pcDNA3.1(−) to generate pcDNA-Oct-1 and pcDNA-Oct-2.38
To detect the activity of the octamer element, four base pairs in the middle of the octamer motif (ATGCAAAT), located in the promoter region of the VH4-59 gene, were deleted using the QuikChange site-directed mutagenesis kit (Stratagene/Agilent Technologies, La Jolla, CA, USA), and the truncated −940bp, −800bp, −610bp and −300bp fragments containing the mutant promoters were inserted into plasmids designated as 940-bp mutant, 800-bp mutant, 610-bp mutant and 300-bp mutant, separately. The sequence of these constructs was confirmed by DNA sequencing.
Transfection and dual luciferase reporter assay
For the transient reporter gene assay, the cells were transfected either with 9 µg of pGL3 luciferase reporter plasmids and 1 µg of renilla luciferase control plasmids (pRL-TK, Promega) or with 4.5 µg of pcDNA3.1(−)-associated plasmids, 4.5 µg of pGL3 luciferase reporter plasmids and 1 µg of pRL-TK.
A 20-ms electrical pulse at 120 V was delivered to A549, HeLa, HeLa MR, HeLa S3 and U2OS cells; 130 V was delivered to HepG2, HT-29 and SW480 cells, and 140 V was delivered to Daudi and Jurkat cells. After standing at room temperature for 10 min, the transfected cells were allowed to recover at 37 °C in DMEM or RPMI-1640 supplemented with 10% fetal bovine serum for approximately 30 h in 24-well culture plates. Firefly and renilla luciferase activities were measured using a dual-luciferase reporter assay system (Promega). Luminescence was measured using a Veritas microplate luminometer (Turner Biosystem, Sunnyvale, CA, USA). Relative luciferase activity was calculated by measuring the ratio of firefly to renilla luciferase activities.
EMSA and gel supershift experiment
The complementary oligonucleotides containing the octamer motif located in the VH4-59 promoter region (5′-ACAAAGGCACCACCCACATGCAAATCCTCACTTAAGCACCC-3′ and 5′-GGGTGCTTAAGTGAGGATTTGCATGTGGGTGGTGCCTTTGT-3′) were synthesized with 3′ overhangs, annealed and end-labeled with DIG (second-generation DIG gel shift kit; Roche Applied Science, Basel, Switzerland). Nuclear extracts were prepared as described previously.39 The binding reaction system was prepared as described in the Roche second-generation DIG gel shift kit. The reaction system was mixed on ice and incubated at 4 °C for 10 min and was then allowed to stand at room temperature for 15 min. For DNA competition experiments, a 125-fold molar excess of unlabeled oligonucleotide was added and was incubated with nuclear extracts and a labeled probe. For supershift experiments, 1 µl of polyclonal anti-Oct-1 or anti-Oct-2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was added to the completed binding reaction at room temperature for 20 min before the DNA probe was added. Electrophoresis was performed with 5% native polyacrylamide gel using 0.5× TBE buffer. After electrophoresis, the gel was transferred to a nylon membrane (Pharmacia/GE Healthcare, Piscataway, NJ, USA), and the DNA–protein complex on the membrane was detected by an enhanced chemifluorescence detection system (Amersham Bioscience/GE Healthcare, Piscataway, NJ, USA).
Chromatin immunoprecipitation
To determine whether Oct-1 binds to or is localized to the octamer motif in the promoter region of VH4-59 in HeLa S3 cells, a ChIP assay was performed. Approximately 1×107 cells were harvested and fixed in 1% formaldehyde at room temperature for 30 min. The cross-linking reaction was stopped by adding glycine to a final concentration of 0.125 M at room temperature for 5 min. The cultures were incubated with gentle agitation at room temperature for 20 min and then at 4 °C overnight. Cells were collected by centrifugation, washed twice with ice-cold Phosphate Buffered Saline, and resuspended in cell lysis buffer (50 mM HEPES-KOH, pH 7.5; 140 mM NaCl; 1 mM EDTA; 1% Triton X-100; 10% glycerol; 0.5% NP-40; freshly added protease inhibitor), and incubated on ice for 10 min. After centrifuging at 4000 r.p.m. at 4 °C for 10 min, the nuclei in the pellet were resuspended in nuclear lysis buffer (200 mM NaCl; 1 mM EDTA; 0.5 mM EGTA; 10 mM Tris, pH, 8.0; freshly added protease inhibitor) on ice for 10 min, disrupted by sonication and centrifuged at 15 000 r.p.m. The supernatant was then sonicated to shear the DNA to an average size of 0.3–1.0 kb. Each sample was pre-cleared by incubating with 20 µl of pre-blocked and pre-washed protein G beads (50% slurry) at 4 °C for 2–4 h. The pre-cleared samples were incubated with 1.5 µg of anti-Oct-1 at 4 °C overnight followed by incubation with 20 µl of pre-blocked protein G beads (50% slurry) at 4 °C for 1–2 h. The complex was washed with washing buffer (50 mM HEPES, pH 7.5; 140 mM NaCl; 1 M EDTA; 1% Triton X-100; 0.1% DOC; freshly added protease inhibitor) five times and was resuspended in 100 µl of TE buffer. Proteinase K (200 µg/ml) and SDS (0.5%) were added to both the bound and the input DNA samples, and samples were incubated at 65 °C overnight to reverse crosslinking. Immune complexes were precipitated, the DNA was purified, and a 2 µl aliquot was used for each PCR reaction using primer sequences that amplify a 220-bp DNA fragment that contains the octamer motif located at the IgH promoter of VH4-59 (forward, 5′-AGACCCCAAGAAGACAACTG-3′ reverse, 5′-TGCACAGGAGGTCCAGGAC-3′). PCR products were separated by 1.0% agarose gel electrophoresis and visualized by ethidium bromide staining.
Transfection of plasmids containing Oct-1 or Oct-2 or antisense oligonucleotides for Oct-1 and analysis of IgG expression by flow cytometry
Plasmids containing Oct-1, Oct-2, antisense oligodeoxynucleotides for Oct-1 ASODN (5′-AACAATCCGTCAGAA-3′) or random oligodeoxynucleotides (ODN) (5′-TTGTTAG GCAGTCTT-3′) were introduced into HeLa S3 cells by electroporation (10 µg for plasmids; 10 mM for oligonucleotides). The cells were harvested 30 h after electroporation and fixed in 70% ethanol at 4 °C for 24 h. The cells were then washed and blocked with 2% fetal bovine serum at room temperature for 30 min, incubated with rabbit antihuman IgG-FITC or anti-human IgG-PE (Sigma, St. Louis, MO, USA) at 4 °C for 30 min, washed with phosphate-buffered saline, and analyzed for IgG expression by flow cytometry (Becton Dickinson, Franklin Lakes, NJ, USA).
Statistical analysis
All results are expressed as mean±s.e.m. Differences between multiple experimental groups were detected using single-factor analysis of variance. P<0.05 was considered to be statistically significant. All data were obtained at least in duplicate from at least three separate experiments.
Acknowledgments
This work was supported by Fundamental Research Grants 30572094 and 30772470 from the Natural Sciences Foundation, China. We thank Dr Dalong Ma and Dr Mingxu Xu (Peking University Center for Human Disease Genomics) for their comments and suggestions. This manuscript was proofread by an English-speaking professional with a science background at Elixigen Corporation.
References
- Henderson A, Calame K. Transcriptional regulation during B cell development. Annu Rev Immunol. 1998;16:163–200. doi: 10.1146/annurev.immunol.16.1.163. [DOI] [PubMed] [Google Scholar]
- Qiu XY, Yang GZ. Study on the characteristics of Ig-like protein from malignant tumor cytoplasm and analysis of Ig gene structure. Chin J Immunol. 1996;5:296. [Google Scholar]
- Qiu XY, Yang GZ. Ig-like protein present in epithelial malignant tumor cells. J Norman Bethune Univ Med Sci. 1996;6:572–575. [Google Scholar]
- Wang DS, Qiu XY, Zhu XH, Lv P, Jiang N, Lv P, et al. Purification and Western-blot analysis of Ig-like protein. J Beijing Med Univ. 2000;32:310–312. [Google Scholar]
- Qiu X, Zhu X, Zhang L, Mao Y, Zhang J, Hao P, et al. Human epithelial cancers secrete immunoglobulin g with unidentified specificity to promote growth and survival of tumor cells. Cancer Res. 2003;63:6488–6495. [PubMed] [Google Scholar]
- Huang J, Sun X, Mao Y, Zhu X, Zhang P, Zhang L, et al. Expression of immunoglobulin gene with classical V-(D)-J rearrangement in mouse brain neurons. Int J Biochem Cell Biol. 2008;40:1604–1615. doi: 10.1016/j.biocel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Zhu X, Li C, Sun X, Mao Y, Li G, Liu X, et al. Immunoglobulin mRNA and protein expression in human oral epithelial tumor cells. Appl Immunohistochem Mol Morphol. 2008;16:232–238. doi: 10.1097/PAI.0b013e31814c915a. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang L, Ma T, Zhang P, Qiu X. Expression of immunoglobulin gene with classical V-(D)-J rearrangement in mouse testis and epididymis. J Histochem Cytochem. 2009;57:339–349. doi: 10.1369/jhc.2008.951434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Huang J, Mao Y, Liu S, Sun X, Zhu X, et al. Immunoglobulin gene transcripts have distinct VHDJH recombination characteristics in human epithelial cancer cells. J Biol Chem. 2009;284:13610–13619. doi: 10.1074/jbc.M809524200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimoto Y. Expression of heavy-chain constant region of immunoglobulin and T-cell receptor gene transcripts in human non-hematopoietic tumor cell lines. Genes Chromosomes Cancer. 1998;22:83–86. doi: 10.1002/(sici)1098-2264(1998)22:1<83::aid-gcc12>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Li M, Feng DY, Ren W, Zheng L, Zheng H, Tang M, et al. Expression of immunoglobulin kappa light chain constant region in abnormal human cervical epithelial cells. Int J Biochem Cell Biol. 2004;36:2250–2257. doi: 10.1016/j.biocel.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Babbage G, Ottensmeier CH, Blaydes J, Stevenson FK, Sahota SS. Immunoglobulin heavy chain locus events and expression of activation-induced cytidine deaminase in epithelial breast cancer cell lines. Cancer Res. 2006;66:3996–4000. doi: 10.1158/0008-5472.CAN-05-3704. [DOI] [PubMed] [Google Scholar]
- Chen Z, Gu J. Immunoglobulin G expression in carcinomas and cancer cell lines. FASEB J. 2007;21:2931–2938. doi: 10.1096/fj.07-8073com. [DOI] [PubMed] [Google Scholar]
- Liu HD, Zheng H, Li M, Hu DS, Tang M, Cao Y. Upregulated expression of kappa light chain by Epstein–Barr virus encoded latent membrane protein 1 in nasopharyngeal carcinoma cells via NF-kappaB and AP-1 pathways. Cell Signal. 2007;19:419–427. doi: 10.1016/j.cellsig.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Zheng H, Li M, Ren W, Zeng L, Liu HD, Hu D, et al. Expression and secretion of immunoglobulin alpha heavy chain with diverse VDJ recombinations by human epithelial cancer cells. Mol Immunol. 2007;44:2221–2227. doi: 10.1016/j.molimm.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Lee G, Laflamme E, Chien CH, Ting HH. Molecular identity of a pan cancer marker, CA215. Cancer Biol Ther. 2008;7:2007–2014. doi: 10.4161/cbt.7.12.6984. [DOI] [PubMed] [Google Scholar]
- Lee G. Cancer cell-expressed immunoglobulins: CA215 as a pan cancer marker and its diagnostic applications. Cancer Biomark. 2009;5:137–142. doi: 10.3233/CBM-2009-0610. [DOI] [PubMed] [Google Scholar]
- Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- Quong MW, Martensson A, Langerak AW, Rivera RR, Nemazee D, Murre C. Receptor editing and marginal zone B cell development are regulated by the helix–loop–helix protein, E2A. J Exp Med. 2004;199:1101–1112. doi: 10.1084/jem.20031180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki M, Ordinario EC, Cummings WJ, Fujii MM, Maizels N. E2A acts in cis in G1 phase of cell cycle to promote Ig gene diversification. J Immunol. 2009;182:408–415. doi: 10.4049/jimmunol.182.1.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlitzky I, Angeles CV, Siegel AM, Stanton ML, Riblet R, Brodeur PH. Identification of a candidate regulatory element within the 5′ flanking region of the mouse Igh locus defined by pro-B cell-specific hypersensitivity associated with binding of PU.1, Pax5, and E2A. J Immunol. 2006;176:6839–6851. doi: 10.4049/jimmunol.176.11.6839. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Suzuki H, Yamamoto-Nagamatsu N, Barman HK, Kikuchi H, Takami Y, et al. HDAC2 controls IgM H- and L-chain gene expressions via EBF1, Pax5, Ikaros, Aiolos and E2A gene expressions. Genes Cells. 2007;12:359–373. doi: 10.1111/j.1365-2443.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- Kim U, Qin XF, Gong S, Stevens S, Luo Y, Nussenzweig M, et al. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 1996;383:542–547. doi: 10.1038/383542a0. [DOI] [PubMed] [Google Scholar]
- Schubart K, Massa S, Schubart D, Corcoran LM, Rolink AG, Matthias P. B cell development and immunoglobulin gene transcription in the absence of Oct-2 and OBF-1. Nat Immunol. 2001;2:69–74. doi: 10.1038/83190. [DOI] [PubMed] [Google Scholar]
- Ju Z, Volpi SA, Hassan R, Martinez N, Giannini SL, Gold T, et al. Evidence for physical interaction between the immunoglobulin heavy chain variable region and the 3′ regulatory region. J Biol Chem. 2007;282:35169–35178. doi: 10.1074/jbc.M705719200. [DOI] [PubMed] [Google Scholar]
- Laumen H, Nielsen PJ, Wirth T. The BOB.1/OBF.1 co-activator is essential for octamer-dependent transcription in B cells. Eur J Immunol. 2000;30:458–469. doi: 10.1002/1521-4141(200002)30:2<458::AID-IMMU458>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Poellinger L, Yoza BK, Roeder RG. Functional cooperativity between protein molecules bound at two distinct sequence elements of the immunoglobulin heavy-chain promoter. Nature. 1989;337:573–576. doi: 10.1038/337573a0. [DOI] [PubMed] [Google Scholar]
- Kunkel GR, Pederson T. Upstream elements required for efficient transcription of a human U6 RNA gene resemble those of U1 and U2 genes even though a different polymerase is used. Genes Dev. 1988;2:196–204. doi: 10.1101/gad.2.2.196. [DOI] [PubMed] [Google Scholar]
- Nakajima N, Horikoshi M, Roeder RG. Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol Cell Biol. 1988;8:4028–4040. doi: 10.1128/mcb.8.10.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Muller-Immergluck MM, Seipel K, Janson L, Westin G, Schaffner W, et al. Both Oct-1 and Oct-2A contain domains which can activate the ubiquitously expressed U2 snRNA genes. EMBO J. 1991;10:2291–2296. doi: 10.1002/j.1460-2075.1991.tb07765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkley C, Perry M. Histone H2B gene transcription during Xenopus early development requires functional cooperation between proteins bound to the CCAAT and octamer motifs. Mol Cell Biol. 1992;12:4400–4411. doi: 10.1128/mcb.12.10.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Kitchingman GR. Bidirectional transcription from the human immunoglobulin VH6 gene promoter. Nucleic Acids Res. 1994;22:861–868. doi: 10.1093/nar/22.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M, Georgiev O, van Leeuwen H, van der Vliet P, Schaffner W. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 1996;15:2781–2790. [PMC free article] [PubMed] [Google Scholar]
- Massa S, Junker S, Schubart K, Matthias G, Matthias P. The OBF-1 gene locus confers B cell-specific transcription by restricting the ubiquitous activity of its promoter. Eur J Immunol. 2003;33:2864–2874. doi: 10.1002/eji.200323882. [DOI] [PubMed] [Google Scholar]
- Bartholdy B, Du Roure C, Bordon A, Emslie D, Corcoran LM, Matthias P. The Ets factor Spi-B is a direct critical target of the coactivator OBF-1. Proc Natl Acad Sci USA. 2006;103:11665–11670. doi: 10.1073/pnas.0509430103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran LM, Karvelas M, Nossal GJ, Ye ZS, Jacks T, Baltimore D. Oct-2, although not required for early B-cell development, is critical for later B-cell maturation and for postnatal survival. Genes Dev. 1993;7:570–582. doi: 10.1101/gad.7.4.570. [DOI] [PubMed] [Google Scholar]
- Corcoran LM, Hasbold J, Dietrich W, Hawkins E, Kallies A, Nutt SL, et al. Differential requirement for OBF-1 during antibody-secreting cell differentiation. J Exp Med. 2005;201:1385–1396. doi: 10.1084/jem.20042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- Delacroix L, Begon D, Chatel G, Jackers P, Winkler R. Distal ERBB2 promoter fragment displays specific transcriptional and nuclear binding activities in ERBB2 overexpressing breast cancer cells. DNA Cell Biol. 2005;24:582–594. doi: 10.1089/dna.2005.24.582. [DOI] [PubMed] [Google Scholar]


