Abstract
Nucleotide-binding and oligomerization domain 2 (NOD2), a member of the NOD protein family, plays an important role in innate immunity. In response to pathogen attack, NOD2 stimulates cytokine and defensin production by activating nuclear factor (NF)-κB, a key transcription factor responsible for mediating downstream reactions. However, the mechanism linking NOD2 regulation and NF-κB activation is poorly understood. Using bioinformatics, we found a completely preserved canonical NF-κB binding site in the NOD2 core promoter (−16 to −25 bp) in both humans and chimpanzees. The functional role of this NF-κB binding site was investigated using the enhanced green fluorescent protein (EGFP) reporter system, site-directed mutagenesis, the NF-κB activation inhibitor (JSH-23) and the chromatin immunoprecipitation (ChIP) assay. The results show that the NF-κB binding site is critical for regulation of the NOD2 gene. Either deletion of the NF-κB binding elements within the NOD2 promoter or treatment with an NF-κB activation inhibitor could lead to a significant loss of NOD2 promoter activity as detected by reporter gene assay. The canonical NF-κB binding site was bound by NF-κB as determined by the ChIP method. Based on these results, we suggest a positive feedback regulation between NF-κB and NOD2, which may represent an efficient mechanism in response to pathogen invasion.
Keywords: innate immunity, NF-κB binding site, NOD2, promoter
Introduction
The innate immune system provides an immediate defense against pathogen attack before establishment of the delayed adaptive immune response. In addition to the skin barrier and the complement system, inflammation and phagocytosis are key defense mechanisms of innate immunity. The cells detect conserved microbial components, such as pathogen-associated molecular patterns, through pattern recognition receptors. According to their specificity, function and localization, pattern recognition receptors are classified into different families. The Toll-like receptors (TLRs) are a group of transmembrane proteins that recognize extracellular and endosomal pathogen-associated molecular patterns.1 The nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs) are intracellular sensors of bacterial cell wall peptidoglycan present in both Gram-positive and -negative bacteria. Studies have shown that the functional role of NLRs is to complement and synergize with TLRs in response to pathogen invasion.2 Following the revelation of their functional roles, recent studies have identified more than 20 NLRs in humans.3 Among them are two well-known intracellular pathogen sensors, NOD1 and NOD2. Each detects different sets of pathogen components.4 NOD1 recognizes diaminopimelic acid from Gram-negative bacteria and NOD2 detects intracellular muramyl dipeptide,5 a peptidoglycan constituent of both Gram-positive and -negative bacteria.6 In addition, the NOD1 protein is found in various tissues, while NOD2 expression is restricted to intestinal epithelial cells, monocytes, macrophages, dendritic cells and T cells.7
NLRs are characterized by three distinct functional domains: the C-terminal leucine-rich repeat (LRR) domain is the microbe detection sensor, the central NACHT domain is required for self-oligomerization and nucleotide binding that leads to protein activation, and the N-terminal caspase activation and recruitment domain (CARD) is an effector-binding region that facilitates protein–protein interaction.8 Systematic mutation studies of NOD2 have indicated that the C-terminal LRR motifs are critical for pathogen recognition.9 The NOD2 protein might undergo conformational rearrangement so that the protein becomes more accessible upon sensing bacteria.10 Subsequently, NOD2 is able to recruit downstream effectors. Through CARD–CARD interaction, NOD2 forms a complex with the receptor-interacting protein 2 (RIP2).11 The NOD2–RIP2 complex further interacts with the NF-κB essential modulator,12 which consequently activates NF-κB by releasing it from the IκB proteins. Activated NF-κB initiates a large number of gene transcripts and triggers the immune response following its translocation into the nucleus.
The importance of NF-κB in the innate immune system has been evidenced by numerous studies demonstrating that TLRs and NLRs could induce cytokine production through NF-κB activation.13, 14, 15 For example, functional binding sites for NF-κB and activator protein-1 in the human β-defensin 2 promoter are essential for NOD2-mediated induction.16 In addition, a mutation of the NOD2 gene has consistently been associated with the reduced response of cytokines and a diminished NF-κB activation.17 Due to the close relationship between NOD2 and NF-κB in immunity, there is an urgent need to understand the regulatory mechanisms governing these two proteins. In this report, we describe our approach to explore the characterization of the canonical NF-κB binding site in the NOD2 promoter region by comparing the promoter sequences among six mammalian species. We further verified the function of this binding site using molecular biological methods. The study will provide important insights for effective inflammation treatment.
Materials and methods
NLR and NOD2 promoter analyses for highly conserved motifs
We intend to identify highly conserved transcription factor binding sites in the promoter regions among NLR genes. There are currently 22 known NLR genes in the human genome.4 We extracted and scanned the sequences of these 22 gene promoters. Then, we focused on the NOD2 promoter for comparative analysis due to its broad sensing capability of pathogens. We examined both DNA and protein sequences. In this report, we consider that the proximal promoter ranges from −1000 to +300 bp relative to the transcription start site (TSS), while the core promoter is limited to upstream −40 bp. The TSS at the most 5′ end is used if the gene has more than one transcript. All DNA sequences were taken from GenBank18 and promoters were extracted using Gene Sorter (UCSC, http://genome.ucsc.edu/index.html). Homologous gene sequences from the NCBI HomoloGene site (http://www.ncbi.nlm.nih.gov/sites/entrez?db=homologene) were used to conduct comparative analysis. Based on our preliminary screening, we narrowed down our target of NOD2 homolog genes (HomoloGene ID 11156) to six mammalian members, including human (Homo sapiens, NM_022162.1), chimpanzee (Pan troglodytes, XM_510962.2), mouse (Mus musculus, NM_145857.2), rat (Rattus norvegicus, XM_226330.4), dog (Canis lupus, XM_544412.2) and cow (Bos taurus, NM_001002889.1).
Promoter sequences were scanned for conserved motifs by Dragon Motif Builder.19 Highly conserved sequences were selected and further matched against both JASPAR (http://jaspar.cgb.ki.se/) and MATCH databases (http://www.gene-regulation.com/cgi-bin/pub/programs/pmatch/bin/p-match.cgi) to verify the known transcription factor binding sites. CLUSTAL W2 (http://www.ebi.ac.uk/Tools/clustalw2/) was used for multiple sequence alignment.
Polymerase chain reaction (PCR)
Human genomic DNA extracted from peripheral blood using a Qiagen kit was used as a template. Four sizes of NOD2 promoter containing an NF-κB binding site consensus (GGGAATTTCC) were amplified by the PCR. The primers were designed using Primer 5.0 software and synthesized by Shanghai Sangon Biological Engineering Technology & Service (Shanghai, China). The primer sequences used were the following: the upstream primer U1 (nt-566), 5′-GGCGGCATTAATGAAGCAGGAGAAACT-3′ U2 (nt-696), 5′-GGCGGCATTAATACTGACAGATTTCGC-3′ U3 (nt-1085), 5′-GGCGGCATTAATAGTGTTAGGGAGGGAGAA-3′ U4 (nt-1336), 5′-GGCGGCATTAATTTGAGCAATGGGAGGTC-3′ and the universal downstream primer D (nt+51), 5′-AAAACTGCAGAGAACGCCAAAAGCC-3′. The underlined nucleotides indicate the recognition sites for restriction endonucleases VspI (5′ end) and PstI (3′ end). The sizes of the PCR products were 617, 747, 1136 and 1387 bp, respectively. The reaction mixture was predenatured at 94 °C for 5 min, 30 cycles at 94 °C for 45 s, 57–61 °C for 45 s, 72 °C for 1–1.5 min and a final extension at 72 °C for 10 min. Each sample was amplified in duplex reactions. The PCR products were separated by 1% (w/v) agarose gel electrophoresis.
Construction of enhanced green fluorescent protein (EGFP) expression vectors controlled by NOD2 promoters
Fragments from the above PCR of interest were excised from an agarose gel and digested with VspI and PstI restriction enzymes (AseI Isoschizomer). The amplified products were directly linked to the pEGFP-N3 vector (Clontech, Mountain View, CA, USA) in which the original cytomegalovirus (CMV) promoter was previously removed. The ligated products were transformed into DH5α competent cells (Invitrogen, Carlsbad, CA, USA) and the recombinants were selected by kanamycin. For verification, all constructs were PCR screened, endonuclease digested and sequenced (ABI3700 sequencer). Finally, the four NOD2 promoter-controlled EGFP expression vectors, pNOD2 (617)-EGFP, pNOD2 (747)-EGFP, pNOD2 (1136)-EGFP and pNOD2 (1387)-EGFP, were constructed.
Construction of a mutated plasmid without an NF-κB binding site
To investigate the function of the NF-κB binding site for gene transcription, the binding site was deleted from the 617-bp NOD2 promoter fragment using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). The primer set was designed as forward, 5′-CCGGCTTTTCCTTTCTTGAAGGTGGGGT-3′ and reverse, 5′-ACCCCACCTTCAAGAAAGGAAAAGCCGG-3′. The mutant promoter (607 bp) without the NF-κB binding site replaced the CMV promoter in pEGFP-N3 using the same method described above. The mutated plasmid was constructed and designated as pmNOD2 (607)-EGFP.
Cell culture and transfection
Human embryonic kidney 293 (HEK293) and HeLa cells were purchased from the Chinese Academy of Sciences (Beijing, China) and cultured in DMEM (Gibco, Invitrogen) supplemented with 10% fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin sulfate. All cell cultures were maintained at 37 °C in a humidified incubator with 5% CO2. The cells were seeded at a density of 106 cells/well on six-well plates 24 h before transfection and grown until confluence of 80–90% at the time of transfection. Two micrograms of the plasmid was added to each well and transfected into HEK293 or HeLa cells using LipofectamineTM 2000 (Invitrogen).
Treatment with NF-κB activation inhibitor
HEK293 cells were transfected with pNOD2 (617)-EGFP as described above. After 24 h of transfection, the cells were treated with 20 µm JSH-23 (NF-κB activation inhibitor; Calbiochem, San Diego, CA, USA). The green fluorescent protein (GFP) expression was measured under a fluorescence microscope 48 h after transfection.
Fluorescence intensity measurement
The expression of GFP driven by NOD2 promoters was evaluated quantitatively based on the fluorescence intensity of cells transfected with various pNOD2-EGFP constructs and pmNOD2-EGFP. The slides were examined under an inverted microscope (Leica Qwin; Leica, Tokyo, Japan) at an excitation wavelength of 488 nm and an emission wavelength of 510 nm. The fluorescence intensity from the cells was measured and normalized by subtracting the background intensity. Each measurable field was 373 800 mm2 for this study. Twenty non-redundant fields for each experimental group were used for intensity calculation. The optimal cytoplasmic region for examination was selected by Scion Image software. Image analysis and processing software from Leica Qwin was used to capture and calculate the intensity. Quantitation of the fluorescence intensity in the entire cell (expressed as a percentage of total cellular content) was performed using the formula:
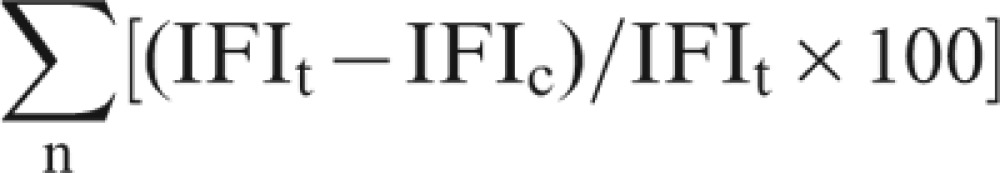 |
where n stands for the number of optical sections required to scan the entire cell, and IFIt and IFIc stand for integrated fluorescence intensity of the entire cell and of the cytoplasm within a given optical section, respectively.20
Chromatin immunoprecipitation (ChIP)
ChIP was performed following the protocol from Millipore. HEK293 cells were cultured in DMEM, supplemented with 10% fetal calf serum, and transfected with the pNOD2 (617)-EGFP using LipofectamineTM 2000 reagent. Plates were returned to the incubator for 40–48 h. Approximately 3×106 cells were used for each immunoprecipitation. The fixation was performed using 1% formaldehyde at 37 °C for 30 min and quenched with glycine to 125 mM. The cells were washed with phosphate-buffered saline and subsequently collected. The cells were resuspended and incubated on ice for 10 min. The resuspended cells were sonicated to shear DNA in order to obtain genomic DNA fragments ranging in size from 200 bp to 1 kb. The sonicated cell extracts were immunoprecipitated using an antibody against the p65 subunit of NF-κB (Santa Cruz Biotechnology, Santa Cruz, CA, USA). As a negative control, an aliquot of cross-linked chromatin was subjected to the same immunoprecipitation reaction in the presence of a normal mouse IgG antibody. Prior to immunoprecipitation, an aliquot of each sample was served as an ‘input' fraction (PCR control). The immunoprecipitates were washed and cross-links were reverted by heating at 65 °C overnight. The samples were then treated with proteinase K and DNA was purified using spin columns. PCR was performed on both genomic input and ChIP DNA. PCR primers were designed covering the NF-κB consensus sequences in the NOD2 promoter (forward: 5-TAGTTCTGGAAGGCTGGT-3; reverse: 5- CCCATCAAAGCCCATTAG-3). As a positive control, the primers were designed for the human glyceraldehyde-3-phosphate dehydrogenase genes. Amplification was performed using titanium Taq polymerase at thermocycler settings of one cycle at 95 °C for 3 min, followed by 32 cycles of 95 °C for 45 s, 58 °C for 30 s and 72 °C for 40 s. The PCR reaction was performed in triplicate for each independent ChIP preparation.
Statistical analysis
Each set of experiments was repeated three times using the same protocol. Statistical analyses were performed using the SPSS 13.0. The results were presented as mean±SD. The significant difference between experimental groups was analyzed by one-way ANOVA. For a comparison of significant differences the Bonferroni post hoc test was applied.
Results
NLR gene promoter analysis
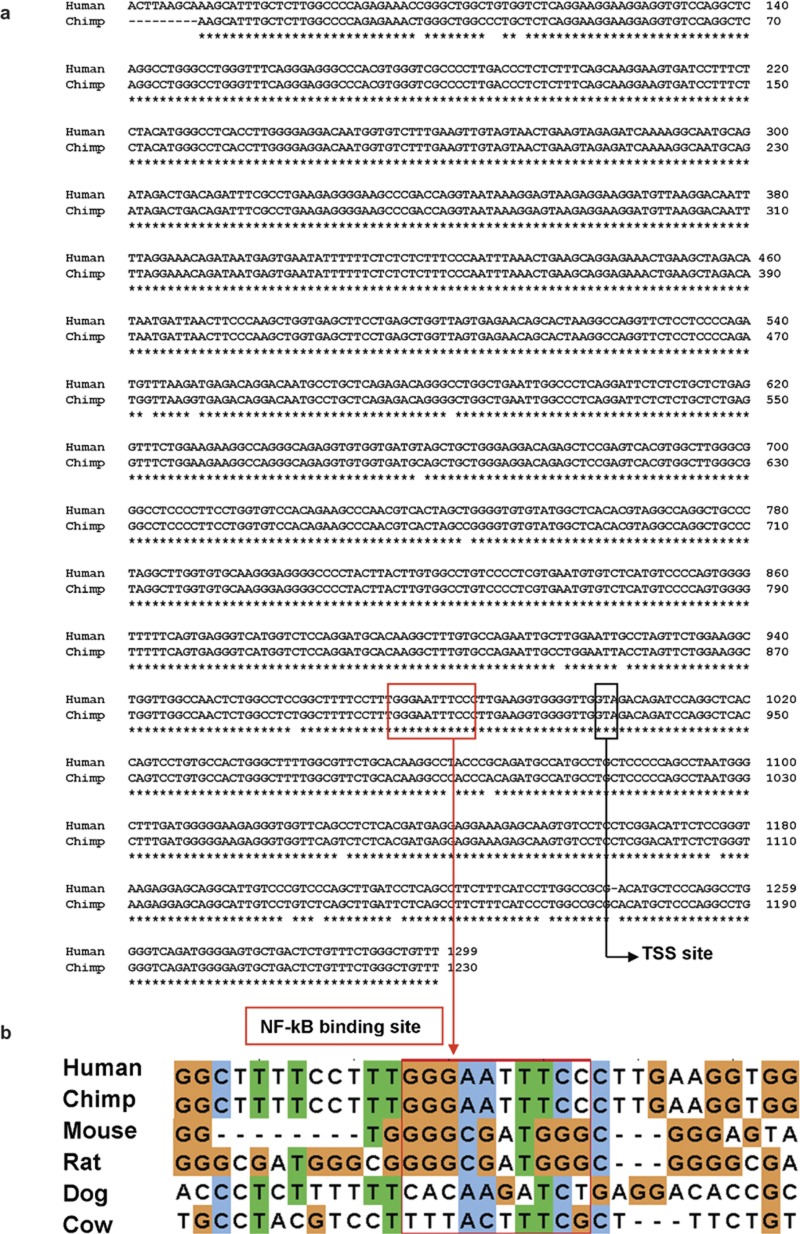
Twenty-two human NLR gene promoters were screened and the SP1 binding site was found in four genes (NLRP1, NLRP2, NLRP13 and NLRC3), while the canonical NF-κB binding site ‘GGGAATTTCC' appeared only in the NOD2 gene. We further examined homologous genes to seek the preservation of the SP1 and NF-κB binding sites among closely related mammals. We could not find the SP1 binding site at a similar location among homologs, but we found a completely preserved NF-κB binding site in the chimpanzee NOD2 promoter. A key finding is that the location of this consensus is in the core promoter range, only 16 bp upstream from the TSS. In addition to the NF-κB binding site, the proximal promoters of the NOD2 gene were also analyzed for more information pertaining to a role in transcription. Although the available sequence of chimpanzee NOD2 is limited to −930 bp (upstream to TSS), the alignment of these two sequences shows a high level of similarity. They are 98% identical and have only a single 1-bp gap in the 1230-bp long sequence (−930 to +300 bp). Figure 1 shows the alignment and preservation of the NF-κB binding site in the human and chimpanzee NOD2 promoter sequences in detail.
Figure 1.
NOD2 promoter sequence analysis. (a) Alignment of NOD2 proximal promoters between humans (−1000 to +300 bp) and chimpanzees (–930 to +300 bp) shows a 98% identical rate with only a 1-bp gap. The canonical NF-κB binding site and the TSS are boxed and labeled. (b) The canonical NF-κB binding site (–16 to –25 bp) is present in both human and chimpanzee NOD2 core promoters, but it is not found in the mouse, rat, dog and cow homologs. NF, nuclear factor; NOD2, nucleotide-binding and oligomerization domain 2; TSS, transcription start site.
NOD2 promoter and protein sequence alignment
We performed multiple sequence alignment for NOD2 promoters among humans, chimpanzee, dog, cow, mouse and rat. The proximal promoter (−1000 to +300 bp) alignment showed poor conservation among the five species. To distinguish potential meaningful motifs from noise, we limited their promoter sequences to the segment of −500 to +300 bp. The overall comparison exhibits enhanced improvement in similarity in the shorter sequences (Table 1). The alignments showed identical rates, ranging from 48 to 53% between humans and dog, cow, mouse or rat. However, over 85% similarity was found between mouse and rat. No NF-κB binding site consensus was found in the NOD2 promoters from dog, cow, mouse and rat (Figure 1b).
Table 1. NOD2 promoter (−500 to +300 bp) sequence comparison (identical rate: %).
| Organism | Human | Chimpanzee | Dog | Cow | Mouse |
|---|---|---|---|---|---|
| Chimpanzee | 97.88 | ||||
| Dog | 48.41 | 48.93 | |||
| Cow | 52.60 | 49.02 | 51.25 | ||
| Mouse | 49.77 | 49.88 | 48.91 | 50.22 | |
| Rat | 49.65 | 49.42 | 50.17 | 50.86 | 85.43 |
Abbreviation: NOD2, nucleotide-binding and oligomerization domain 2.
The divergence of promoter sequences among the homologs prompted us to conduct further investigation with the coding sequence of NOD2 genes. The similarity of protein sequences among the homologs is significantly correlated to the promoter sequence alignment, as shown in Table 2. The highest similarity among these protein sequences is found between humans and chimpanzee (98% identical rate), followed by mouse and rat (93% identical rate). Functional structures, such as CARD, LRR and NATCH domains, are well conserved in all NOD2 proteins, while the length of the peptides varies individually.
Table 2. NOD2 protein sequence comparison.
| Organism | Accession number | Number of amino acids | Identical rate compared to human NOD2 (%) |
|---|---|---|---|
| Human | NP_071445.1 | 1040 | – |
| Chimpanzee | XP_510962.2 | 1040 | 98 |
| Dog | XP_544412.2 | 1311 | 80 |
| Cow | NP_001002889.1 | 1013 | 81 |
| Mouse | NP_665856.2 | 1013 | 79 |
| Rat | NP_001099642 | 932 | 80 |
Abbreviation: NOD2, nucleotide-binding and oligomerization domain 2.
Validation of NOD2 promoter construction
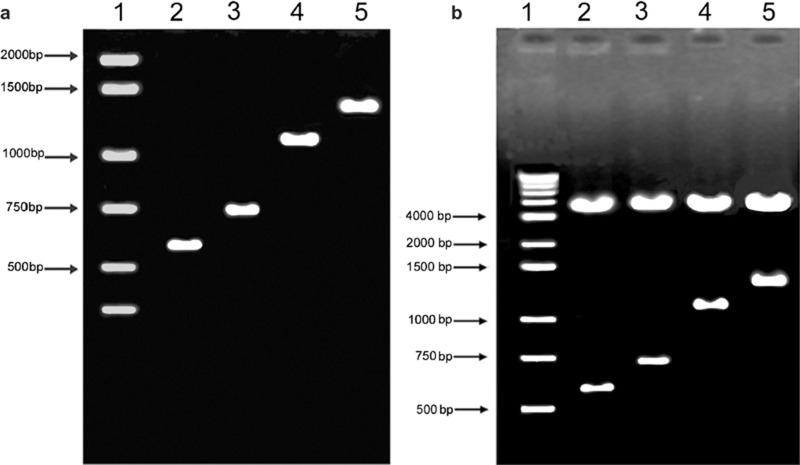
To verify the identities and correct orientation of the promoter sequences, the recombinants were purified, digested, amplified and sequenced. Our experiments confirmed that four sizes of NOD2 promoter fragments were constructed correctly into the pEGFP-N3 vector. The PCR results are illustrated in Figure 2a and the recombinants after restriction enzyme digestion are shown in Figure 2b. All recombinants contain the canonical NF-κB binding site in their promoters.
Figure 2.
Verification of NOD2 promoter recombinants. (a) The promoter fragments obtained from PCR were separated by 1% agarose gel electrophoresis. Lane 1: marker; lane 2: 617 bp; lane 3: 747 bp; lane 4: 1136 bp; lane 5: 1387 bp. (b) Recombinant vectors were digested by VspI and PstI restriction enzymes. Lane 1: marker; lane 2: pNOD2 (617)-EGFP; lane 3: pNOD2 (747)-EGFP; lane 4: pNOD2 (1136)-EGFP; lane 5: pNOD2 (1387)-EGFP. EGFP, enhanced green fluorescent protein; NOD2, nucleotide-binding and oligomerization domain 2.
Detection of GFP-positive cells
The GFP expression was observed under inverse fluorescence microscopy in HEK293 cells transfected with pEGFP-N3, pNOD2 (617)-EGFP, pNOD2 (747)-EGFP, pNOD2 (1136)-EGFP, pNOD2 (1387)-EGFP, pmNOD2 (607)-EGFP, pNOD2 (617)-EGFP treated with JSH-23 and deleted promoter (negative control), respectively (Figure 3a). The fluorescence intensity was measured to infer NOD2 promoter activity and is shown in Figure 3b.
Figure 3.
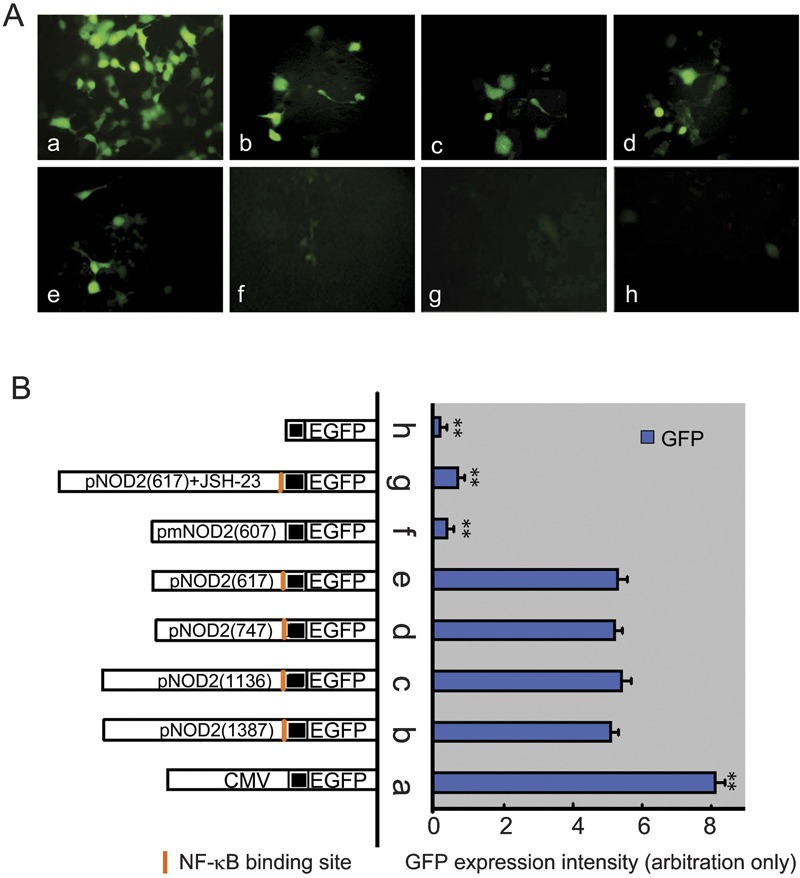
Detection of GFP by fluorescence microscopy 48 h after transfection (×400). (A) Expression of GFP in transfected HEK293 cells driven by various NOD2 promoters. (B) Recombinant vector structure and the corresponding fluorescence intensity in HEK293 cells. **P<0.01 versus groups b–e. GFP, green fluorescent protein; HEK293, human embryonic kidney 293; NOD2, nucleotide-binding and oligomerization domain 2.
The GFP expression was obvious in the pNOD2-EGFP groups and the pEGFP-N3 group (CMV promoter control) but was not found in the negative control group. No significant difference was observed among the pNOD2-EGFP groups with various sizes of NOD2 promoter fragments, although their expression was significantly weaker compared with the CMV control group. In addition, the substitution of NOD2 promoter fragments did not substantially change the profiles of reporter gene expression. Similar morphology and behavior were seen in pNOD2-EGFP- and pEGFP-N3-transfected cells, as well as in HeLa cells (data not shown). These findings confirm that the constructed pNOD2-EGFP system is functional. It also suggests that the shortest promoter fragment (617 bp) contains essential sequences for transcription.
The GFP expression in cells with the NF-κB binding site deletion and treatment with JSH-23
Given the results in Figure 3 that showed that GFP expression did not vary significantly with the various lengths of NOD2 promoters, we constructed pmNOD2-EGFP by deleting the 10-bp NF-κB binding site from the 617-bp fragment. The NF-κB knockout recombinant was identified by restriction enzyme digestion and sequencing (Figure 4). As expected, a dramatic decline in GFP expression was observed in cells containing the mutated plasmid (NF-κB binding site deletion). The fluorescence intensity was reduced by nearly sixfold in cells transfected with pmNOD2-EGFP compared to the pNOD2-EGFP groups, and no significant difference was seen between the pmNOD2-EGFP group and the pNOD2 (617)-EGFP treated with JSH-23 group (Figure 3). Similar results were found in HeLa cells (data not shown).
Figure 4.
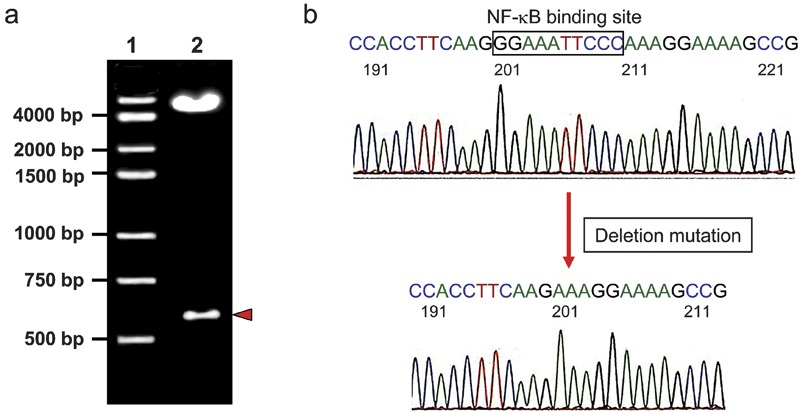
Deletion of the NF-κB binding site from the NOD2 promoter is confirmed by restriction enzyme digestion and sequencing. (a) The mutated recombinant vector pmNOD2 (607)-EGFP was digested by VspI and PstI restriction enzymes and separated by 1% agarose gel electrophoresis. Lane 1: Marker; lane 2: pmNOD2 (607)-EGFP. (b) The sequencing result showed the deletion of the NF-κB binding site (GGAAATTCCC). EGFP, enhanced green fluorescent protein; NF, nuclear factor; NOD2, nucleotide-binding and oligomerization domain 2.
Identification of NF-κB binding to NOD2 promoter by ChIP method
A ChIP experiment was performed to determine the binding of NF-κB to the NOD2 promoter. The promoter sequences were amplified specifically from the samples and the abundant enrichment for these sequences was seen when an anti-p65 antibody was used (Figure 5). A mouse IgG antibody served as a control for non-specific precipitation of protein–DNA complexes and no PCR signal of the target transcript was observed. The samples were also subjected to a PCR reaction using primers for the glyceraldehyde-3-phosphate dehydrogenase promoter to ensure that the immunoprecipitation was successful.
Figure 5.
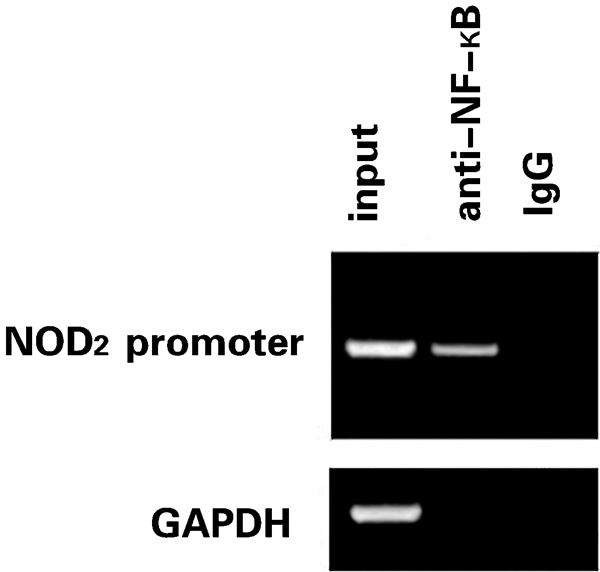
Binding of NF-κB to the NOD2 promoter was confirmed by ChIP. HEK293 cells were transfected with the promoter construct. ChIP was performed with an antibody specific for p65 as well as a non-specific antimouse IgG antibody. PCR was carried out using primers specific for the NF-κB binding site in the NOD2 promoter. Prior to immunoprecipitation, an aliquot of each sample was saved as an ‘input' fraction (PCR control). The samples were also subjected to a PCR reaction using primers for GAPDH promoter. ChIP, chromatin immunoprecipitation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HEK293, human embryonic kidney 293; IgG, immunoglobulin G; NF, nuclear factor; NOD2, nucleotide-binding and oligomerization domain 2.
Discussion
Gene expression is regulated at multiple levels, and the formation of the functional initiation complex in the core promoter is a key step for transcription. The transcription factors are a group of proteins that activate or block gene transcription by binding to specific sequences in a promoter where clusters of functional motifs are assembled for vigorous regulation. Although promoters are extremely divergent and difficult to characterize,21 they do share some common features. In fact, promoter sequences with comparable patterns for protein binding and structure conformation are considered indicators of functional association.22 Experimental evidence has shown that the frequency and location of NF-κB/Rel binding sites in proximal promoters can be used to successfully classify a subset of genes.23 In this study, we attempted to identify common regulatory motifs among human NLR genes to understand their regulatory mechanism. In our promoter screening exercise, we observed SP1 binding sites in four of 22 NLR genes, but none of them was located in the core promoter. Attempts to detect them in mammalian homologs were also unsuccessful. These results are not unexpected considering the complimentary roles of NLR genes and the genetic diversity among homologs.
To our best knowledge, this is the only report of the extreme similarity between the NOD2 promoters of humans and chimpanzees. Our key finding is that this high similarity covers the proximal promoter region (−930 to +300 bp) with only a 1-bp gap. If we assume that the core promoter is responsible for instructing the initiation of transcription, the proximal promoter covers primary elements and the distal promoter embeds selective components for transcription regulation, then the 1230 bp proximal promoter should contain almost all of the essential motifs and the instructions for transcription. Therefore, it is reasonable to believe that the human and chimpanzee NOD2 genes are regulated by a highly similar transcriptional mechanism. This hypothesis is supported by a genome-wide regulatory evolution study between humans and chimpanzees.24 On the other hand, a lack of conserved promoter structure between humans and other closely related mammals, such as mouse, rat, dog and cow, may indicate divergent mechanisms used in their NOD2 regulation. Thus, experimental evidence from these mammals may not be appropriate to infer the regulation of the human NOD2 gene and the subsequent innate immune response.
In this study, we did not find the typical TATA box, GT box and CT box in NOD2 promoters, but instead we positively identified a canonical NF-κB binding site near the TSS. Considering the core promoter position and sequence conservation between humans and chimpanzees, we believe that this binding site plays a vital role in the regulation of the NOD2 gene. The NF-κB binding site in the human NOD2 promoter had been identified, and its deletion resulted in a loss of gene activation induced by tumor-necrosis factor (TNF)-α and TNF-α/interferon-γ.25 Our results further confirm that this NF-κB binding site is an essential element driving transcription without cytokine stimulation. Interestingly, a canonical NF-κB binding site near the TSS was found to affect Cyp2c11 gene transcription.26
Transcriptional regulation is complicated in that no universal core promoter model fits all genes, and no identical set of transcription factors is present in all cells. In order to validate the active role of the NF-κB binding site in living cells, we examined the constructed NOD2 promoter in HEK293 cells and HeLa cells (data not shown). An EGFP reporter system was used to minimize functional disruption of the cells, and fluorescence intensity was analyzed to infer NOD2 promoter activity. Similar fluorescence intensity from cells transfected with various sizes of NOD2 promoter indicates that the minimum size fragment (617 bp) contains sufficient instruction for transcription. NOD2 promoter activity was markedly reduced in cells containing the deleted NF-κB binding site promoter or the shortest promoter fragment (617 bp) treated with NF-κB activation inhibitor and the ChIP method confirmed that NF-κB could bind to the NOD2 promoter. These results imply that this binding site has an essential role in the regulation of the NOD2 gene. In addition, NOD2 promoter activity did not require the presence of TNF-α and TNF-α/interferon-γ. This cytokine-independent regulatory mechanism may enable living cells to rapidly promote NOD2 expression and initiate innate immune response.
The innate immune system is the first layer of defense against pathogen invasion. It features a fast reaction to complement the delayed adaptive immune response. This fast response relies on the ability of the cells to recognize pathogens through extracellular and intracellular sensors, namely, TLRs and NLRs. NOD2, a key member of NLR family, activates NF-κB via the CARD–CARD interaction with RIP2 and other molecules. As a result, active NF-κB induces cascades of reactions to regulate inflammatory molecules in response to infection.27 After translocation to the nucleus, NF-κB binds to specific DNA sequences in a target gene's promoter or enhancer. As a central event in innate immunity, the NOD2-induced NF-κB activation pathway has been a focus in research, but little attention has been paid to how NF-κB affects NOD2 by feedback control. Our results in this study suggest a positive feedback regulation between NF-κB and NOD2. Responding to bacterial components, NOD2 triggers an innate immune response through NF-κB activation. In return, NF-κB may amplify the cascade reaction by upregulating the NOD2 gene. This positive feedback may represent an efficient mechanism during a response to pathogen invasion.
In summary, we have confirmed a functional NF-κB binding site in the human NOD2 core promoter. This binding site is critical for NOD2 regulation and its deletion or the disruption of NF-κB activation significantly reduced NOD2 promoter activity. In addition, the NOD2 promoter construct can drive gene expression without cytokine stimulation. These results may reveal a novel mechanism by which NF-κB regulates NOD2 to respond efficiently to invading pathogens. We also suggest that a similar NOD2 regulatory mechanism may exist in chimpanzees due to the presence of proximal promoter sequences and the NF-κB binding site, which is highly similar to that of humans. The variation in NOD2 promoters among other mammals indicates diverse regulatory mechanisms. Thus, the experimental results and the models derived from these mammals may not be appropriate to our understanding of the innate immune response in humans.
Acknowledgments
We thank Professor Yubing Zhou (Medical College of Jinan University) for her guidance and technical assistance. Our authors also appreciate the valuable discussion and amendment made by Dr Kum-heng Poon (Health Canada). This work was supported by grants from the Natural Science Foundation of Guangdong Province (No. 06025159) and the Natural Science Foundation from Department of Education of Guangdong Province (No. 126 (2005)).
References
- Heine H, Ulmer AJ. Recognition of bacterial products by Toll-like receptors. Chem Immunol Allergy. 2005;86:99–119. doi: 10.1159/000086654. [DOI] [PubMed] [Google Scholar]
- Murray PJ. NOD proteins: an intracellular pathogen-recognition system or signal transduction modifiers. Curr Opin Immunol. 2005;17:352–358. doi: 10.1016/j.coi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Proell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS One. 2008;3:e2119. doi: 10.1371/journal.pone.0002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Travassos LH, Hervé M, Blanot D, Boneca IG, Philpott DJ, et al. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003;278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- Kufer TA, Banks DJ, Philpott DJ. Innate immune sensing of microbes by Nod proteins. Ann NY Acad Sci. 2006;1072:19–27. doi: 10.1196/annals.1326.020. [DOI] [PubMed] [Google Scholar]
- Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- Inohara N, Nuñez G. The NOD: a signalling molecular that regulates apoptosis and host defense against pathogens. Oncogene. 2001;20:6473–6481. doi: 10.1038/sj.onc.1204787. [DOI] [PubMed] [Google Scholar]
- Tanabe T, Chamaillard M, Ogura U, Zhu L, Qiu S, Masumoto J, et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004;23:1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmanski JM, Petnicki-Ocwieja T, Kobayashi KS. NLR proteins: integral members of innate immunity and mediators of inflammatory diseases. J Leukoc Biol. 2008;83:13–30. doi: 10.1189/jlb.0607402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufer TA, Fritz JH, Philpott DJ. NACHT-LRR proteins (NLRs) in bacterial infection and immunity. Trends Microbiol. 2005;13:381–388. doi: 10.1016/j.tim.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Abbott DW, Wilkins A, Asara JM, Cantley LC. The Crohn's disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol. 2004;14:2217–2227. doi: 10.1016/j.cub.2004.12.032. [DOI] [PubMed] [Google Scholar]
- Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Shaw MH, Reimer T, Kim YG, Nuñez G. NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Curr Opin Immunol. 2008;20:377–382. doi: 10.1016/j.coi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss E, Wehkamp J, Wehkamp K, Stange EF, Schröder JM, Harder J. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J Biol Chem. 2006;281:2005–2011. doi: 10.1074/jbc.M511044200. [DOI] [PubMed] [Google Scholar]
- Rosenstiel P, Jacobs G, Till A, Schreiber S. NOD-like receptors: ancient sentinels of the innate immune system. Cell Mol Life Sci. 2008;65:1361–1377. doi: 10.1007/s00018-008-7502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E, Yang L, Chowdhary R, Kassim A, Bajic VB. Proceedings of the Information Processing and Living Systems. Singapore: World Scientific Press; 2005. An algorithm for ab initio DNA motif detection; pp. 611–614. [Google Scholar]
- Janecki AJ, Janecki M, Akhter S, Donowitz M. Quantitation of plasma membrane expression of a fusion protein of Na/H exchanger NHE3 and green fluorescence protein (GFP) in living PS120 fibroblasts. J Histochem Cytochem. 2000;48:1479–1492. doi: 10.1177/002215540004801105. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter – the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Ng P, Zhao M, Wong TK, Yiu SM, Lau YL. Promoter-sharing by different genes in human genome – CPNE1 and RBM12 gene pair as an example. BMC Genomics. 2008;9:456. doi: 10.1186/1471-2164-9-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting K, Rao S, Hardy K, Woltring D, Denyer GS, Wang J, et al. Genome-wide analysis of gene expression in T cells to identify targets of the NF-kappa B transcription factor c-Rel. J Immunol. 2007;178:7097–7109. doi: 10.4049/jimmunol.178.11.7097. [DOI] [PubMed] [Google Scholar]
- Uddin M, Wildman DE, Liu G, Xu W, Johnson RM, Hof PR, et al. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc Natl Acad Sci USA. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstiel P, Fantini M, Bräutigam K, Kühbacher T, Waetzig GH, Seegert D, et al. TNF-alpha and IFN-gamma regulate the expression of the NOD2 (CARD15) gene in human intestinal epithelial cells. Gastroenterology. 2003;124:1001–1009. doi: 10.1053/gast.2003.50157. [DOI] [PubMed] [Google Scholar]
- Iber H. Chen Q, Cheng PY, Morgan ET. Suppression of CYP2C11 gene transcription by interleukin-1 mediated by NF-kappaB binding at the transcription start site. Arch Biochem Biophys. 2000;377:187–194. doi: 10.1006/abbi.2000.1772. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2000;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]