Abstract
We describe a novel assay that permits measurement of entry of murine leukemia virus and pseudotypes with greater sensitivity and more rapidly than previously possible. To achieve this, we encapsulated a sensitive reporter enzyme, luciferase, directly into fully infectious, intact viral particles. The enzyme is specifically targeted to the viral lumen, as a C-terminal fusion on the viral envelope protein. Only when the incorporated luciferase is released from the viral lumen and gains access to its substrates is light emitted and readily detected. When cells are perfused with luciferin, quantitative measurements of entry can be made in real time on live cells. Uniquely, the amount of cell-bound virus can be determined in the same assay by addition of detergent to expose the luciferase. We demonstrate that virus carrying a mutation in the fusion peptide binds normally to cells but is unable to infect them and gives no entry signal. Using this assay, we show that inhibitors of endosomal acidification inhibit signal from vesicular stomatitis virus pseudotypes but not murine leukemia virus, consistent with a pH-independent mode of entry for the latter virus. Additionally, the fusion kinetics are rapid, with a half-life of 25 min after a delay of 10 to 15 min. The future use of this assay will permit a detailed examination of the entry mechanism of viruses and provide a convenient platform to discover novel entry inhibitors. The design also permits packaging of potential therapeutic protein cargoes into functional virus particles and their specific delivery to cellular targets.
To infect cells, viruses must overcome the substantial barrier imposed by the cell membrane and deliver the virus core, containing the genome, into the cell cytoplasm. For enveloped viruses, this requires catalyzing fusion of the viral and cell lipid membranes, an energetically unfavorable event. Conformational changes in the envelope proteins are required for this and typically result in exposure of previously buried hydrophobic peptides that insert themselves into the cell membrane and possibly the viral membrane. Fusion peptide insertion and other conformational changes destabilize the bilayer, resulting in formation of a pore that connects the cytoplasm of the virus and cell. The pore then widens until the core can be released. Much of our understanding of this process has been obtained through the identification and use of specific inhibitors and the development of sufficiently sensitive entry assays.
Our knowledge of enveloped virus entry mechanisms has been dominated by pH-dependent models such as influenza A virus and Semliki Forest virus. This is principally because, for these viruses, it is possible to induce an en masse fusion event by dropping the pH of the medium. Under these circumstances, fluorescence dequenching, or FRET (fluorescence resonance energy transfer), assays effectively measure the kinetics of fusion and have been used to understand the effects of mutations and antiviral drugs (3, 8). For this, fluorescent probes incorporated into the virus membrane mix with and become diluted into the target cell or liposome membrane. The resulting change in fluorescence gives a real-time measure of fusion. Unfortunately, the application of these classical entry assays to the pH-independent viruses has been difficult, as fusion events are infrequent and cannot be coordinated and receptors tend to be integral, multi-transmembrane span-containing proteins that are difficult to manipulate. In such studies, passive diffusion of the fluorophore contributes significantly to the signal and complex analysis of the data is required to observe signal due to fusion events.
The need for more sensitive measurement of pH-independent virus entry has led to the development of assays to detect cell-cell fusion and early genome replication events and assays in which recombinant viral protein-green fluorescent protein (GFP) fusions are used (12, 18, 27). In the cell-cell fusion assay, cells made to express virus envelope proteins on their surfaces are labeled with one fluorophore and mixed with target cells bearing receptor and a second fluorophore. Fusion is measured by observing syncytium formation. Independent labeling of the cell membrane and cytoplasm provides information on membrane and cytoplasm mixing. This assay has been used to confirm the role of factors important in fusion. However, syncytium formation is slow and does not correlate to infection kinetics. Additionally, for human immunodeficiency virus (HIV), the chemokine receptor Bonzo promotes syncytia and yet plays no significant role in entry (11, 25).
Other assays detect virus infection. Most commonly, infection is measured by using reporter gene expression in the infected cell. Obviously, to obtain expression, a virus must penetrate the cell membrane, the core must be trafficked to the correct subcellular location and then the genome becomes exposed, and finally the reporter must be expressed. This process is complex and as for retroviruses may require the cell to be at a specific stage in the growth cycle. For retroviruses, gene expression requires at least 24 h after contact with cells and is far removed from the initial entry event.
Virus-protein fusions to GFP have been useful to follow virus after it has entered the cell, and the use of fluorescently labeled dUTP has even permitted visualization of genomes undergoing reverse transcription (18). However, these assays cannot be easily used to examine entry as cell-bound virus cannot be differentiated from that which has just entered, and for retroviruses at least, particle/infectious particle ratios typically exceed 10 to 100. This means that most virus is either defective or trafficked to nonproductive pathways within the cell.
Content-mixing assays demonstrate great potential for the rapid measurement of virus entry. This type of assay measures the release of virus contents into the cell or target vesicle. For retroviruses, the most commonly used method takes advantage of the fact that synthesis of viral cDNA is limited by access to deoxyribonucleotides. When the viral genome is exposed by capsid disassembly after entry, deoxynucleoside triphosphates (dNTPs) can access the viral polymerase and synthesis proceeds. Transcripts can then be detected by PCR, typically around 4 h after cell contact. This assay has been used to demonstrate that avian leukosis virus entry may be inhibited by lysosomotropic agents (19) and that extraction of cholesterol from virion membranes inhibits HIV entry (14). However, it is not known at what point within this 4-h window genome uncoating takes place and when the dNTP pool is contacted. To make a more quantitative assay, a method was recently developed in which the enzyme β-lactamase was fused to the HIV protein, Vpr (5). Vpr is packaged into HIV particles as part of virus assembly and provided a means of targeting a marker enzyme into the particle. Caged substrate was perfused into cells to give signal. In practice, this assay lacked sensitivity, as detection of entry required 12 h of cell culture for production of sufficient reaction product. A multiplicity of infection (MOI) of more than 10 to 100 is required to achieve shorter measurement times, but this challenge load is not physiological.
Here, we report the use of luciferase as a marker of entry. The use of this enzyme has allowed much greater sensitivity over its predecessors at an MOI of less than 1. Uniquely, the enzyme is targeted to virus particles by fusion to the carboxyl terminus of the envelope glycoprotein. This places it into the lumen of the virus, in a pocket between the viral membrane and the matrix, which forms the outer shell of the core. By controlling the amount of enzyme that is incorporated into each virus particle, it was possible to completely encapsidate the marker, eliminating signal due to defective particles. The enzyme is immediately exposed upon fusion of the cell and viral membranes and detected by rapidly perfusing the cells with luciferin. We validate the assay by demonstrating that signal is receptor dependent, virus with a point mutation in the fusion peptide binds but gives no entry signal, and inhibitors of endosomal acidification do not affect murine leukemia virus (MLV) entry. We show that entry kinetics are rapid, with events being detected as early as 20 min after cell contact.
MATERIALS AND METHODS
Chemicals.
Bafilomycin A1 and chloroquine were from Calbiochem. Luciferase and reaction buffer were from Promega. All other chemicals were Sigma Ultragrade.
Plasmid constructs.
All plasmids were prepared by using Qiagen kits or by cesium gradient centrifugation following standard methods. The envelope-luciferase (env-luc) fusion vector was made by modifying the 3′ end of the Friend 57 MLV envelope gene to replace the native stop codon with an EcoRI restriction endonuclease site. This was achieved using PCR. The primers used were 5′ CCATCGATTAGTTCAATTTGTTAAAGACAG 3′ and 5′ GATCGAATTCTGGCTCGTATTCTAGTGGTTTTAGC 3′. The firefly luciferase gene was modified to gain an EcoRI restriction endonuclease site at its 5′ end. A short linking peptide (Glu-Phe) was also added in the same reaction through the EcoRI site. The primers used were 5′ GATCGAATTCGAAGACGCCAAAAACATAAAGAAAG 3′ and 5′ GATGCGGCCGCTTACACGGCGATCTTTCCGCCCTT 3′. The latter primer also gave two tandem stop codons, followed by a NotI restriction endonuclease site at the 3′ end of the gene. The recombinant MLV envelope gene was then cloned into pCDNA3 (Invitrogen) using a native HindIII site at its 5′ end and the artificial EcoRI site. The modified luciferase gene was then added using EcoRI and NotI sites. The construct was sequenced and had the predicted nucleotide sequence.
The fusion peptide mutant was made by PCR-mediated site-directed mutagenesis. This was the same as described previously (29). The sense strand oligonucleotide for the T471P mutation was 5′ CGCCGCGGGAGTAGGGCCCGGAACTACCGCC 3′ with an ApaI endonuclease site (underlined) added. The fragment of DNA encoding the changes was cloned into native KpnI and ClaI sites in the Friend 57 ecotropic envelope gene and into the env-luc construct. Base changes were confirmed by restriction enzyme cleavage using ApaI and sequencing.
Production of pseudotyped MLV and viruses containing envelope-luciferase fusion protein.
293 HEK cells were grown to 80% confluence on 10-cm-diameter plates in Dulbecco's modified Eagle's medium (DMEM) containing 8% fetal calf serum (Gemini Bio-products). The cells were transfected by the calcium phosphate method of Chen and Okayama (6). Five micrograms of each plasmid was used: (i) pGAG-POL (encoding the MLV gag and polymerase), (ii) pEnv (Friend 57 envelope protein in pCDNA3) or pVSV-G (G protein of vesicular stomatitis virus), (iii) pψβ-gal or pψΕGFP (encoding β-galactosidase or enhanced GFP [EGFP]; Clontech, respectively, under control of the MLV long terminal repeat [LTR] and packaging sequence). To make virus containing the env-luc fusion protein, 1 μg of this construct was added to the mixture unless stated otherwise. After overnight incubation, the medium was replaced with fresh medium and incubated for a total of 36 h. At this time, the virus titer peaked and the supernatants were collected and filtered through a 0.45-μm-pore-size cellulose acetate filter. The filtrate was then either used directly, or virus was pelleted by 1 h of centrifugation at 16,000 × g and the pellet was used. In some experiments, virus was collected by pelleting through a cushion of 20% (wt/vol) sucrose-10 mM Tris-HCl (pH 7.4).
Entry assay.
293 HEK cells were used for all assays. A previously characterized clone, expressing recombinant mCAT-1 (MLV ecotropic virus receptor) fused to a C-terminal hemagglutinin (HA) tag (293-CAT), was used for infection with the ecotropic constructs (9). Vesicular stomatitis virus (VSV) pseudotypes were assayed on wild-type 293 cells. Virus titers were determined by end-point dilution by serial fivefold dilution of stocks and application to 293-CAT cells. After 36 to 48 h, cells stained for β-galactosidase activity (for virus made with pψ β-gal) or fluorescent cells expressing EGFP (for pψEGFP) were counted using an inverted epifluorescence microscope.
For the entry assay, cells (typically 105/sample) were incubated for 1 h (unless stated otherwise) with env-luc-containing virus at an MOI of 0.1 to 0.5. Excess virus was washed free of cells by pelleting by centrifugation at 200 × g for 5 min and resuspension in DMEM. The cells were pelleted again and resuspended in 0.1 ml of luciferase assay buffer (Promega). Luciferase activity was measured after 1 min in a Turner Designs TD 20/20 luminometer and expressed as counts per second. We also performed the assay in a 96-well plate using 104 cells per well. While the signal was reduced proportionally, it remained at least 10-fold above the background of the detector (Perkin-Elmer plate reader).
For pulse-chase experiments, virus was made as described above. To monitor binding to cells, [35S]methionine-labeled virus was produced. One day after transfection with plasmids, cells were washed twice in DMEM and then incubated overnight in methionine-free DMEM containing 0.25 mCi of cell-labeling-grade [35S]methionine (Amersham). Virus-containing supernatants were collected and filtered, and particles were pelleted through a 20% sucrose cushion by centrifugation at 50,000 × g for 3 h. The pellet was collected in phosphate-buffered saline and used. To ensure that radioactivity was associated with virus particles, some of the supernatant was applied to a Sepharose CL-4B column (Sigma) and the amount eluting with the void volume and retained volumes was measured by scintillation counting. We found that 80 to 90% of radioactivity eluted in the void for VSV and Friend MLV, indicating that it was likely associated with virus particles and not free methionine. Cells were then incubated for 5 min with virus and rapidly washed three times by gentle pelleting at 200 × g for 3 min and resuspension in DMEM. They were then incubated at 37°C with gentle agitation and used for measurements at the times given.
Treatment of cells with lysosomotropic agents.
Chloroquine, bafilomycin A1, and ammonium chloride were used. Ammonium chloride and chloroquine were dissolved directly in DMEM and incubated with cells for 1 h before and during incubation with virus. Bafilomycin A1 was first dissolved in dimethyl sulfoxide as a 50 μM stock and diluted in DMEM before use. Cells were incubated with virus for 1 h, and luciferase activity was then measured as above. To check that the drugs did not alter the permeability of cells to luciferin, cells were infected with a luciferase-expressing retrovirus made by using the plasmid pFB-ψluc (Stratagene), and 2 days later, luciferase activity was measured. Further, cells were incubated with luciferase-containing virus and then drug was added for 1 h. Luciferase activity was then measured.
RESULTS
Our understanding of pH-independent virus entry has been impeded by the lack of sensitive but simple assays to measure entry. Fluorescence dequenching methods are the most direct and have been successfully applied to measure membrane fusion of pH-dependent viruses. However, these are not easily adapted to pH-independent viruses, mainly due to the low frequency of membrane fusion events. We wanted to develop an assay that would not only quantitatively measure entry but would be sensitive enough to measure very early virus-cell membrane fusion events at low MOI. Firefly luciferase is an enzyme that reacts with ATP, oxygen, and luciferin to emit light. This is an efficient reaction, and as few as 2,000 molecules of luciferase can be detected by current measuring devices (7). The substrates ATP and oxygen are present in cells in nonlimiting quantities, and luciferin can rapidly and evenly enter live cells by an undefined mechanism, possibly a metabolite transporter (7). Membrane-permeable caged luciferin is also commercially available. We reasoned that if firefly luciferase could be encapsidated into virions and be released upon fusion of cell and virus membranes, it would then provide a superior indicator of virus entry. To do this, we needed to deliver the enzyme into the virus lumen while maintaining the integrity of the viral membrane. Ideally, the enzyme should also be permitted to freely diffuse after entry and not be dependent on dissociation or trafficking of the virus core.
Others have indicated that enzymes, such as β-galactosidase, can nonspecifically enter MLV particles and be delivered to infected cells (15). We attempted to take advantage of this observation, substituting luciferase for β-galactosidase. MLV particles were made by simultaneous transfection of 293 cells with plasmids encoding the MLV structural proteins and polymerase, Friend 57 MLV envelope, and pFB-luc (packageable luciferase expression vector; Stratagene). We found that luciferase expression in these cells peaked at the same time as virus titer. Unfortunately, while luciferase activity was detected in the culture supernatants, this material was not pelleted with virus, indicating that it had likely been released or secreted independently of virus (Table 1). We therefore sought another strategy for incorporation of luciferase into the virus.
TABLE 1.
Encapsidation of luciferase into virus particlesa
| Plasmid transfected
|
Luciferase activity (counts/s/10 μl)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| pFB-ψluc | pψEGFP | pGag-pol | pEnv | pEnv-luc1 | pEnv-luc2 | Cell lysate | Supernatant | Pellet | Lysed/unlysed ratio |
| − | + | + | + | − | − | 0 | 10 | 8 | NDb |
| + | − | + | + | − | − | 574,000 | 1,101 | 22 | ND |
| − | + | + | − | + | − | 2,212,000 | 556 | 975 | 15 |
| − | + | + | − | − | + | 2,022,000 | 350 | 243 | 1 |
| − | + | + | + | + | − | ND | 2,145 | 2,891 | 11 |
First, 293 cells were transfected with the plasmids indicated, and after 36 h, the supernatants were harvested, passed through a 0.45-μm filter, and tested for luciferase activity. Part of the supernatant (0.25 ml) was overlaid onto a 0.5-ml 20% (wt/vol) sucrose cushion, and pelleted material was collected after 1 h at 16,000 × g. Cells and pellets were lysed by resuspension in 1.0 or 0.1 ml of 1% NP-40, respectively; 10 μl was then used to determine luciferase activity. The lysed/unlysed ratio was determined by dividing the luciferase activity obtained from particles treated with 1% NP-40 by the activity for intact particles.
ND, not determined.
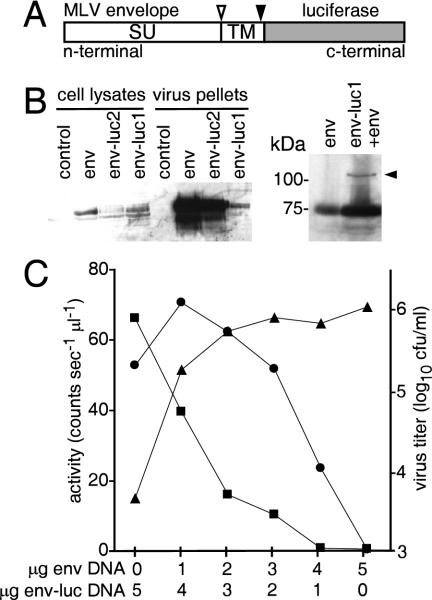
Specific targeting of foreign proteins to virus particles has been achieved for HIV. Both GFP and β-lactamase fused to the HIV Vpr protein can be incorporated at low levels (5, 18). As the timing of Vpr release from particles is not known and may occur well after entry, we tested envelope protein as an alternative targeting protein. The envelope protein of the MLV is made as a single polypeptide that is rapidly cleaved in the endoplasmic reticulum into two subunits, SU (70 kDa) and TM (15 kDa). TM anchors the complex to the cell and eventually to virus membranes. Immediately before or just after budding from the surface of the cell, TM is further cleaved by a viral protease to release a C-terminal peptide, p2e. Rein and others have shown that this is important for infection competency of the virus (24) and may act through gross alteration of envelope structure (1). Otherwise, a role for the released peptide remains unclear. Fusion of proteins to the C terminus of TM would provide a novel method of delivering recombinant protein to the viral lumen, between the membrane and matrix shell of the viral core. The cleavage of the protein by viral protease after budding would also release the luciferase, permitting it to diffuse into the cell cytoplasm after membrane fusion. To test this, we made two constructs, env-luc1 and env-luc2, which fused the Friend 57 MLV envelope to the N terminus of the luciferase gene (Fig. 1A). env-luc1 and env-luc2 differed by the length of the spacer peptide between the envelope protein C terminus and the luciferase N terminus: these were a Glu-Phe and a Glu-Phe-Gly-Ser peptide, respectively. We again made MLVs but added the plasmids encoding the env-luc1 and -2 constructs. The pψEGFP plasmid substituted for the pFB-luc vector, permitting direct determination of virus titer by infecting cells and counting colonies expressing EGFP. We found that for both constructs, luciferase was present in the supernatant. Unlike the previous experiment, most of this activity was pelleted by centrifugation and penetrated a 20% sucrose cushion at 50,000 × g, a characteristic of intact MLV particles. The pelleted material was also associated with infectious virus and gave titers of 3 × 103 and 4 × 104 CFU/ml for env-luc1 and env-luc2, respectively. In later experiments, sucrose gradients (5 to 60% [wt/vol]) were also used to purify virus. We found that >90% of the luciferase activity comigrated with the infectious virus peak. On Western blots probed with anti-SU antibodies, differences were seen in the levels of production of each construct in cells and incorporation into particles (Fig. 1B). Each was rapidly processed to the native SU band. The env-luc1 protein was produced at similar levels to wild-type envelope protein, while smaller amounts of env-luc2 were detected. Surprisingly, env-luc2 was incorporated more efficiently into particles than env-luc1. By adding more plasmid encoding wild-type envelope protein to the transfection mixture, it was possible to observe an uncleaved precursor for env-luc1 in cell lysates migrating above 100 kDa (Fig. 1B, right panel); this band was absent in pelleted material. This indicated that the env-luc protein was processed correctly by cellular proteases into SU and TM. Attempts to detect the luciferase-TM product on Western blots were unsuccessful as commercially available antiluciferase antibodies were insensitive (detection limit was >100 ng/ml using recombinant luciferase; data not shown). However, the fact that luciferase activity was present in the infectious pellets indicates that enzyme had been successfully incorporated.
FIG. 1.
Construct design, production, and optimization of MLV incorporating luciferase in the viral lumen. (A) Schematic showing design of MLV envelope-luciferase fusion protein (env-luc). The open and solid arrowheads indicate native cleavage site by furin (at SU-TM junction) and viral proteases (at TM-p2e junction), respectively. (B) Western blots of lysates from cells transfected with plasmids and pelleted particles collected from the culture supernatants. Cells were transfected with plasmids encoding wild-type Friend MLV envelope alone (env) or with the env-luc1 and env-luc2 constructs. The control was the expression vector pCDNA3 alone. After 2 days, cells were lysed in 1% NP-40 and applied to 10% polyacrylamide-sodium dodecyl sulfate gels. After transferring to nitrocellulose, blots were probed with anti-Rauscher MLV envelope antibody, which cross-reacts with Friend MLV envelope protein. The left panel shows cell lysates and particles isolated by pelleting through a 20% sucrose cushion. The right panel shows lysate of cells transfected with plasmids encoding env (5 μg) or env+env-luc1 (4 and 1 μg respectively). The arrowhead indicates the band of the predicted size for env-luc fusion protein; size markers are at left. (C) Optimization of virus production. The ratio of env-luc1 to wild-type envelope protein was varied by changing the amount of DNA used for transfection. All virus was pelleted through a 20% sucrose cushion, and luciferase activity (left axis) was determined for intact (squares) or lysed (circles; 1% NP-40 added) particles. Virus titer (triangles, right axis) was determined by infecting 293-CAT cells in serial fivefold dilutions and staining them for β-galactosidase activity after 2 days.
To be useful as a measure of entry, luciferase would need to be encapsidated into particles impermeable to the luciferase substrates. MLV are normally impermeable to small solutes such as dNTPs (19) but can be permeabilized by addition of detergents such as 1% NP-40, which strip away the viral lipid membrane and envelope proteins (22). To measure if the luciferase was encapsidated in intact virions, luciferase activity was measured in the presence or absence of 1% NP-40. Virus made with env-luc1 gave a threefold-higher luciferase activity than virus made with env-luc2. For both, addition of 1% NP-40 increased the signal. With env-luc1, the ratio of luciferase activity for lysed versus unlysed virus was 15, whereas env-luc2 gave a ratio of 1. This indicated that while both constructs successfully targeted luciferase into virus, incorporation of env-luc2 was more disruptive, resulting in a greater number of membrane breaches. In contrast, most virus made with env-luc1 was intact (Table 1). This indicated that the particles have a finite capacity for luciferase and that env-luc1 limits this by being poorly incorporated. In further experiments, we used only env-luc1 (referred to as env-luc hereafter).
We then attempted to raise the titer of virus made with env-luc (103 CFU/ml compared to 106 CFU/ml for wild-type virus) by addition of wild-type envelope encoding plasmid in the transfection mixture. The ratio of env-luc to wild-type envelope was adjusted, and the lysed/unlysed luciferase signal ratio and virus titer were measured by marker gene expression (Fig. 1). A ratio of 4:1 for env-luc to wild-type envelope (env) plasmid gave virus of low titer and a high lysed/unlysed signal ratio. In contrast, the reciprocal ratio of 1:4 improved virus titer to 106 CFU/ml, which was similar to env alone and a lysed/unlysed signal ratio of 10:1. In other experiments, less env-luc did not improve this ratio. Over a series of six separate experiments, the average ratio was 11.2 ± 3.7. Together, these data indicated that these virus particles should act as a molecular beacon, with signal being produced after the luciferase is released by fusion of cell and virus membrane and a signal/noise ratio of at least 10-fold.
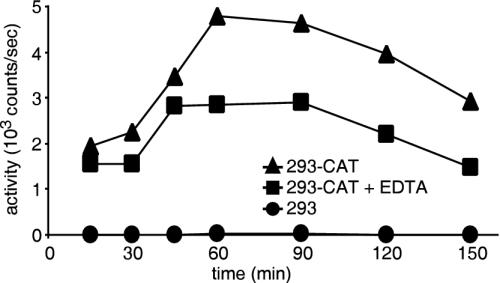
We next determined if these particles would produce a receptor-dependent entry signal when incubated with cells. 293 cells are not permissive for ecotropic MLV infection until they are made to express mCAT-1, the ecotropic MLV receptor. These served as a negative control, while a previously characterized clone of 293 cells expressing a recombinant mCAT-1 with a C-terminal HA tag, termed 293-CAT, was used as a susceptible target (9). Cells were incubated in the presence of 0.5 ml of virus containing culture medium for times up to 2 h at 37°C with gentle mixing. The cells were then pelleted, washed free of unbound virus, and assayed for luciferase activity by incubating the cells directly in luciferase buffer. We were able to detect signal on cells bearing virus receptor. The signal peaked 1 min after addition of the assay buffer, consistent with rapid uptake and equilibration of the luciferin into the cells. Indeed, similar kinetics were observed with cells transfected with a luciferase-encoding plasmid (pFB-luc; data not shown). We found that 293 cells gave no activity above the background signal of the luminometer (approximately 10 counts/s). In contrast, cells expressing receptor gave signal that peaked after 1 to 1.5 h at 5,000 counts/s (Fig. 2, triangles). In other experiments, we found that the magnitude of the signal changed in direct proportion to the number of receptor-bearing cells (104 to 107) or amount of virus used (MOI of 0.01 to 10; data not shown). These observations demonstrated that we were successful in targeting particles containing luciferase to receptor-bearing cells and that a signal was produced, consistent with receptor-dependent exposure of the enzyme.
FIG. 2.
Luciferase activity of virus mixtures incubated over time with 293 cells bearing (293-CAT; triangles) or lacking receptor (293; circles). Luciferase activity was determined on intact cells as described in Materials and Methods. To determine the amount of luciferase that was exposed on the cell surface, EDTA (100 mM; squares) was added immediately before addition of the luciferase substrates.
To determine the portion of the signal that was from virus having entered the cell from that of particles that had broken open on the cell surface, in suspension or residual defective particles, we added 100 mM EDTA (isotonic), pH 7.4, to the sample. Luciferase requires MgATP to function, and this treatment effectively inhibits activity by sequestering the Mg2+ present in the supernatant. We found that the signal dropped, on average, by 30%, with the remainder being resistant (Fig. 2, squares). In other experiments, EDTA inhibited activity to 10 to 20%. This is consistent with the virus-associated luciferase having been taken into an EDTA-inaccessible cellular compartment by either receptor-dependent endocytosis or fusion of the virus membrane with cell membranes. The EDTA-sensitive portion may be due to the residual permeable, defective particles bound to cells or virus in early stages of entry that may be more easily disrupted.
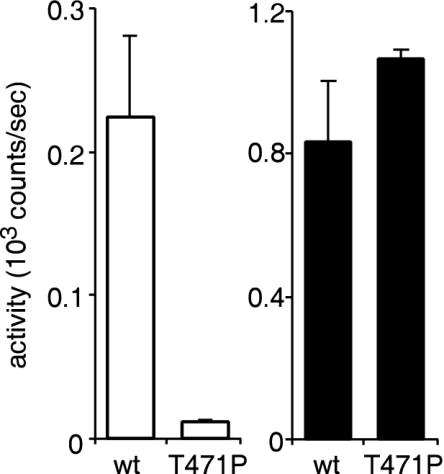
To further test that the signal measured was due to penetration of the virus-associated luciferase into cells through virus-mediated membrane fusion, we made an envelope protein containing a point mutation in the fusion peptide, T471P. This change was previously characterized and shown to bind to cells but not infect them (29). The mutation was placed in both the envelope and in the env-luc construct. Virus was produced, and cells were challenged. The supernatant containing virus with the T471P change gave similar luciferase activity to that of the wild type, 40 counts/s/μl, and a typical lysed/unlysed ratio of at least 5. After 1 h of incubation, the T471P mutant gave a signal that was close to the background signal of the luminometer and 20-fold-lower than that for wild-type virus (Fig. 3, left panel). Detergent was used to expose the encapsidated luciferase and permit determination of total cell-associated virus. Addition of 1% NP-40 detergent gave similar activities for both the wild type and the T471P mutant (Fig. 3, right panel). In a separate experiment, little activity was present in 293 cell lysates (data not shown). This indicated that the T471P mutant virus had bound to cells normally but had not exposed the encapsidated luciferase. Together these data strongly support the conclusion that the assay is measuring receptor-mediated fusion of virus to cells and delivery of the luciferase enzyme into the cell cytoplasm. We then applied the assay to study MLV entry mechanism(s) and kinetics.
FIG. 3.
Analysis of fusion peptide point mutation, T471P, using the entry assay. A single-amino-acid substitution, T471P, was made in the Friend MLV envelope protein and in the env-luc construct. Virus was produced, lysed in 1% NP-40, and matched to the luciferase activity of virus-bearing wild-type (wt) envelope protein and env-luc. This was applied to cells, and luciferase activity was measured after 1 h on intact cells (left panel) or in the presence of 1% NP-40 (right panel).
In general, enveloped viruses can be divided into those with a pH-dependent or -independent mechanism of entry. The pH-dependent viruses require trafficking to acidified endosomal compartments. A block to infection by inhibitors of endosomal acidification has been used as evidence for pH dependence. For retroviruses like HIV and MLV, it has been difficult to determine if a pH-dependent component exists. Indeed, it was recently reported that the avian retrovirus avian leukosis virus has a receptor- and pH-dependent mechanism of entry that was only revealed by using a semiquantitative PCR assay to detect early virus replication (19). However, this finding is still controversial, as the inhibitors used are typically cytotoxic and may affect stages of infection other than entry, such as uncoating and trafficking to the cellular dNTP pool (10). For MLV, both pH-dependent (2, 17) and pH-independent (19) mechanisms have been proposed. Because the luciferase-based assay is a more rapid and quantitative measure of entry than others, it should permit a less ambiguous determination of whether MLVs have a pH-dependent component of entry and allow us to quantitatively measure the effect of inhibitors.
To demonstrate that the luciferase assay could measure inhibition of entry for a pH-dependent virus, we constructed an MLV pseudotype bearing the envelope protein of VSV (VSV-G) together with env-luc. VSV pseudotypes of MLV have been previously reported and enter cells through a VSV-G-dependent mechanism (4, 23), possibly by clathrin-mediated endocytosis. VSV has also been shown to have a pH-dependent fusion mechanism in vitro (3, 21). We reasoned that the env-luc protein would not participate in entry if assays were performed on 293 cells lacking the ecotropic receptor, as supported by the data in Fig. 2. We therefore made a VSV-G/env-luc chimeric virus by replacing the Friend envelope expression plasmid with that of VSV-G. Virus was collected and gave a lysed/unlysed ratio of 8.7 ± 2.2, which was slightly lower than the original Friend envelope-containing virus but still demonstrated that most particles encapsidated the luciferase. Similarly, the overall activity of the lysed particles was approximately one-half of that of the Friend virus, which is 10 counts/s/μl of culture supernatant.
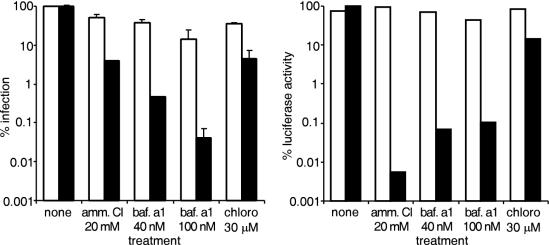
We then tested the activity of three inhibitors of endosomal acidification on the VSV-G and Friend MLV luciferase-containing viruses. Due to the short time frame of the assays, we were able to maintain cells in the continued presence of the drugs without affecting the viability of the cells. Ammonium chloride and chloroquine are weak bases that accumulate in and buffer the endosomal compartment. Bafilomycin A1 is an inhibitor of the endosomal ATP-dependent proton pump. Each was compared by measuring the effect on infection efficiency, as measured by staining in a conventional reporter gene expression assay after 2 days or in the luciferase entry assay after 1 h of incubation (Fig. 4). In this experiment, we increased the number of cells (106) and virus used per sample by 10-fold. This gave a proportional increase in signal and sensitivity. Using the β-galactosidase infection assay, with reporter enzyme expression after 36 h, we confirmed that infection of the VSV-G pseudotype was sensitive to each drug, with bafilomycin being the most potent, reducing infection by 100-fold at 40 nM (Fig. 4, solid bars). By comparison, the luciferase assay using the VSV-G pseudotypes was more sensitive. We were able to show that 20 mM and 40 nM for ammonium chloride and bafilomycin respectively, decreased the signal by 1,000- to 10,000-fold (Fig. 4, right panel, solid bars). Chloroquine was not as effective and inhibited the infection and the entry signal by only 20- and 7-fold respectively. When the Friend pseudotype was used, infection was weakly inhibited by all of the drugs by up to threefold. This represents a relatively small decrease in virus titer from 106 to 3 × 105 CFU/ml. Similarly, the luciferase activity for this virus was decreased by no more than twofold (Fig. 4, open bars). In comparison to the VSV-G pseudotype, this change is small and likely reflects slight cytotoxic effects of each drug on the cells. To ensure that the inhibitors did not alter the entry of luciferin into cells and access to ATP, we measured luciferase activity in cells stably transduced with a luciferase-expressing retrovirus (made using pFB-luc; Stratagene). At the concentrations of inhibitor used, we found little change in signal after the cells were incubated in luciferase buffer. In addition, we applied luciferase-containing Friend or VSV-G pseudotyped virus to cells and after 1 h treated them with each drug. When assayed for activity, the cells gave similar small changes in signal compared to cells preincubated in drug (not shown). Taken together, these data strongly indicate that the assay is measuring entry of virus and confirm that the entry of ecotropic MLV is not significantly affected by inhibitors of endosomal acidification.
FIG. 4.
Effect of endosomal acidification inhibitors on infection and luciferase entry assay signal. Cells were treated with ammonium chloride (amm. Cl.), bafilomycin A1 (baf. a1), or chloroquine (chloro) at 20 mM, 40 nM + 100 nM, and 30 μM, respectively, and then Friend MLV (open bars) or VSV-G (solid bars) pseudotyped virus encoding β-galactosidase and containing env-luc was applied. Colonies were then counted after 2 days by staining for β-galactosidase activity (left panel), or luciferase activity was determined (right panel) after 1 h.
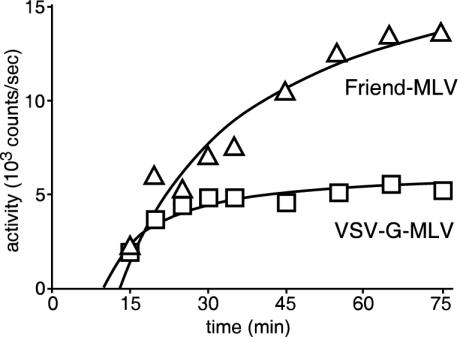
The kinetics of entry for MLV are poorly understood. The best approximations have been made by stripping particles off cells by treatment with acidic (pH 3 or less) buffers or extensive protease treatment and then measuring infection by staining surviving cells for marker gene expression. These treatments are likely to have adverse affects on the cell itself and affect the measurements made. To better understand virus entry kinetics, we performed a pulse-labeling experiment using the luciferase assay to measure virus entry of the Friend MLV and VSV-G pseudotypes. The particles were incubated with 293 or 293-CAT cells for 5 min, and unbound virus was removed by rapid washing. Cells were then incubated for up to 75 min, and luciferase activity was measured (Fig. 5). After a 5-min binding step, the earliest time at which measurements could be practically made was 10 min after excess virus had been removed by washing. For both pseudotypes, some activity was already detectable at this time. For the Friend MLV, this signal grew steadily and began to plateau at 75 min. Nonlinear regression analysis gave a fit to a simple hyperbolic binding isotherm with R2 = 0.94 and indicated that half of the particles had fused with cells by 40 ± 1.3 min postbinding. For the VSV-G MLV pseudotype, the kinetics were more rapid, reaching a plateau at 30 min. A similar linear regression analysis showed that half of signal was reached by 18 ± 1.2 min (R2 = 0.87). The signal obtained from the VSV MLV was approximately threefold lower than that for Friend MLV.
FIG. 5.
Pulse-chase analysis of entry kinetics using the luciferase assay. Cells (2 × 106 per assay point) were incubated with luciferase-containing Friend-MLV (triangles) or VSV-G MLV (squares) pseudotyped virus at an MOI of 0.5 for 5 min at 37°C. They were then washed free of unbound virus and further incubated at 37°C. Samples were then collected at the times indicated (calculated from time of virus addition), and activity was measured as described previously. Curves (rectangular hyperbola) were fitted to data by nonlinear regression analysis using GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego, Calif.).
To test if this difference was due to smaller amounts of VSV MLV virus being bound to cells and to ensure that similar amounts of virus had remained bound to cells during the chase phase of the experiment, [35S]methionine-labeled, density gradient-purified virus was included in the assay. Over the entire time course, cell-associated virus remained relatively constant with an average of 4.3% ± 0.4% of the input VSV-G-containing particles being bound to cells, corresponding to a final MOI of 0.002 (calculated from the titer of virus on 293-CAT cells, which was 1.3 × 106 CFU/ml). For Friend MLV, 10.4% ± 1% of the input virus was cell associated, giving an MOI of 0.006 (titer of 1.5 × 106 CFU/ml). These results indicate that the observed difference in the VSV-MLV and Friend-MLV plateaus was due to the amount of virus that had bound during the 5-min preincubation and that a similar proportion of each was able to yield a signal. In other experiments with extended time courses, we observed that by 2.5 h, the signal had dropped to 60% of the peak at 75 min, similar to Fig. 2. This indicated that the signal was labile with a half-life of >2.5 h. However, this slow decay should have little effect on the above analysis.
DISCUSSION
We demonstrate a novel, sensitive, and rapid assay for detection of virus entry. This assay is based on the principle of content mixing, whereby a marker is released into the target to produce a signal. Previous content-mixing assays have used soluble fluorophores incorporated into virosomes, detection of genome exposure, and enzyme activities such as β-lactamase and virus polymerases in intact virus. For retroviruses, detection of reverse transcription provides sufficient signal to measure entry events in <4 h after membrane fusion. However, the PCR-based assay is difficult to perform and is semiquantitative at best. Real-time PCR permits better quantitation but reduces throughput and requires extensive controls. All of these assays suffer from considerable background signal, making interpretation of results difficult. Unlike previous content-mixing methods, the data presented demonstrate that the current assay has greater sensitivity and permits detection of entry, for MLV, within 20 min after incubation with the cell.
We tested the assay by showing that only receptor-bearing cells gave signal, demonstrating that we were measuring specific adherence to cells and not nonspecific breakdown of the luciferase-containing particles. Furthermore, a mutant containing the amino acid substitution T471P in the fusion peptide of the MLV envelope protein gave no signal but bound to cells normally. This substitution had previously been shown to block infection but gave normal binding to the cell (29). These data indicate that the assay is measuring entry of virus and not other events following virus attachment, such as endocytosis or nonspecific degradation of the particles that might expose the luciferase. For these experiments, binding was determined by lysing the cells and measuring total luciferase activity. This is a unique and highly advantageous feature of the assay that permitted evaluation of the amino acid substitution for both virus-receptor interaction and entry in the same sample. This should also permit the influence of other amino acid substitutions on entry to be easily evaluated.
The work using EDTA further supports the conclusion that the assay is measuring processes important in virus entry. We reasoned that if virus were breaking open on the surface, then sequestration of magnesium, a necessary cofactor of the luciferase enzyme, should block the signal. EDTA cannot penetrate the cell membrane, and therefore luciferase within the cell would not be affected. Indeed, if virus particles were deliberately lysed with detergent and EDTA was added, the signal was lost. We expect that the portion of the signal that was blocked by EDTA addition (30%) may be due to defective viruses that are present. As the bulk of the signal remained resistant to EDTA, this indicated that the luciferase had reached an EDTA-inaccessible compartment or likely was inside the cell, where it has access to its substrates.
The data are consistent with a rapid process of entry that follows receptor-dependent virus binding to cells. A recent report on the avian retrovirus avian leukosis virus, using a lipid-mixing assay, indicated that cell-virus membrane fusion may be rapid: within 10 min after cell contact with virus (10). A similar study with HIV indicated that fluorescent fatty acid transfer occurs within 5 min of incubation with receptor-bearing cells (26). However, such fluorophores, particularly R18 and similar fatty acids, are subject to passive diffusion into the target membrane, especially when the virus is brought into close juxtaposition by receptor interaction. Complex analysis has been recommended to separate signals resulting from passive diffusion and specific membrane fusion (20). Furthermore, these studies required high particle/cell ratios, exceeding 20-fold. This is a nonphysiological amount of virus that may adversely affect cell function. Another recent report on amphotropic MLV infection indicated that entry begins as early as 30 min after cell contact (16). In this study, cells were treated with buffer at pH 3 to destroy virus that had not entered, but it is not known how this affected other cellular processes important for infection. The current assay gives an independent measure of virus entry kinetics for a retrovirus at MOIs of much less than 1. Under these conditions, signal could be detected between 5 and 20 min after cell contact and followed simple kinetics for over an hour. From analysis of pulse-chase experiments, a delay of approximately 15 min after virus binding to the cell occurred before significant fusion was seen. The importance of this delay remains unclear, but it is very similar to lags seen for HIV entry (13). It may represent the time required to form a fusion pore and deliver the luciferase into the cell cytoplasm and then access its substrates. Indeed, the data are consistent with the relatively slow formation of the fusion pore seen for other enveloped viruses such as influenza A virus and VSV, which can take many minutes to grow large enough to release the virus capsid (8, 21). However, the lag may also indicate that cellular signaling, reorganization, or trafficking events may be required for entry, as suggested for HIV (13). This will require further investigation to resolve. Nevertheless, the findings are consistent with a rapid mechanism of entry with similar kinetics to that seen for the pH-dependent enveloped viruses such as influenza A virus and VSV.
The rapid execution of the assay will permit an extensive dissection of the virus-entry pathway using inhibitors that are otherwise toxic to cells on prolonged exposure. Extended use of inhibitors can also affect pathways downstream of the point of action, such as the blockage of late-stage endosomal trafficking by inhibitors of endosomal acidification (28). Indeed, for MLV, reports have indicated both pH-dependent (2, 17) and pH-independent (19) mechanisms of entry. This discrepancy may reflect differences in treatment method. Here, the luciferase-based assay demonstrated that MLV entry was not significantly inhibited by inhibitors of endosomal acidification. The ability to produce a VSV-G pseudotype with similar luciferase activity to that of MLV provided a control for these experiments and emphasizes the flexibility of the assay system. Chloroquine and ammonium chloride are weak bases that accumulate in and buffer the change in pH in the endosome. Bafilomycin A1 is a specific inhibitor of the endosomal proton pump (19). Each compound only weakly affected the signal for the Friend MLV particles. However, they potently inhibited the signal observed with the VSV-G pseudotype, which is known to have a pH-dependent mechanism of entry. When optimized, we were able to observe a 1,000- to 10,000-fold decrease in signal for the VSV-G pseudotype in the presence of bafilomycin and ammonium chloride, as compared to at most a 2-fold change for MLV. While we confirm the previous report that MLV does not require acidification in endosomes to trigger entry (19), the current assay yielded quantitative data with a sensitivity that far exceeded this and other previous assays. These observations also provide further support that the assay recapitulates the entry pathway of the virus studied and will be useful for future study of entry mechanism and inhibitors.
The method used to deliver the luciferase to the virus particle may also facilitate the packaging of other protein cargoes. This has been a goal of virus-mediated therapies and nanotechnology. We estimate from titration of recombinant luciferase (Quantilum; Promega) in buffer that we are able to detect approximately 2,000 molecules. The inclusion of medium used to harvest virus did not affect this number greatly. Given that the virus titer was 106 infectious particles/ml with an activity of approximately 105 counts/s/ml, this gives a specific activity of 0.1 count/s/infectious particle. Assuming a particle/infectious particle ratio of 10 to 100, an even distribution of enzyme, and efficient lysis of virus particles, we estimate that between 2 and 20 molecules of luciferase were packaged per particle. This is likely to be the maximum tolerated by the virus, as attempts to increase the amount of env-luc protein resulted in exposure of the enzyme to the suspension buffer and loss of virus titer (Fig. 1). Consistent with this is the observation that the env-luc1 construct, while produced at slightly higher levels than env-luc2 in cells (Fig. 1), by itself incorporated poorly into particles. In comparison, env-luc2, which was incorporated well, produced particles that were permeable to luciferin. However, by combining wild-type envelope and the env-luc1 construct to make virus, titers were restored to normal, with most virus being intact. Proteins of a similar size to luciferase (61 kDa) may also be tolerated. This then provides a simple system to package a therapeutic protein, deliver it specifically to a target cell by efficient receptor-dependent targeting, and have it released free of the virus into the cytoplasm of the target cell.
Luciferase-containing virus pseudotypes may also be readily adapted for diagnostic assays. The demonstration that a VSV-G chimeric virus can be efficiently made may mean that other virus pseudotypes will behave the same way. Luciferase-based assays are common in most diagnostic facilities and provide sensitive and high-throughput analyses. Indeed, we adapted the assay to work in a 96-well format without difficulty. Luciferase-containing virus could be used to screen for neutralizing antibodies in patient sera or for discovery of compounds that inhibit virus entry or receptor binding. The rapid execution of the assay would reduce a 1- to 2-day diagnostic assay into several hours. This, of course, requires that the envelope protein of the donor virus and the env-luc protein accumulate on the membrane at the same locale, to be incorporated into the same particle. Given that the number of MLV pseudotypes that have been successfully produced is continually on the increase, the potential use for this assay will also expand.
Luciferase-containing viruses may also permit the visualization of entry events. With a sufficiently sensitive camera, it would be possible to detect the production of light upon combination of the released luciferase and substrates. Others have shown that imaging of cells expressing luciferase is possible but requires the use of image-intensifying cameras and exposure of the sample for tens of seconds (7). As camera sensitivity increases, this may become more practical.
Acknowledgments
We thank Mardelle Susman and Kathryn Davey for careful reviews of the manuscript and comments.
This work was supported by funding from the University of Texas Medical Branch.
REFERENCES
- 1.Aguilar, H. C., W. F. Anderson, and P. M. Cannon. 2003. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R peptide. J. Virol. 77:1281-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, K. B., and B. A. Nexo. 1983. Entry of murine retrovirus into mouse fibroblasts. Virology 125:85-98. [DOI] [PubMed] [Google Scholar]
- 3.Blumenthal, R., A. Bali-Puri, A. Walter, D. Covell, and O. Eidelman. 1987. pH-dependent fusion of vesicular stomatitis virus with Vero cells. Measurement by dequenching of octadecyl rhodamine fluorescence. J. Biol. Chem. 262:13614-13619. (Erratum, 263:588, 1988.) [PubMed] [Google Scholar]
- 4.Burns, J. C., T. Friedmann, W. Driever, M. Burrascano, and J. K. Yee. 1993. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA 90:8033-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavrois, M., C. De Noronha, and W. C. Greene. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:1151-1154. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig, F. F., A. C. Simmonds, D. Watmore, F. McCapra, and M. R. White. 1991. Membrane-permeable luciferin esters for assay of firefly luciferase in live intact cells. Biochem. J. 276:637-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danieli, T., S. L. Pelletier, Y. I. Henis, and J. M. White. 1996. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J. Cell Biol. 133:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey, R. A., Y. Zuo, and J. M. Cunningham. 1999. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J. Virol. 73:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earp, L. J., S. E. Delos, R. C. Netter, P. Bates, and J. M. White. 2003. The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH. J. Virol. 77:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edinger, A. L., T. L. Hoffman, M. Sharron, B. Lee, B. O'Dowd, and R. W. Doms. 1998. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology 249:367-378. [DOI] [PubMed] [Google Scholar]
- 12.Erlwein, O., C. J. Buchholz, and B. S. Schnierle. 2003. The proline-rich region of the ecotropic Moloney murine leukaemia virus envelope protein tolerates the insertion of the green fluorescent protein and allows the generation of replication-competent virus. J. Gen. Virol. 84:369-373. [DOI] [PubMed] [Google Scholar]
- 13.Gallo, S. A., C. M. Finnegan, M. Viard, Y. Raviv, A. Dimitrov, S. S. Rawat, A. Puri, S. Durell, and R. Blumenthal. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614:36-50. [DOI] [PubMed] [Google Scholar]
- 14.Guyader, M., E. Kiyokawa, L. Abrami, P. Turelli, and D. Trono. 2002. Role for human immunodeficiency virus type 1 membrane cholesterol in viral internalization. J. Virol. 76:10356-10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, M. L., B. L. Winther, and M. A. Kay. 1996. Pseudotransduction of hepatocytes by using concentrated pseudotyped vesicular stomatitis virus G glycoprotein (VSV-G)-Moloney murine leukemia virus-derived retrovirus vectors: comparison of VSV-G and amphotropic vectors for hepatic gene transfer. J. Virol. 70:2497-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu, C. W., L. O'Reilly, and M. J. Roth. 2003. G100R mutation within 4070A murine leukemia virus Env increases virus receptor binding, kinetics of entry, and viral transduction efficiency. J. Virol. 77:739-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClure, M. O., M. A. Sommerfelt, M. Marsh, and R. A. Weiss. 1990. The pH independence of mammalian retrovirus infection. J. Gen. Virol. 71:767-773. [DOI] [PubMed] [Google Scholar]
- 18.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679-689. [DOI] [PubMed] [Google Scholar]
- 20.Ohki, S., T. D. Flanagan, and D. Hoekstra. 1998. Probe transfer with and without membrane fusion in a fluorescence fusion assay. Biochemistry 37:7496-7503. [DOI] [PubMed] [Google Scholar]
- 21.Paternostre, M. T., R. J. Lowy, and R. Blumenthal. 1989. pH-dependent fusion of reconstituted vesicular stomatitis virus envelopes with Vero cells. Measurement by dequenching of fluorescence. FEBS Lett. 243:251-258. [DOI] [PubMed] [Google Scholar]
- 22.Pinter, A., R. Kopelman, Z. Li, S. C. Kayman, and D. A. Sanders. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J. Virol. 71:8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Que, X., D. Kim, A. Alagon, K. Hirata, H. Shike, C. Shimizu, A. Gonzalez, J. C. Burns, and S. L. Reed. 1999. Pantropic retroviral vectors mediate gene transfer and expression in Entamoeba histolytica. Mol. Biochem. Parasitol. 99:237-245. [DOI] [PubMed] [Google Scholar]
- 24.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharron, M., S. Pohlmann, K. Price, E. Lolis, M. Tsang, F. Kirchhoff, R. W. Doms, and B. Lee. 2000. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood 96:41-49. [PubMed] [Google Scholar]
- 26.Sinangil, F., A. Loyter, and D. J. Volsky. 1988. Quantitative measurement of fusion between human immunodeficiency virus and cultured cells using membrane fluorescence dequenching. FEBS Lett. 239:88-92. [DOI] [PubMed] [Google Scholar]
- 27.Spitzer, D., K. E. Dittmar, M. Rohde, H. Hauser, and D. Wirth. 2003. Green fluorescent protein-tagged retroviral envelope protein for analysis of virus-cell interactions. J. Virol. 77:6070-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Weert, A. W., K. W. Dunn, H. J. Gueze, F. R. Maxfield, and W. Stoorvogel. 1995. Transport from late endosomes to lysosomes, but not sorting of integral membrane proteins in endosomes, depends on the vacuolar proton pump. J. Cell Biol. 130:821-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu, N. L., P. M. Cannon, D. Chen, and W. F. Anderson. 1998. Mutational analysis of the fusion peptide of Moloney murine leukemia virus transmembrane protein p15E. J. Virol. 72:1632-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]