Abstract
Chronic infection by hepatitis C virus (HCV) can lead to severe hepatitis and cirrhosis and is closely associated with hepatocellular carcinoma. The replication cycle of HCV is poorly understood but is likely to involve interaction with host factors. In this report, we show that NS5B, the HCV RNA-dependent RNA polymerase (RdRp), interacts with a human RNA helicase, p68. Transient expression of NS5B alone, as well as the stable expression of all the nonstructural proteins in a HCV replicon-bearing cell line (V. Lohmann, F. Korner, J.-O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager, Science 285:110-113), causes the redistribution of endogenous p68 from the nucleus to the cytoplasm. Deletion of the C-terminal two-thirds of NS5B (NS5BΔC) dramatically reduces its coimmunoprecipitation (co-IP) with endogenous p68, while the deletion of the N-terminal region (NS5BΔN1 and NS5BΔN2) does not affect its interaction with p68. In consistency with the co-IP results, NS5BΔC does not cause the relocalization of p68 whereas NS5BΔN1 does. With a replicon cell line, we were not able to detect a change in positive- and negative-strand synthesis when p68 levels were reduced using small interfering RNA (siRNA). In cells transiently transfected with a full-length HCV construct, however, the depletion (using specific p68 siRNA) of endogenous p68 correlated with a reduction in the transcription of negative-strand from positive-strand HCV RNA. Overexpression of NS5B and NS5BΔN1, but not that of NS5BΔC, causes a reduction in the negative-strand synthesis, indicating that overexpressed NS5B and NS5BΔN1 sequesters p68 from the replication complexes (thus reducing their replication activity levels). Identification of p68 as a cellular factor involved in HCV replication, at least for cells transiently transfected with a HCV expression construct, is a step towards understanding HCV replication.
Hepatitis C virus (HCV) is a positive-stranded RNA virus belonging to the family Flaviviridae. It infects more than 170 million people worldwide and poses an important medical problem in developed and underdeveloped countries. HCV replicates in the liver, and chronic infection often leads to cirrhosis, steatosis, and hepatocellular carcinoma. Practically all processes in the life cycle of viruses, including viral entry, translation, processing and modification of viral proteins, maturation, and release of viral particles from host cells, are dependent on the host cell machinery and involve intimate interaction between viral and host proteins. The role of host proteins in the replication cycle of HCV and the effect of virus-host interaction have remained poorly understood. This is due to a lack of cell culture and animal model systems for studying the biology of this virus. The study of the interaction of HCV with cellular proteins has been limited to the identification of host proteins interacting with individual viral proteins.
The proteins encoded by the HCV genome are three structural proteins (core protein and envelope proteins E1 and E2) and seven nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). NS5B is the viral RNA-dependent RNA polymerase (RdRp) that transcribes the positive- and negative-strand RNA genome of the virus. The level of RdRp activity of the NS5B measured by in vitro assays is low (9). This could be due to the lack of cofactors, either viral and/or host proteins, and of the appropriate cellular environment for optimal activity of the HCV RdRp. Viral nonstructural proteins interact with one another: NS3 and NS4A form a complex with NS5B (17), and NS4B and NS5A (35) have been suggested to be cofactors for NS5B. When the nonstructural proteins are coexpressed in stable cell lines harboring replicons, they are colocalized to the estrogen receptor (ER) membranes, indicating that they might form a complex (27, 31). These nonstructural viral proteins, together with host factors, form the viral replicase, the complex in which viral replication is thought to take place. NS5B has been shown to interact with a SNARE-like protein (38) and with human eIF4AII, an RNA-dependent ATPase/helicase and a component of the translation initiation complex (19). Interactions of RdRp's of other viruses with host proteins have also been reported. The poliovirus RdRp interacts with and causes the relocalization of human Sam68 (a protein that associates with Src during mitosis) (25), the tobacco mosaic virus RdRp forms a complex with a plant protein that has homology with a yeast RNA-binding subunit of eIF3 (30), and the brome mosaic virus RdRp associates with a plant eIF3 subunit (32).
Little is known about the role of cellular proteins in promoting HCV replication. An important stumbling block in HCV research is the lack of a cell culture and small animal model for studying viral replication and pathogenesis. Recently, several HCV replicon cell lines have been made available (3, 16, 23); these cell lines are widely used as models for studying HCV RNA replication. These systems have been selected for their ability to self-replicate the HCV subgenomic RNA at high levels.
Here, we describe the identification of p68, an RNA helicase, as an NS5B-interacting protein. p68 was first identified by its immunological cross-reactivity to a monoclonal antibody to the large T antigen of simian virus 40 (10). It belongs to a large family of DEAD box RNA helicases that have well-conserved orthologues from yeasts (15) to humans. Its intrinsic RNA helicase and ATPase activity can be measured in vitro (13, 14). It is localized to the nucleus, is largely excluded from nucleoli in interphase cells, and is only transported to nascent nucleoli at late telophase (28). The biological function of p68 may be diverse; it has been suggested to be involved in modulating RNA structures for RNA splicing, processing, transcription, and translation. Mutations in Dbp2, the yeast homologue, causes an increase in levels of nonsense-containing transcripts and a reduction in levels of mature ribosomal RNAs (4). Recently, p68 and a closely related homologue, p72, have been shown to act as specific coactivators for ER alpha (ERα)-mediated transcription (8, 39). Increased levels of poly-ubiquitinylated p68 are observed in preinvasive and invasive lesions in colorectal tumors, suggesting that p68 might have a role in early tumor development (6).
In this study, we showed that NS5B interacts with endogenous p68 and that the C terminus of NS5B is essential for this interaction. Overexpression of NS5B causes the redistribution of endogenous p68 from the nucleus to the cytoplasm. The expression of all nonstructural proteins of HCV in a replicon cell line also causes endogenous p68 to relocalize. Deletion of the C terminus of NS5B abolishes its ability to cause p68 to redistribute from the nucleus to the cytoplasm. In cells transiently expressing full-length HCV RNA genome, knockdown (using small interfering RNA [siRNA]) of endogenous p68 reduces the transcription of negative-strand HCV RNA from its positive-strand template. A reduction of negative-strand synthesis was also observed upon overexpression of NS5B and NS5BΔN, which are able to bind to p68 and possibly sequester p68 from active viral replicases. We were, however, unable to detect any change in positive- and negative-strand RNA levels in a replicon cell line when endogenous p68 was knocked down using siRNA.
MATERIALS AND METHODS
Construction of plasmids.
HCV sequences were derived from a HCV-S1 cDNA of a type 1b strain (21). For yeast two-hybrid screens and assays, NS5B was the bait and the library was a spleen cDNA library cloned into pACT2 (Clontech). Vectors derived from pXJ40 (40) were used for expressing and tagging proteins at the N termini in mammalian cells. The HCV full-length genome (including the 5′ and 3′ untranslated region [UTR]) was cloned into the EcoRV site of pcDNA3.1(+) (Invitrogen). This construct, pcDNA-HCV(Q19), contains the full-length S1 HCV cDNA (similar to that found in a tetracycline-inducible vector) (22) except that the transcription is driven by the cytomegalovirus promoter in pcDNA3.1(+). Deletion of NS5B (amino acids [aa] 106 to 494) from this construct was achieved by digesting it with HpaI (unique site in HCV genome) and XbaI (unique site in vector) and ligating it with a 0.5-kb fragment from the KpnI site (blunted) to the XbaI site released from this construct. The characteristics of the plasmids used in this study are summarized in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pFG111 | pXJ40flag-NS5B | This work |
| pFG116 | pXJ40myc-NS5B | This work |
| pFG261 | pcDNA-HCV(Q19) | This work |
| pFG346 | pXJ40flag-GST | This work |
| pFG352 | pXJ40myc-p68 (1-614) | This work |
| pFG367 | pXJ40myc-GST | This work |
| pFG371 | pXJ100flag-NS5BΔC (1-214) | This work |
| pFG372 | pXJ100flag-NS5BΔN1 (215-590) | This work |
| pFG374 | pXJ100flag-NS5BΔN2 (298-590) | This work |
| pFG384 | pSG-myc-p68 (D248N) | F. Fuller-Pace |
| pFG402 | pXJ40-5′ UTR + 3′ UTR | This work |
| pFG435 | pXJ40-NS5BΔN1 (215-590) | This work |
| pFG436 | pXJ40-NS5BΔN2 (298-590) | This work |
| pFG439 | pcDNA-HCVΔ5B | This work |
| pXJ100-flag | Mammalian expression vector for tagging proteins with flag at N terminus | This work |
Tissue culture.
HeLa (human cervical cancer cells), 293 (derived from human kidney fibroblasts), and Huh-7 (a human hepatocarcinoma cell line) cells were maintained in standard Dulbecco's minimal Eagle's medium supplemented with 10% fetal calf serum (HyClone Laboratories) and antibiotics, penicillin at 10 U/ml, and streptomycin at 100 μg/ml (Sigma, St. Louis, Mo.). HeLa and 293 cells were obtained from the American Type Culture Collection, while Huh-7 cells were obtained from Japan Health Sciences Foundation (Chou-ku, Osaka, Japan). The Huh-7-containing subgenomic HCV RNA replicon, 9-13 (kindly provided by R. Bartenschlager) (Universitätsklinikum Heidelberg, Germany), was maintained in Dulbecco's minimal Eagle's medium with 10% fetal calf serum and 1 mg of neomycin (G418)/ml.
Antibodies for immunofluorescence (IF) and Western blotting (WB).
Anti-p68 rabbit polyclonal antibodies (2130 and 2906) were generated against a peptide corresponding to the C-terminal 15 aa of p68. PAb204 was originally raised against the simian virus 40 large T antigen, but it cross-reacts with p68 (aa 506 to 523) (10, 20).
The procedure for IF was performed as described previously (12). Monoclonal and polyclonal anti-Flag (Sigma) and anti-myc (Santa Cruz Biochemicals, Santa Cruz, Calif.) antibodies, and anti-PARP1 polyclonal (Santa Cruz Biochemicals), anti-NS5B monoclonal, and 5B-3B1 (26) antibodies were used at a 1:100 dilution, anti-p68 polyclonal 2906 at was used at a 1:500 dilution, and anti-p68 monoclonal PAb204 was used undiluted. Secondary antibodies (Santa Cruz Biochemicals) were applied at 1:100.
Coimmunoprecipitation (co-IP) and WB analyses were performed as described previously (18). WBs were probed with anti-Flag (Sigma) and anti-myc (Santa Cruz Biochemicals) antibodies, anti-p68 polyclonal antibodies (2130) were used at 1:1,000 to 1:2,000 dilution, anti-NS3 monoclonal antibody (Devaron, Inc.) was used at 1:1,000 dilution, and anti-NS5B (5B-3B1) was used at 1:500 dilution. To show a more obvious difference in p68 levels in RNA interference experiments, WBs were probed with anti-p68 polyclonal 2130 for 1 h at 4°C.
RNA interference assays.
Cells were transfected (using Effectene transfection reagents) (Qiagen) with siRNA (synthesized by Dharmacon Research Inc.) with or without DNA constructs. Generally, for each microgram of RNA/DNA the reagents were added in these proportions: 40 μl of EC buffer, 4 μl of Enhancer, and 20 μl of Effectene. The amounts of p68 siRNA were adjusted, and the total amount of siRNA in each transfection was normalized with control siRNAs (siRNA to bacterial gene glutathione-S-transferase [GST] or a scrambled siRNA from Dharmacon Inc.). The sequence targeted for p68 was nucleotide (nt) 135 to 153 (p68-135), while that for GST was nt 117 to 138 of the respective coding regions. At 6 to 7 h after transfection, the medium was replaced with fresh medium. The cells were incubated for 2 days and then harvested and extracted for proteins and RNA.
RNA extraction and RT-PCR.
Total RNA from transfected cells was extracted using an RNeasy kit (Qiagen). The RNA template was treated with DNase I (Roche) to remove any contaminating DNA. RNA (0.5 μg) was used per reverse transcription (RT) reaction with Superscript II (Invitrogen). To obtain PCR products below saturation points, the cDNAs synthesized in RT reactions were diluted 1:100 for positive strand, 1:10 for GAPDH, and undiluted for negative strand. A total of 0.5 μl of RT product was used as a template for the first PCR, after which 0.5 μl of the first PCR product was used as a template for the second PCR; all PCRs were done in 50-μl volumes in accordance with HotStarTaq polymerase (Qiagen) protocols. Data regarding the primers (Genset [Singapore] or BioAsia [Shanghai, China]) used for RT, the first PCR, and the second PCR are summarized in Table 2. PCR cycles were as follows: 5 min at 95°C (to activate the Taq polymerase) followed by 35 cycles of 45 s at 95°C, 30 s at 55°C (for positive- and negative-strand PCR) or 68°C (for GAPDH), and 45 s at 72°C followed by a final step of extension at 72°C for 7 min. The primers for positive- and negative-strand RT-PCR are directed to the 5′ UTR of the HCV genome, while primers for GAPDH are at nt 193 to 214 and nt 664 to 680 of the coding region. TAGFNC1 (GCATCATGGTGGCCAATGACTCCACCATAGATCACTCCC) (non-HCV sequences are underlined), 209TAGR (CATGCTCGCGATACTCGAGGTGCACGGTCTACGAGACCT), and TAGF (GCATCATGGTGGCCAATG) were designed in this work. The sequences for primers 939 and 940 (29) and primers 209 and 211 (11) were derived from previous reports.
TABLE 2.
Primers for RT, first PCR, and second PCR for positive-strand, negative-strand and GAPDH products
| Product | Primer for RT | Primers for first PCR (forward/reverse) | Primers for second PCR (forward/reverse) |
|---|---|---|---|
| Positive strand | 209TAGR | 939/209 | 940/211 |
| Negative strand | TAGFNC1 | TAGF/209 | TAGF/211 |
| GAPDH | GAPDrev | GAPDfor/rev | GAPDfor/rev |
Northern blotting.
RNA extracted with an RNeasy kit (Qiagen) was run on 0.8% formaldehyde agarose gels and transferred onto Hybond N membranes (Amersham) by capillary blotting. The blots were baked at 80°C for 1 h and then stained with methylene blue to visualize the 18S and 28S rRNA bands. The blot was cut between the two rRNA bands; the bottom part was probed with an actin-specific probe, while the top part was probed with either positive- or negative-strand-specific probes.
Labeling of probes for Northern blotting.
Strand-specific probes were labeled by single-strand synthesis using HotStarTaq polymerase (Qiagen). To label the positive-strand-specific probe, the template was pFG402 cut with BamHI and primed with the reverse primer 209 (3′-GGTGCACGGTCTACGAGACCT-5′). To synthesis the negative-strand-specific probe, the same plasmid was cut with BglII and primed with the forward primer 939 (28). pFG402 contains the 5′ UTR and 3′ UTR of HCV joined together and cloned between BamHI and BglII sites in the following orientation: BamHI-5′ UTR-3′ UTR-BglII. After 10 cycles of polymerase reaction on a thermal cycler (Perkin-Elmer), unincorporated [32P]dGTP was removed using a nucleotide removal kit (Qiagen). The actin probe was similarly labeled using a PCR product of the entire β-actin coding sequence as a template and the reverse primer for synthesizing the antisense DNA strand.
RESULTS
NS5B interacts with cellular p68 and causes it to relocalize from the nucleus to the cytoplasm.
With NS5B as bait in yeast two-hybrid screens, we screened 500,000 clones from a human spleen cDNA library. A spleen cDNA library was used instead of a liver library, as screenings using a liver library repeatedly yielded many candidate genes encoding secreted proteins and false-positive candidates. The liver is known to express a large number of secreted proteins that are not likely to interact with NS5B, a cytoplasmic protein. Besides infecting the liver, HCV has been reported to infect B cells as well (36, 42), and the spleen is the organ in which B cells originate. A partial cDNA clone isolated from this screen encodes an RNA helicase, p68 (aa 90 to 614). The 5′ end of the cDNA for p68 was amplified from the spleen cDNA library by PCR and ligated with the partial clone to complete the open reading frame. The full-length p68 interacts with NS5B in a yeast two-hybrid test, and the interaction was confirmed by in vitro binding assays (data not shown).
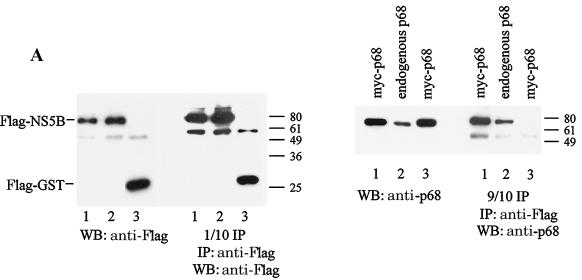
To show that NS5B interacts specifically with p68 in cells, HeLa cells were transfected with constructs expressing Flag-NS5B and myc-p68, Flag-NS5B alone, or Flag-GST and myc-p68. Flag-tagged proteins were precipitated with anti-Flag beads, and exogenous and endogenous p68 that resulted in co-IP with Flag-NS5B was detected with anti-p68 antibodies (Fig. 1A). p68 resulted in co-IP with NS5B but not with GST, and the amount of p68 precipitated was proportional to the total amount of p68 in the cells.
FIG. 1.
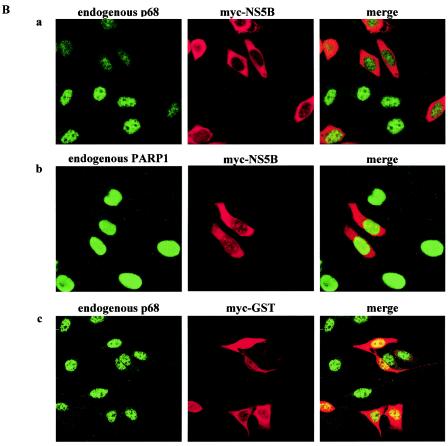
NS5B coimmunoprecipitates specifically with p68 and causes the redistribution of endogenous p68 from the nucleus to the cytoplasm. (A) HeLa cells were transfected with constructs expressing Flag-NS5B and myc-p68 (lanes 1), Flag-NS5B only (lanes 2), or Flag-GST and myc-p68 (negative control; lanes 3). Flag-tagged proteins were immunoprecipitated (IP) with anti-Flag beads (1/10 IP) and the coimmunoprecipitated myc-p68 or endogenous p68 (9/10 IP) was detected with anti-p68 antibodies (2130). Expression of transfected constructs is shown in WBs of 10 μg of total protein with the indicated antibodies. Molecular weight markers are indicated on the right of the blots. (B) HeLa cells were transfected with constructs expressing myc-NS5B (panels a and b) or myc-GST (panels c) and stained with anti-p68 monoclonal PAb204 (panels a and c) or anti-PARP (panels b) antibodies (left panels) and anti-myc antibodies (middle panels). The merged signals of fluorescein isothiocyanate- and rhodamine-conjugated secondary antibodies are shown in the right panels.
p68 has been shown to be localized to the nucleus and nascent nucleoli, while NS5B is primarily localized to the cytoplasm and ER membranes. To understand how these two proteins (which are present in different compartments of the cell) can interact, we transfected HeLa cells with a construct expressing myc-NS5B and visualized the staining pattern of NS5B and endogenous p68 by indirect IF using anti-myc and anti-p68 antibodies. Strikingly, overexpression of NS5B causes p68 to relocalize from the nucleus to the cytoplasm (Fig. 1Ba). This phenomenon was seen with PAb204 (Fig. 1B) and 2906 (data not shown), two anti-p68 antibodies directed against different epitopes in p68 (Materials and Methods), indicating that the protein that was detected was indeed p68 and not a cross-reacting antigen. The p68 signal in the nucleus was reduced, while faint staining was seen in the cytoplasm. p68 is normally found in the nucleus and is released into the cytoplasm when the nuclear envelope dissociates during mitosis (Fig. 2Dc). This mitotic phase presumably provides the situation in which p68 and NS5B can exist in the same compartment. When the nuclear envelope reforms, p68 is transported back to the nucleus. In the presence of NS5B, some p68 is bound and retained in the cytoplasm by NS5B. The reduction in p68 signal in mitotic cells may be due to the effect of the general dispersion of p68 protein into the whole cell or it may reflect a reduction in protein levels during mitosis. Nascent synthesized p68 could also be retained in the cytoplasm by NS5B; that would prevent it from being translocated into the nucleus. Less than 10% of NS5B-expressing cells did not show a reduction in p68 in the nucleus (Fig. 1Ba; cell in lower left corner), possibly because these cells have not undergone mitosis at the point at which NS5B has accumulated to sufficient levels. NS5B-expressing cells were also stained with anti-PARP1 to visualize the localization of another nuclear protein, PARP1, in the presence of NS5B (Fig. 1Bb). PARP1 remains in the nucleus in the presence or absence of NS5B, indicating that NS5B specifically causes nuclear p68 to be redistributed. As another control, myc-GST (Fig. 1Bc) and NS5A (data not shown), both cytoplasmic proteins, were overexpressed in HeLa cells. These two proteins did not elicit the relocalization effect of p68, showing that p68 does not become relocalized upon overexpression of other cytoplasmic proteins.
FIG. 2.
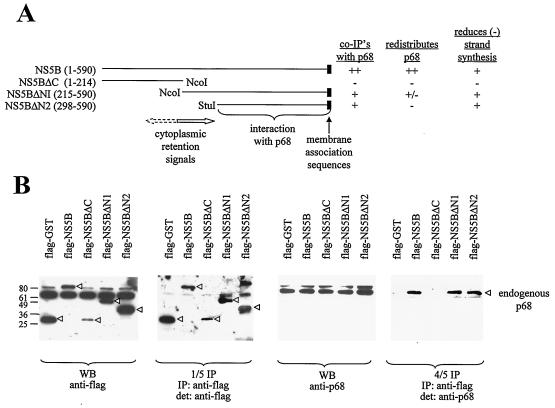
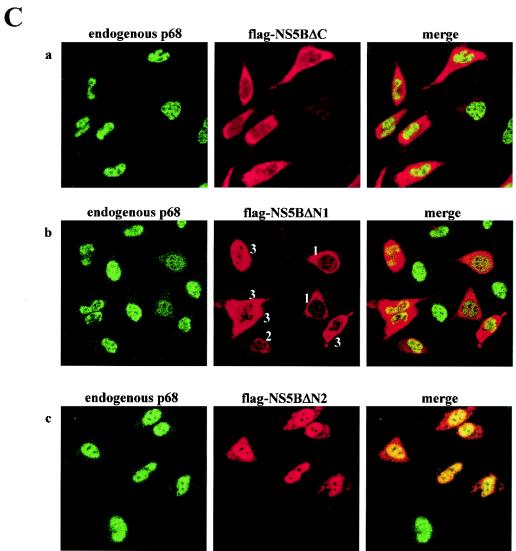
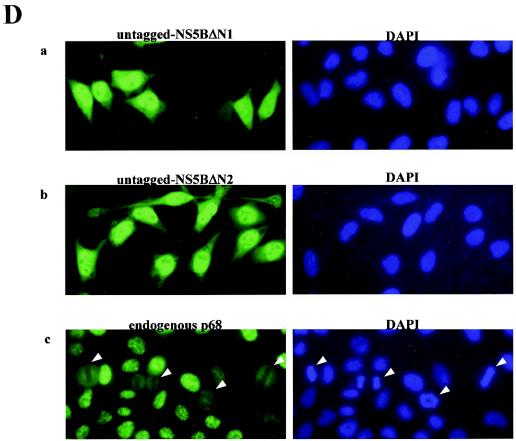
Deletion of the C terminus of NS5B abolishes its ability to interact with p68 and its relocalization effect with respect to p68. (A) Schematic diagram of NS5B deletion mutants used in this study. The regions containing cytoplasmic retention signals, membrane association sequences, and the p68-interacting domain are indicated. The ability to undergo co-IP with endogenous p68 and the effect on p68 cellular localization and on negative-strand [(−) strand] transcription of these proteins at a high or low level are summarized. ++, +, +/−, and − indicate very strong, strong, weak, and negative effects, respectively. (B) HeLa cells were transfected with plasmids expressing Flag-tagged GST or full-length NS5B, NS5BΔC, NS5BΔN1, or NS5BΔN2. The Flag proteins were immunoprecipitated (IP) with anti-Flag antibodies (1/5 IP), and the coimmunoprecipitated p68 (4/5 IP) was detected (det) with anti-p68 antibodies. A WB of 10 μg of total protein shows the expression of Flag-tagged proteins (indicated by arrowheads) and endogenous p68. Numbers on the left indicate the molecular weight markers. (C) HeLa cells were transfected with constructs expressing Flag-tagged NS5B deletion mutants NS5BΔC (a), NS5BΔN1 (b), and NS5BΔN2 (c). NS5BΔN1 could be found in the cytoplasm alone (cells labeled with a 1), mainly in the nucleus (2), or in both compartments (3). Flag-tagged NS5B proteins were detected with anti-Flag antibodies (middle panels), and the endogenous p68 was detected with PAb204 (left panels). Right panels, merged images. (D) HeLa cells were transfected with constructs expressing untagged NS5BΔN1 (a) and NS5BΔN2 (b) and stained with 5B-3B1 (an anti-NS5B monoclonal) (left panels) and DAPI (4′,6′-diamidino-2-phenylindole) (a nuclear DNA stain) (right panels). Actively dividing HeLa cells were stained with anti-p68 polyclonal 2906 (c) to show the distribution of endogenous p68 in mitotic and in interphase cells. Mitotic cells with condensed chromosomes are indicated by arrowheads. Images were visualized with a Zeiss Axioplan microscope and captured with a Zeiss camera (AxioCam) and the program AxioVision.
To define the region of NS5B that interacts with p68, various truncations were made (Fig. 2A) and tested for their interaction with endogenous p68 in co-IP experiments (Fig. 2B). HeLa cells were transfected with constructs expressing Flag-tagged NS5B and NS5B deletion mutants. The Flag-tagged proteins were immunoprecipitated with anti-Flag beads, and the coimmunoprecipitated p68 was detected in WBs with anti-p68 antibody. Deletion of the C terminus of NS5B (NS5BΔC) weakens its binding to p68, while deletion of the N-terminus region of NS5B (NS5BΔN1 and NS5BΔN2) does not affect its co-IP with p68 (compared to the results seen with full-length NS5B) (Fig. 2B).
The cellular localization of p68 was examined in the presence of NS5B deletion mutants (Fig. 2C). NS5BΔC mutant is localized to the cytoplasm, but it does not have the ability to sequester endogenous p68, which remains in the nucleus (Fig. 2Ca). This is consistent with the reduced binding activity of NS5BΔC with p68. Deletion of the N-terminal 214 aa causes NS5BΔN1 be localized to the nucleus or cytoplasm; in most cells, however, it is found in both compartments (Fig. 2Cb). A further deletion up to aa 298 in NS5BΔN2 abolishes its cytoplasmic localization (Fig. 2Cc). The complex localization pattern of Flag-NS5BΔN1 could be due to a competition between cytoplasmic localization signals in NS5B and nuclear localization signals in p68. The interaction between NS5BΔN1 and p68 then produces a range of effect on p68 localization, depending on where NS5BΔN1 is found. Generally, when NS5BΔN1 is in the cytoplasm p68 is relocalized to the cytoplasm and when NS5BΔN1 is in the nucleus or in both nucleus and cytoplasm the nuclear p68 staining is reduced (compared to the results seen with untransfected cells); however, the reduction is not as pronounced as in cells with NS5BΔN1 in the cytoplasm. A larger deletion of the N-terminal region (aa 1 to 297) results in the localization of NS5BΔN2 in the nucleus; not surprisingly, endogenous p68 remains in the nucleus (Fig. 2Cc). Deletion of the N terminus of NS5B (up to aa 298) may have caused NS5B to lose its cytoplasmic anchoring properties. Interaction with p68 may then bring NS5BΔN mutants into the nucleus. We cannot rule out another possibility: that the N-terminal deletion mutants are passively diffusing into the nucleus because they are not large enough to be excluded. Once in the nucleus they are retained by p68, while the small C-terminal truncation is not retained because it cannot associate with p68.
To ascertain that the change in cellular localization of NS5BΔN is not the effect of the insertion of a Flag tag at the N termini of these truncated proteins, we observed the localization of untagged NS5BΔN1 and NS5BΔN2 proteins with an anti-NS5B antibody, 5B-3B1 (26) (Fig. 2D). This monoclonal antibody was reported to be incapable of detecting its epitope in the C-terminal region (aa 372 to 382) in IF experiments. We were able to stain cells with it, however, possibly because of the milder cell-fixing conditions (using only methanol) that were used. Untagged NS5BΔN1 protein is predominantly concentrated in the nucleus, with some visible in the cytoplasm (Fig. 2Da). About 20% of the cells show a general distribution of the untagged NS5BΔN1 in the whole cell (Fig. 2Da). This localization pattern is somewhat different from that of the N-terminally Flag-tagged protein. The insertion of a Flag tag seemed to diminish the amount of protein translocated to the nucleus (Fig. 2Cb). Although Flag-tagged NS5BΔN1 did not reflect its bona fide localization, its presence in the cytoplasm incidentally provided a situation in which the redistribution of p68 could be seen (Fig. 2Cb). Both tagged and untagged NS5BΔN2 show nuclear localization, indicating that the loss of cytoplasmic retention is due to the deletion of cytoplasmic retention signals in the N-terminal region (around aa 215 to 298) in NS5B (Fig. 2A) and is not an artifact of the insertion of a Flag epitope. Additional membrane association sequences have been defined in the C-terminal 21 aa of NS5B (34), but the presence of these sequences may not be sufficient to retain NS5BΔN mutants in the cytoplasm.
Relocalization of p68 and analysis of RNA replication in a replicon cell line, 9-13.
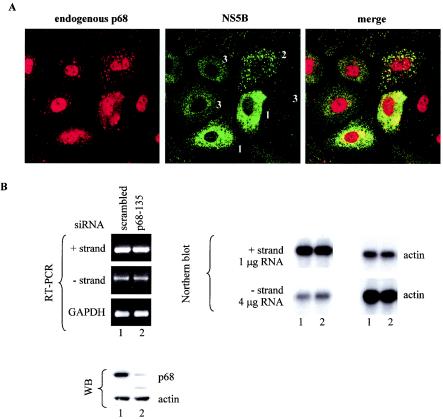
With a HCV RNA replicon that expresses all the nonstructural proteins, we showed that the relocalization of p68 was also visible. 9-13 cells were plated for 2 days, and NS5B and endogenous p68 localization were visualized with anti-NS5B monoclonal 5B-3B1 and anti-p68 polyclonal 2906, respectively. In 9-13 cells the level of NS5B differs from cell to cell, which may reflect the cell cycle fluctuation of cellular factors on which HCV replication and/or translation is dependent (24). NS5B is localized around the nucleus and in punctate structures in the cytoplasm in cells with high levels of expression, while in others they are found in dispersed punctate structures in the cytoplasm (Fig. 3A and B). Cells in which NS5B is present in high levels have p68 in the cytoplasm as well as in the nucleus. In some cells, the p68-positive spots colocalize with NS5B staining. In cells expressing low levels or undetectable levels of NS5B, the redistribution of p68 is not obvious. These cells act as internal controls, showing that the redistribution is dependent on the presence of NS5B in sufficient levels in the cytoplasm. The redistribution of p68 in a Huh-7 background as well as in HeLa cells (Fig. 2C) indicates that it is not cell line specific. It was difficult to show co-IP of p68 with NS5B in these cells, because NS5B is present in much lower quantities than in HeLa cells overexpressing NS5B.
FIG. 3.
Knockdown of p68 in replicon-bearing cell line 9-13 has no effect on the replication of the subgenomic replicon. (A) 9-13 cells were plated for 2 days and processed for IF. The results of localization of p68 with polyclonal anti-p68 (2130) are shown in the left panel and those of NS5B with monoclonal NS5B-3B1 are shown in the middle panel, while the merged images of the left and middle panels are shown in the right panel. Cells labeled 1, 2, and 3 show different levels of NS5B expression. (B) Replicon cell line 9-13 was transfected with scrambled siRNA (lanes 1) or p68-135 (lanes 2). The final concentration of siRNAs in the culture medium was 50 nM. Cells were harvested 2 days after transfection. Nine-tenths of the cells were extracted for RNA, while 1/10 of the cells were extracted for proteins for WB. The RNA was amplified by RT-PCR for positive- and negative-strand HCV RNA and cellular GAPDH RNA. Northern blots of total RNA was probed with strand-specific probes as follows: for positive strands, 4 μg of RNA was loaded; for negative strands, 1 μg of RNA was loaded. The level of actin mRNA was used as a loading control. A total of 10 μg of total proteins was probed for p68 (with polyclonal antibody 2130) and for β-actin to show equal loading.
The observations described above suggest that the interaction of NS5B with p68 might serve to recruit the cellular RNA helicase into the viral replicase to assist in viral RNA replication. To test this possibility, we used siRNA to specifically inhibit the expression of endogenous p68 (7) and assayed for the synthesis of negative-strand HCV RNA. siRNA p68-135 was a very potent inhibitor of p68 expression (Fig. 3B and Fig. 4B and C). At 2 days after transfection of p68-135 siRNA, the endogenous p68 level was reduced to less than 10% of the level seen with cells transfected with control siRNA. The HCV replicon is a robust system that has been selected for its ability to self-replicate a subgenomic RNA at high levels and has been widely accepted as a model for studying HCV replication (3, 23). We used the replicon 9-13 (23) to see whether HCV RNA replication is affected upon knocking down endogenous p68. 9-13 cells were transfected with a scrambled siRNA as a control (Fig. 3B, lane 1) or p68-135, the p68 siRNA (Fig. 3B, lane 2). Both siRNAs did not elicit any change in the level of positive or negative strand, as shown by RT-PCR and Northern blot assays (Fig. 3B). Note that four times as much RNA was loaded for the negative-strand blot and that the signal of the negative-strand HCV RNA was about four times less than that of the positive-strand RNA. The difference in positive- versus negative-strand levels is in the range of 10- to 20-fold. The RT-PCR results, however, indicate that the difference between the positive and negative strand levels is about 400- to 500-fold, considering that the template for PCR for positive strand was diluted 1:100.
FIG. 4.
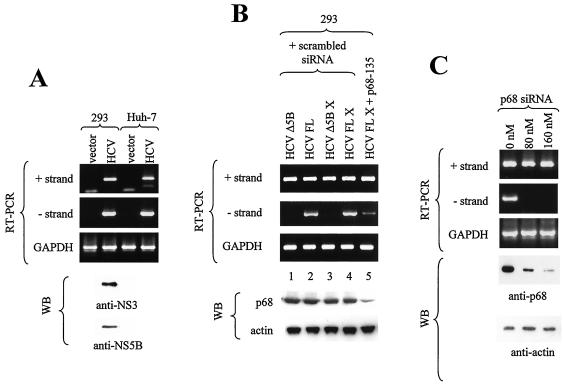
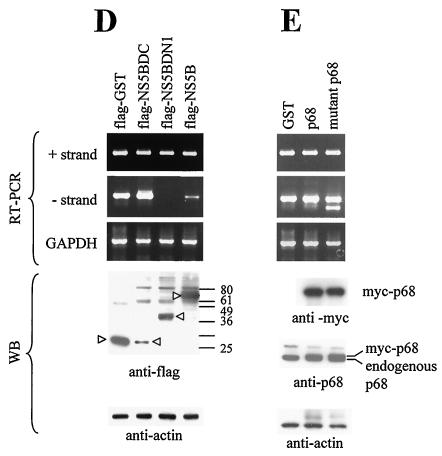
p68 could play a role in HCV RNA replication, as assayed in a transient expression system. (A) A replication assay using a full-length HCV construct was established. 293 and Huh-7 cells were transfected with vector or pcDNA-HCV(Q19) (HCV) plasmid DNA. At 2 days after transfection, the cells were harvested and extracted for RNA and proteins. The RNAs were assayed for the presence of positive- and negative-strand HCV RNA and for endogenous GAPDH RNA as a control. A total of 40 μg of total proteins was detected for NS3 and NS5B with anti-NS3 and anti-NS5B monoclonal antibodies. (B) 293 cells were transfected with uncut pcDNA-HCVΔNS5B (HCVΔ5B; lane 1), pcDNA-HCV(Q19) (HCV FL; lane 2), pcDNA-HCVΔNS5B cut with XbaI (HCVΔ5B X; lane 3), or pcDNA-HCV(Q19) cut with XbaI (HCV FL X; lanes 4 and 5) together with control scrambled siRNA (lanes 1 to 4) or p68 siRNA (lane 5). pcDNA-HCVΔNS5B has most of the NS5B coding sequences removed, and XbaI digestion removes the poly(A) tail from the RNA produced. 293 cells were cotransfected with pcDNA-HCV(Q19) and increasing amounts of p68 siRNA (C) or Flag-GST, Flag-NS5BΔC, Flag-NS5BΔN1, and Flag-NS5B (D) or Flag-GST, myc-p68, and myc-mutant p68 (E). A total of 9/10 of the cells were extracted for RNA, and 1/10 of the cells were extracted for proteins for WB. WB (using anti-p68 antibody) of 10 μg of total protein showed the level of endogenous p68; β-actin levels served as a loading control. The amount of positive- and negative-strand viral RNA was amplified through RT-PCR and visualized on 1.7% agarose gels. RT-PCR of GAPDH RNA served as a control for the quantity and quality of RNA used.
Knockdown of p68 reduces the negative-strand synthesis of HCV genome in a transient expression system.
The HCV replicon system was selected for very high levels of HCV RNA self-replication. Such high levels may be difficult to knock down. For this reason, we established another system in which full-length HCV RNA was expressed transiently in a cell line.
pcDNA-HCV(Q19), the full-length HCV construct used for producing positive-strand RNA template, was cloned into a mammalian expression vector, pcDNA3.1(+), which expresses the positive strand from the cytomegalovirus promoter constitutively. For unknown reasons the positive-strand transcript was produced at very low levels (as it could barely be detected), while negative-strand transcripts was not detectable by Northern blotting of RNA from cells at 2 days posttransfection. This could have been due to a low percentage of cells being transfected or to sequences in the HCV genome that reduced its transcription. The first possibility could not be tested, as the level of NS3 or NS5B translated from the full-length construct was too low to be detected by IF. Two cell lines, 293 and Huh-7, were transfected with pcDNA-HCV(Q19) and assayed for viral protein expression and positive- and negative-strand production by RT-PCR. The expression of NS3 and NS5B could only be detected by WB by using large amounts of protein and a high concentration of antibodies in 293 cells, although both cell lines showed equal levels of negative-strand and positive-strand RNA (Fig. 4A). The 293 cell line was chosen for this study, because it consistently yielded a high concentration of RNA and protein.
To establish the assay system, we first showed that the replication of negative-strand RNA from pcDNA-HCV(Q19) was dependent on the presence of HCV NS5B. The deletion of a large portion of NS5B abolished negative-strand production (Fig. 4B, lanes 1 and 3). The positive-strand HCV RNA transcribed from pcDNA vector would contain a poly(A) tail at the 3′ end, but this still allows the production of the negative strand (Fig. 4A and B). Deletion of the poly(A) tail by cutting pcDNA-HCV(Q19) at XbaI, a unique site at the end of the HCV cDNA, did not affect the efficiency of transcription of negative-strand RNA (Fig. 4B, lane 4) compared to the results seen with transcription of negative-strand from positive-strand RNA with a poly(A) tail (Fig. 4B, lane 2). The negative strand produced from a poly(A)-less RNA was also knocked down with p68 siRNA (lane 5). Cell lines stably transfected with the full-length HCV genome under the control of a tetracycline-inducible promoter were able to generate positive- and negative-strand viral RNA as well as viral-like particles (21).
To assay for the production of negative-strand HCV RNA, 293 cells cotransfected with p68 siRNA and pcDNA-HCV(Q19) were harvested and extracted for RNA 2 days after transfection. Negative-strand synthesis from a positive-strand template is presumably driven by NS5B, probably with the help of viral and cellular factors. The amount of negative-strand RNA was measured after amplification through semiquantitative RT-PCR. RT-PCRs were done on RNA samples from at least three independent experiments to ensure that the results were consistent. A reduction in negative strand was correlated with the knockdown of cellular p68, while positive-strand and GAPDH transcripts were not affected (Fig. 4A). This indicates that p68 plays a role in the transcription of negative-strand RNA from the positive-strand RNA template.
Since the NS5BΔN1 mutant binds p68, the presence of this fragment may compete with NS5B expressed from the full-length HCV genome for p68 and thus reduce the amount of p68 available for viral RNA replication. To test this hypothesis, 293 cells were cotransfected with plasmids expressing wild-type NS5B, NS5B deletion mutants (NS5BΔC or NS5BΔN1), or GST (Fig. 4D). The negative-strand synthesis was not affected in the presence of NS5BΔC but was decreased when NS5BΔN1 was overexpressed (Fig. 4D), correlating well with the relative binding efficiencies of NS5BΔC and NS5BΔN1 with p68 (Fig. 2B). Excess NS5B binds p68 but is not fully active, because a full complement of viral and host factors is not present in the complex. The reduction in negative-strand transcripts could be explained by the sequestration of cellular p68 by NS5BΔN1 and NS5B, which thus reduces the available p68 for binding with functional viral replicases. NS5BΔN1 can also bring p68 into the nucleus, which further reduces the amount of p68 in the cytoplasm (where viral replicases are thought to reside).
In a converse experiment, we expressed mutant p68 to compete with endogenous p68 for binding NS5B. The expression of mutant as well as wild-type p68 did not affect the production of negative strand from the positive-strand RNA (Fig. 4E). This mutant contains a point mutation which changed the DEAD box to NEAD, is inactive with respect to its ATPase/RNA helicase activity (unpublished observations), and is able to interact with NS5B (data not shown). This lack of any effect on negative-strand synthesis could imply that (i) there is sufficient endogenous p68 in the cell to promote negative-strand transcription and that (ii) an active p68 is not necessary for an active viral replicase. The second possibility is not unprecedented; p68 helicase activity was not necessary for p68 to act as a transcriptional coactivator of ERα (8). Overexpression of NS5B, NS5BΔN, p68, and mutant p68 in 9-13, the replicon cell line, did not elicit any change in the positive- and negative-strand HCV RNA.
DISCUSSION
We showed that the overexpression of NS5B, the HCV RdRp, induces the redistribution of p68 (cellular RNA helicase) from the nucleus to the cytoplasm. The relocalization of cellular proteins in the presence of viral proteins or viral RNA has been reported: the poliovirus RdRp interacts with and causes the relocalization of human Sam68 (25), and the export element (constitutive transport element) of type D retroviruses is bound by a human nuclear protein, RNA helicase A, which then shuttles to the cytoplasm (37). The function of these interactions and the subsequent relocalization of cellular proteins may serve to mediate viral replication processes.
NS5B may recruit p68 into the viral replication complex to enhance HCV replication, as a reduction of endogenous p68 reduces the synthesis of negative strand from the viral positive-strand RNA. The logical assumption would be that p68 helps to unwind the viral RNA so that the RdRp can transcribe the RNA more efficiently. The recruitment of a cellular RNA helicase is not expected to be necessary, since the viral genome encodes its own RNA helicase within the NS3 protein. However, the viral helicase may not be sufficiently efficient. It is therefore not inconceivable that the virus also recruits other cellular proteins, such as a cellular RNA helicases, to enhance its replication. The binding of cellular helicases could also bring in additional cellular factors to the viral replicase. In this case, the helicase activity of p68 does not appear to be required, suggesting that p68 is not acting as a bone fide helicase but perhaps is acting more as a transcription factor. A similar situation has been reported for the coactivation of ER by p68 (8).
We showed that p68 potentially plays a role in HCV RNA transcription in 293 cells transiently expressing the full-length HCV RNA. The use of 293, a non-liver-cell line, for studying HCV RNA replication is not totally inappropriate, as the replication of negative-strand RNA from a positive-strand HCV RNA template has been reported so far for liver, kidney, and B- and T-cell-derived cell lines (summarized in references 2 and 33). Recently, 293, the human embryonic kidney cell line, was shown to be permissive for replicon amplification (1). The capacity of a variety of cell types to replicate HCV RNA suggests that the cellular cofactors (such as p68) for viral RNA replication are present in most cell types. The tissue tropism of HCV in liver and lymphoid cells in vivo is thus not limited by cellular cofactors but more likely is limited by processes that precede HCV RNA replication, such as the viral internalization step.
The idea of a role of p68 in negative-strand synthesis was strengthened by the observation that expression of NS5B and the NS5BΔN1 mutant that can bind p68 and can therefore compete for endogenous p68 also inhibits negative-strand production. These data suggest that p68 is recruited to the viral replicase through its interaction with NS5B and that p68 enhances viral RNA transcription. The sequestration of p68 from the nucleus, reducing the amount of protein available for its normal cellular function in the long term, may lead to abnormal cellular function.
The HCV RNA replicon cell lines provide a robust model system for studying RNA replication of the HCV genome. However, we were not able to detect a change in positive- and negative-RNA levels in the 9-13 replicon cell line when endogenous p68 was knocked down by p68 siRNA. We cannot explain the discrepancy in the role of p68 in the two systems tested except by speculating on two possibilities. First, genetic differences in the host genomes and the viral RNAs between the two systems may contribute to these differences. Characterization of a replicon-expressing cell line suggests that the high-level efficiency in HCV replication in cell culture is determined both by adaptation of the viral sequence and by the host cell (24). The accumulated adaptive mutations in the HCV RNA that allow the replication of the subgenomic HCV RNA at high levels may have somehow circumvented the dependence on p68 for its replication. Another study, however, has shown that a subgenomic replicon RNA contained no mutations in the NS3-5B polyprotein, confirming that adaptive mutations are not required for efficient replication in cells (16). It would be interesting to see whether the depletion of endogenous p68 in the cell line carrying this replicon affects its replication. This would indicate whether the lack of dependence on p68 for HCV RNA replication is due to mutations in the viral or host genome.
The second possible explanation for the differences in the role of p68 on HCV replication is that in the transient transfection system, the full-length HCV genome was expressed, while in the replicon, only the nonstructural proteins were expressed. Replication of the full-length genome has been found to be very inefficient in all systems studied so far; indeed, some reports have suggested that core, a structural protein, may regulate the balance between transcription and translation (41). A recent study has shown that the introduction of adaptive mutations into a full-length HCV genome renders it defective in replication in chimpanzees (5).
The recruitment of a cellular helicase by a HCV RdRp may have important implications in the understanding of viral replication. The identification of cellular factors that are critical for viral replication could be useful for the development of specific anti-viral therapies.
Acknowledgments
We thank Sam Nicol (University of Dundee, United Kingdom), Y. W. Choi, H. Hong, and C. Lai for technical assistance. D. Moradpour (University of Freiburg, Freiburg, Germany) kindly provided 5B-3B1, the anti-NS5B monoclonal antibody. The 9-13 cell line was obtained from R. Bartenschlager (Universitätsklinikum Heidelberg, Heidelberg, Germany).
This work was supported by grants from the Agency for Science, Technology and Research, Singapore.
REFERENCES
- 1.Ali, S., C. Pellerin, D. Lamarre, and G. Kukolj. 2004. Hepatitis C virus subgenomic replicons in the human embryonic kidney 293 cell line. J. Virol. 78:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2001. Novel cell culture systems for the hepatitis C virus. Antivir. Res. 52:1-17. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Bond, A. T., D. A. Mangus, F. He, and A. Jacobson. 2001. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol. Cell. Biol. 21:7366-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukh, J., T. Pietschmann, V. Lohmann, N. Krieger, K. Faulk, R. E. Engle, S. Govindarajan, M. Shapiro, M. St. Claire, and R. Bartenschlager. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Causevic, M., R. G. Hislop, N. M. Kernohan, F. A. Carey, R. A. Kay, R. J. Steele, and F. V. Fuller-Pace. 2001. Overexpression and poly-ubiquitylation of the DEAD-box RNA helicase p68 in colorectal tumours. Oncogene 20:7734-7743. [DOI] [PubMed] [Google Scholar]
- 7.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 8.Endoh, H., K. Maruyama, Y. Masuhiro, Y. Kobayashi, M. Goto, H. Tai, J. Yanagisawa, D. Metzger, S. Hashimoto, and S. Kato. 1999. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor α. Mol. Cell. Biol. 19:5363-5372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Ferrari, E., J. Wright-Minogue, J. W. S. Fang, B. M. Baroudy, J. Y. N. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ford, M. J., I. A. Anton, and D. P. Lane. 1988. Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature 332:736-738. [DOI] [PubMed] [Google Scholar]
- 11.Garson, J. A., C., Ring, P., Tuke, and R. S. Tedder. 1990. Enhanced detection by PCR of hepatitis C viral RNA. Lancet 336:878-879. [DOI] [PubMed] [Google Scholar]
- 12.Goh, P.-Y., Y.-J. Tan, S. P. Lim, S. G. Lim, Y. H. Tan, and W. J. Hong. 2001. The hepatitis C virus core protein interacts with NS5A and activates its caspase-mediated proteolytic cleavage. Virology 290:224-236. [DOI] [PubMed] [Google Scholar]
- 13.Hirling, H., M. Scheffner, T. Restle, and H. Stahl. 1989. RNA helicase activity associated with the human p68 protein. Nature 339:562-564. [DOI] [PubMed] [Google Scholar]
- 14.Iggo, R. D., and D. P. Lane. 1989. Nuclear protein p68 is an RNA-dependent ATPase. EMBO J. 8:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iggo, R. D., D. J. Jamieson, S. A. MacNeill, J. Southgate, J. McPheat, and D. P. Lane. 1991. p68 RNA helicase: identification of a nucleolar form and cloning of related genes containing a conserved intron in yeasts. Mol. Cell. Biol. 11:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genomic-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishido, S., T. Fujita, and H. Hotta. 1998. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun. 244:35-40. [DOI] [PubMed] [Google Scholar]
- 18.Khu, Y.-L., E. Koh, S. P. Lim, Y. H. Tan, S. Brenner, S. G. Lim, W. J. Hong, and P.-Y. Goh. 2001. Mutations that affect dimer formation and helicase activity of the hepatitis C virus helicase. J. Virol. 75:205-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyono, K., M. Miyashiro, and I. Taguchi. 2002. Human eukaryotic initiation factor 4AII associates with hepatitis C virus NS5B protein in vitro. Biochem. Biophys. Res. Commun. 292:659-666. [DOI] [PubMed] [Google Scholar]
- 20.Lane, D. P., and W. K. Hoeffler. 1980. SV40 large T shares an antigenic determinant with a cellular protein of molecular weight 68,000. Nature 288:167-170. [DOI] [PubMed] [Google Scholar]
- 21.Lim, S. P., Y. L. Khu, W. J. Hong, A. Tay, A. E. Ting, S. G. Lim, and Y. H. Tan. 2001. Identification and molecular characterization of the complete genome of a Singapore isolate of hepatitis C virus: sequence comparison with other strains and phylogenetic analysis. Virus Genes 23:89-95. [DOI] [PubMed] [Google Scholar]
- 22.Lim, S. P., H. M. Soo, Y. H. Tan, S. Brenner, H. Horstmann, J. M. MacKenzie, M. L. Ng, S. G. Lim, and W. Hong. 2002. Inducible system in human hepatoma cell lines for hepatitis C virus production. Virology 303:79-99. [DOI] [PubMed] [Google Scholar]
- 23.Lohmann, V., F. Korner, J.-O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 24.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride, A. E., A. Schlegel, and K. Kirkegaard. 1996. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. USA 93:2296-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moradpour, D., E. Bieck, T. Hugle, W. Wels, J. Z. Wu, Z. Hong, H. E. Blum, and R. Bartenschlager. 2002. Functional properties of a monoclonal antibody inhibiting the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:593-601. [DOI] [PubMed] [Google Scholar]
- 27.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 28.Nicol, S. M., M. Causevic, A. R. Prescott, and F. V. Fuller-Pace. 2000. The nuclear DEAD box RNA helicase p68 interacts with the nucleolar protein fibrillarin and colocalizes specifically in nascent nucleoli during telophase. Exp. Cell Res. 257:272-280. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto, H., S. Okada, Y. Sugiyama, T. Tanaka, Y. Sugai, Y. Akahane, A. Machida, S. Mishiro, H. Yoshizawa, Y. Miyakawa, and M. Mayumi. 1990. Detection of hepatitis C virus RNA by a two-stage polymerase chain reaction with two pairs of primers deduced from the 5′-noncoding region. Jpn. J. Exp. Med. 60:215-222. [PubMed] [Google Scholar]
- 30.Osman, T. A., and K. W. Buck. 1997. The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIR-3. J. Virol. 71:6075-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quadt, R., C. C. Kao, K. S. Browning, R. P. Hershberger, and P. Ahlquist. 1993. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 90:1498-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schinazi, R. F., E. Ilan, P. L. Black, X. Yao, and S. Dagan. 1999. Cell-based and animal models for hepatitis B and C viruses. Antivir. Chem. Chemother. 10:99-114. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 35.Shirota, Y., H. Luo, W. Qin, S. Kaneko, T. Yakashita, K. Kobayashi, and S. Murakami. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRp) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277:11149-11155. [DOI] [PubMed] [Google Scholar]
- 36.Sung, V. M., S. Shimodaira, A. L. Doughty, G. R. Picchio, H. Can, T. S. Yen, K. L. Lindsay, A. M. Levine, and M. M. Lai. 2003. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J. Virol. 77:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, H., G. M. Gaietta, W. H. Fischer, M. H. Ellisman, and F. Wong-Staal. 1997. A cellular cofactor for the constitutive transport element of type D retrovirus. Science 276:1412-1415. [DOI] [PubMed] [Google Scholar]
- 38.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. Wen, A. E. Gorbalenya, S. B. Hwang, and M. M. Lai. 1999. Hepatitis C Virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe, M., J. Yanagisawa, H. Kitagawa, K. Takeyama, S. Ogawa, Y. Arao, M. Suzawa, Y. Kobayashi, T., Yano, H. Yoshikawa, Y. Masuhiro, and S. Kato. 2001. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor a coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 20:1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Xiao, J. H., I. Davidson, H. Matthes, J. M. Garnier, and P. Chambon. 1991. Cloning, expression and transcriptional properties of the human enhancer factor TEF-1. Cell 65:551-568. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, J., O. Yamada, H. Yoshida, T. Iwai, and H. Araki. 2002. Autogenous translational inhibition of core protein: implication for switch from translation to RNA replication in Hepatitis C virus. Virology 293:141-150. [DOI] [PubMed] [Google Scholar]
- 42.Zignego, A. L., D. Macchia, M. Monti, V. Thiers, M. Mazzetti, M. Foschi, E. Maggi, S. Romagnani, P. Gentilini, and C. Brechot. 1992. Infection of peripheral mononuclear blood cells by hepatitis C virus. J. Hepatol. 15:382-386. [DOI] [PubMed] [Google Scholar]