Recruitment of ribosomes to the mRNA 5′ terminus involves the activity of a large number of functionally discrete eukaryotic translation initiation factors (eIFs). However, how multiple eIFs coordinate during translation initiation remains poorly understood. Walsh and Mohr now identify an eIF3 subunit that regulates eIF4F modification and show that eIF3e is required for inducible eIF4E phosphorylation. This study establishes a mechanism by which 40S ribosome loading imparts a phosphorylation mark on the cap-binding eIF4F complex that regulates selective mRNA translation.
Keywords: eIF4E phosphorylation, Mnk1, eIF3, ribosome recruitment, translation initiation
Abstract
40S ribosomes are loaded onto capped mRNAs via the multisubunit translation initiation factors eIF3 and eIF4F. While eIF4E is the eIF4F cap recognition component, the eIF4G subunit associates with 40S-bound eIF3. How this intricate process is coordinated remains poorly understood. Here, we identify an eIF3 subunit that regulates eIF4F modification and show that eIF3e is required for inducible eIF4E phosphorylation. Significantly, recruitment of the eIF4E kinase Mnk1 (MAPK signal-integrating kinase 1) to eIF4F depended on eIF3e, and eIF3e was sufficient to promote Mnk1-binding to eIF4G. This establishes a mechanism by which 40S ribosome loading imparts a phosphorylation mark on the cap-binding eIF4F complex that regulates selective mRNA translation and is synchronized by a specific eIF3 subunit.
Recruitment of 40S ribosome subunits to the mRNA 5′ terminus in eukaryotes requires a large number of translation initiation factors (eIFs) (Sonenberg and Hinnebusch 2009). Eukaryotic mRNAs have a 5′ methyl-7-GTP (m7-GTP) cap that is recognized by eIF4F, a multisubunit complex consisting of a cap-binding protein (eIF4E) and an RNA helicase (eIF4A) assembled on a large scaffold protein (eIF4G) (Fig. 1A). eIF4F assembly is regulated by eIF4E-binding proteins (4E-BPs) that competitively inhibit eIF4E from interacting with eIF4G. Phosphorylation of 4E-BPs by the kinase mTOR frees eIF4E, making it available to form an eIF4F complex. Once part of the complex, eIF4E is phosphorylated by the eIF4G-associated kinase MAPK signal integrating kinase 1 (Mnk1) or Mnk2 (Buxade et al. 2008). While Mnk2 accounts for basal eIF4E phosphorylation, Mnk1 mediates inducible phosphorylation in response to upstream p38MAPK or extracellular signal-regulated kinase (ERK) activation (Scheper et al. 2001). Phosphorylation of eIF4E regulates translation of specific mRNAs involved in cellular transformation, immune responses, and viral infection (Furic et al. 2010; Walsh and Mohr 2011; Herdy et al. 2012). eIF4G also binds polyA-binding protein (PABP), which binds the polyA tail at the mRNA 3′ end to stimulate translation of fully processed, intact mRNAs (Sonenberg and Hinnebusch 2009).
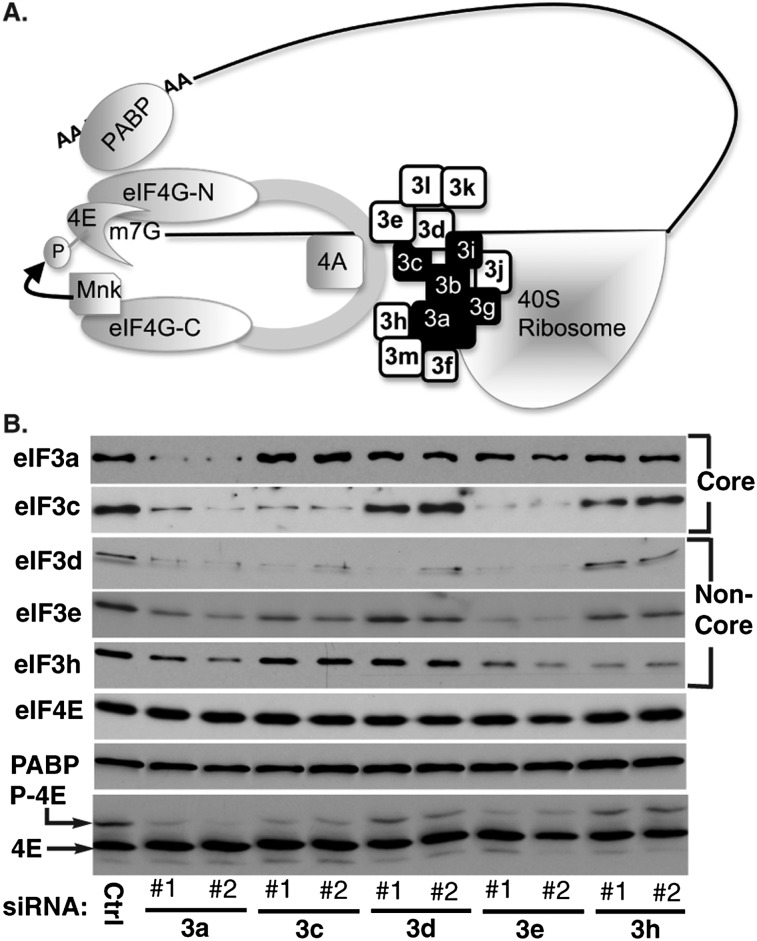
Figure 1.
eIF3 subunits regulate eIF4E phosphorylation. (A) eIF3 tethers the 40S ribosome to eIF4F. The mRNA 5′-m7-GTP cap is recognized by eIF4E, which binds the N terminus of eIF4G (-N). Mnk1 binds the C terminus of eIF4G (-C). eIF4G folding positions Mnk1 proximal to eIF4E, phosphorylating eIF4E as part of the eIF4F complex. eIF4G also binds eIF4A and PABP, which binds the mRNA 3′ polyA tail. Finally, eIF4G interacts with eIF3, consisting of conserved core (black) and noncore (white) subunits, to recruit the 40S ribosome. (B) NHDFs were treated with control (Ctrl), nonsilencing siRNA, or two independent siRNAs targeting eIF3 subunits a, c, d, e, or h. Whole-cell extracts were analyzed by immunoblotting with the indicated antibodies. (Bottom panel) Samples were resolved by IEF to separate phosphorylated (P-4E) and nonphosphorylated (4E) forms of eIF4E, detected by immunoblotting with anti-eIF4E antibody.
eIF4F recruits ribosomes indirectly through bridging interactions with a 40S ribosome-associated complex, eIF3 (Fig. 1A; Hinnebusch 2006). Mammalian eIF3 consists of 10–13 subunits (a–m) with a core comprised of five to eight subunits, of which a, b, c, g, and i have yeast homologs (Zhou et al. 2008; Sun et al. 2011; Querol-Audi et al. 2013), although a “functional core” of subunits a, b, c, e, f, and h has been suggested (Masutani et al. 2007). As a translation initiation factor, eIF3 stimulates ternary complex (TC) recruitment to the 40S ribosome and prevents premature 60S ribosome joining, both of which require noncore eIF3 subunits (Hinnebusch 2006). However, while eIF3f binds mTOR (Harris et al. 2006) and eIF3j interacts with eIF1A in the ribosomal A site (Fraser et al. 2007), the full spectrum of functions performed by subunits of this large complex remain unknown. Here, we demonstrate that eIF3e controls eIF4E phosphorylation by regulating recruitment of the eIF4E kinase Mnk1 to the eIF4F complex. This occurs independently of upstream signaling to Mnk1 and demonstrates that eIF3 not only tethers the 40S ribosome to cap-bound eIF4F but, surprisingly, also regulates eIF4F activity. Moreover, it suggests a mechanism that allows temporal synchronization of eIF4E phosphorylation with 40S ribosome recruitment during translation initiation.
Results and Discussion
eIF3 subunits regulate eIF4E phosphorylation and Mnk1-stimulated protein synthesis
To test whether eIF3 influences eIF4F activity, an siRNA screen was executed to investigate how eIF3 subunit depletion impacts eIF4E phosphorylation, which occurs as part of the eIF4F complex (Pyronnet et al. 1999). Primary normal human dermal fibroblasts (NHDFs) were transfected with independent siRNAs targeting conserved core (a or c) or noncore (d, e, or h) eIF3 subunits, and effects on eIF4E phosphorylation were determined by isoelectric focusing (IEF) (Fig. 1B). Importantly, depletion of individual eIF3 subunits differentially suppressed eIF4E phosphorylation to varying extents. Depleting noncore subunit d or h had relatively little effect on the abundance of other eIF3 subunits examined and only weakly reduced phosphorylated eIF4E steady-state levels. In contrast, depleting the core subunit eIF3a significantly decreased eIF4E phosphorylation but also reduced the abundance of other eIF3 subunits, including eIF3c and eIF3e. This demonstrated that eIF3 as a whole influenced eIF4E phosphorylation. Evidence that eIF3a was not directly responsible for this came from depleting subunit c or e, neither of which detectably reduced eIF3a abundance but both of which suppressed eIF4E phosphorylation. Depletion of eIF3c reduced eIF3d levels, which had little effect on eIF4E phosphorylation, as well as eIF3e. Notably, eIF3c recruits eIF3e to the eIF3 complex (Morris-Desbois et al. 1999; Zhou et al. 2008), while eIF3e directly binds the eIF4F scaffold eIF4G (LeFebvre et al. 2006). Indeed, eIF3c and eIF3e abundance correlated closely with changes in eIF4E phosphorylation (Fig. 1B). Finally, eIF4E and PABP abundance remained unchanged in all samples (Fig. 1B), suggesting that the impact of depleting individual eIF3 subunits on levels of other eIF3 subunits may reflect specific effects on subunit association within the eIF3 complex that might influence their stability or synthesis. Overall, this established that eIF3 controlled phosphorylation of the cap-binding protein eIF4E and was consistent with a direct role for the eIF4G-binding subunit eIF3e and its recruitment by eIF3c.
While different cellular mRNAs have highly variable requirements for phosphorylated eIF4E (Furic et al. 2010), the poxvirus vaccinia virus (VacV) has a genetically and chemically well-defined dependence on both Mnk1 and eIF4E phosphorylation to synthesize its proteins and replicate (Walsh et al. 2008; Herdy et al. 2012). This provided a powerful biological readout to test whether eIF3 could control translation by regulating eIF4E phosphorylation. First, the impact of depleting eIF3 subunits on viral protein synthesis was determined by treating NHDFs with siRNAs and then mock-infecting them or infecting them with VacV. Metabolic labeling of samples showed that in control siRNA-treated cells, VacV suppressed most host protein synthesis, while predominately viral polypeptides accumulated (Fig. 2A). Depleting eIF3a, eIF3c, or eIF3e reduced 35S-Met/Cys incorporation into protein in mock- and VacV-infected cells by ~40%–60% compared with control siRNA (Figs. 2A; Supplemental Fig. S1A). In contrast, depleting eIF3d or eIF3h modestly elevated 35S incorporation by 10%–15% in both mock- and VacV-infected cells (Fig. 2A; Supplemental Fig. S1A). Notably, comparing cellular versus viral proteins synthesized in infected cells following eIF3c or eIF3e depletion, which inhibits eIF4E phosphorylation, revealed a greater reduction of 35S-Met/Cys incorporation into viral polypeptides (Fig. 2A). Significantly, eIF3c or eIF3e depletion inhibited viral protein accumulation by at least 80%, with eIF3e depletion being modestly more effective and achieving a 95% reduction (Fig. 2B; Supplemental Fig. S1B). Finally, while growth and spread of a fluorescent reporter virus was reduced in cells upon depleting eIF3 subunits a, c, or e versus d or h, cell morphology indicated that these defects in VacV replication did not result from differential cell viability (Supplemental Fig. S2). Taken together, this established that eIF3 subunits a, c, and e were particularly important for efficient VacV protein synthesis and virus spread.
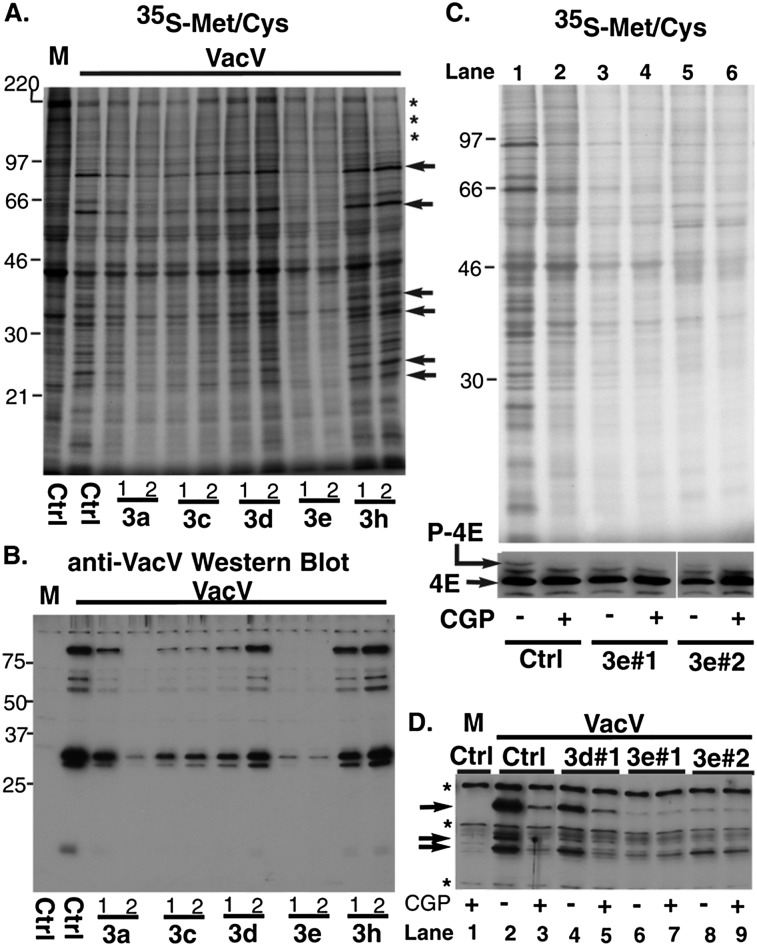
Figure 2.
Depletion of eIF3e renders VacV protein synthesis refractory to Mnk1 inhibition. (A) NHDFs treated with control siRNA (Ctrl) or siRNAs targeting eIF3 subunits were mock-infected (M) or infected with VacV (multiplicity of infection [MOI] = 5) for 16 h. After metabolic labeling for 20 min, cell lysates were fractionated by SDS-PAGE and fixed, and dried gels were exposed to X-ray film. Molecular weight markers (in kilodaltons) are indicated at the left. Representative radiolabeled host polypeptides (*) for comparison with select viral proteins (arrows) are indicated at the right of the panel. (B) Lysates from A were analyzed by immunoblotting with anti-VacV antibody. (C) NHDFs transfected with control (Ctrl), nonsilencing siRNA, or eIF3e siRNAs were treated for 1 h with DMSO (−) or 20 μM CGP57380 (+). Cells were then infected with VacV as in A in the presence of DMSO or GCP57380. (Top panel) 35S-Met/Cys labeling illustrating effects on VacV protein synthesis. (Bottom panel) IEF confirmed the efficacy of CGP57380 treatment and the effects of eIF3e depletion on eIF4E phosphorylation. (D) NHDFs were transfected with control (Ctrl), eIF3d, or eIF3e siRNAs. Samples treated with DMSO (−) or 20 μM CGP57380 (+) were mock-infected (M) or infected with VacV as in A and analyzed by immunoblotting with anti-VacV antibody. Arrows indicate viral proteins; asterisks indicate background bands present in mock-infected samples.
To determine whether suppression of VacV replication by eIF3e depletion was in part due to reduced eIF4E phosphorylation, NHDFs were treated with control or eIF3e siRNAs and then infected with VacV in the presence of DMSO or CGP57380, a Mnk1 inhibitor. In control siRNA-treated NHDFs, CGP57380 reduced eIF4E phosphorylation and suppressed VacV protein synthesis (Fig. 2C, cf. lanes 1 and 2). In contrast, eIF4E phosphorylation in NHDFs depleted of eIF3e was already reduced in DMSO-treated samples (Fig. 2C, lanes 3,5) and was not further reduced upon treatment with CGP57380 (Fig. 2C, cf. lanes 3 and 4 and lanes 5 and 6). In line with this, viral protein synthesis was not detectably reduced by CGP57380 in eIF3e-depleted cells (Fig. 2C, cf. lanes 3 and 4 and lanes 5 and 6). Similarly, while CGP57380 reduced VacV protein accumulation in control siRNA-treated samples (Fig. 2D, cf. lanes 2 and 3), Mnk1 inhibition had no effect on viral protein levels in cells treated with eIF3e siRNAs (Fig. 2D, cf. lanes 6 and 7 and lanes 8 and 9). In contrast, while eIF3d depletion reduced VacV protein production, consistent with its function as an eIF3 subunit, CGP57380 suppressed viral protein synthesis even further in these cells (Fig. 2D, cf. lanes 4 and 5). Notably, eIF3d depletion did not reduce eIF4E phosphorylation (Fig. 1B), serving as a specificity control. Thus, eIF3e depletion suppressed eIF4E phosphorylation and rendered VacV protein synthesis refractory to Mnk1 inhibition. The effects of eIF3e depletion on VacV protein production were greater than those of CGP57380 alone, in line with its roles as (1) an eIF3 subunit and (2) a regulator of eIF4E phosphorylation.
eIF3e regulates Mnk1 recruitment to eIF4F
eIF4E is derepressed by mTORC1-mediated inactivation of 4E-BPs, while upstream p38MAPK and ERK signaling stimulate the eIF4E kinase Mnk1 (Sonenberg and Hinnebusch 2009). To test whether eIF3e affected these upstream signal pathways that control eIF4E phosphorylation, NHDFs were treated with control or eIF3e siRNAs, and activation of mTORC1, p38MAPK, and ERK was monitored. Western blot analysis revealed that eIF3e depletion increased phosphorylation of two mTOR substrates, 4E-BP1 and p70S6K, as evidenced by reduced mobility in SDS-PAGE (Fig. 3A). Surprisingly, eIF4E phosphorylation was reduced in eIF3e-depleted NHDFs despite an increase in p38MAPK and ERK phosphorylation (Fig. 3A). Equivalent results were observed in an established monkey cell line upon eIF3e depletion, demonstrating that this was not unique to NHDFs (Fig. 3A). As such, although upstream signal pathways normally expected to increase eIF4E phosphorylation were activated, these signals were not detectably transmitted to eIF4E in eIF3e-depleted cells.
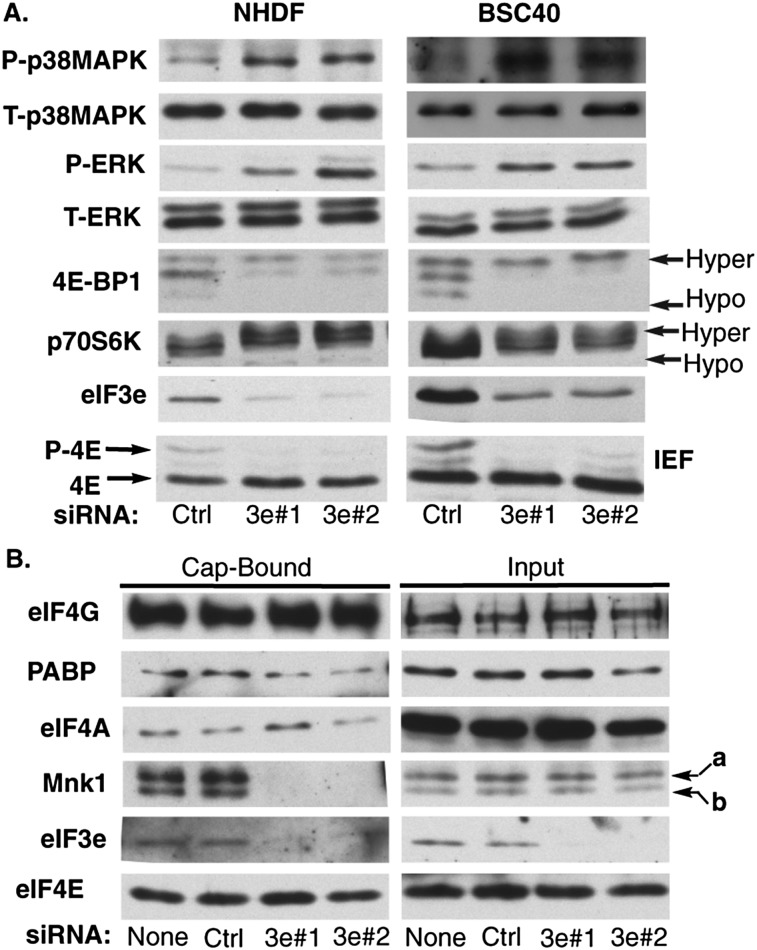
Figure 3.
eIF3e is required for Mnk1 binding to eIF4F. (A) NHDFs (left panels) or African green monkey kidney cells (BSC40; right panels) were transfected with control (Ctrl) nonsilencing or eIF3e siRNAs. Cell extracts were analyzed by immunoblotting with the indicated antibodies. (T) Total; (P) phosphorylated. (B) NHDFs were transfected with no siRNA, control (Ctrl) nonsilencing, or eIF3e siRNAs. Soluble cell-free extracts were prepared, and eIF4E and associated proteins were recovered on m7-GTP-sepharose. Cap-bound and input samples were analyzed by immunoblotting with the indicated antibodies. Mnk1a and Mnk1b are indicated with arrows.
eIF3e binds to eIF4G (LeFebvre et al. 2006) and could potentially exert direct effects on eIF4F formation or activity. To test this, the composition of eIF4F complexes in NHDFs transfected with no siRNA, control nonsilencing siRNA, or eIF3e siRNAs was assessed by recovering eIF4E and associated proteins from soluble cell extracts using m7-GTP chromatography. Compared with controls, eIF3e depletion did not substantially affect association of eIF4G or eIF4A with m7-GTP-bound eIF4E (Fig. 3B). Therefore, eIF3e did not detectably alter eIF4F complex levels, which might have explained reductions in eIF4E phosphorylation. Furthermore, depletion of eIF3e did not detectably affect recovery of eIF4G-associated PABP. Remarkably, neither of the Mnk1 isoforms, a or b, was detected in eIF4F complexes isolated from eIF3e-depleted cells (Fig. 3B). This suggested that eIF3e exerted a highly specific effect on eIF4F complexes to control binding of Mnk1 to eIF4G, implying a potential mechanism to explain how eIF3e controls phosphorylation of the cap-binding protein eIF4E.
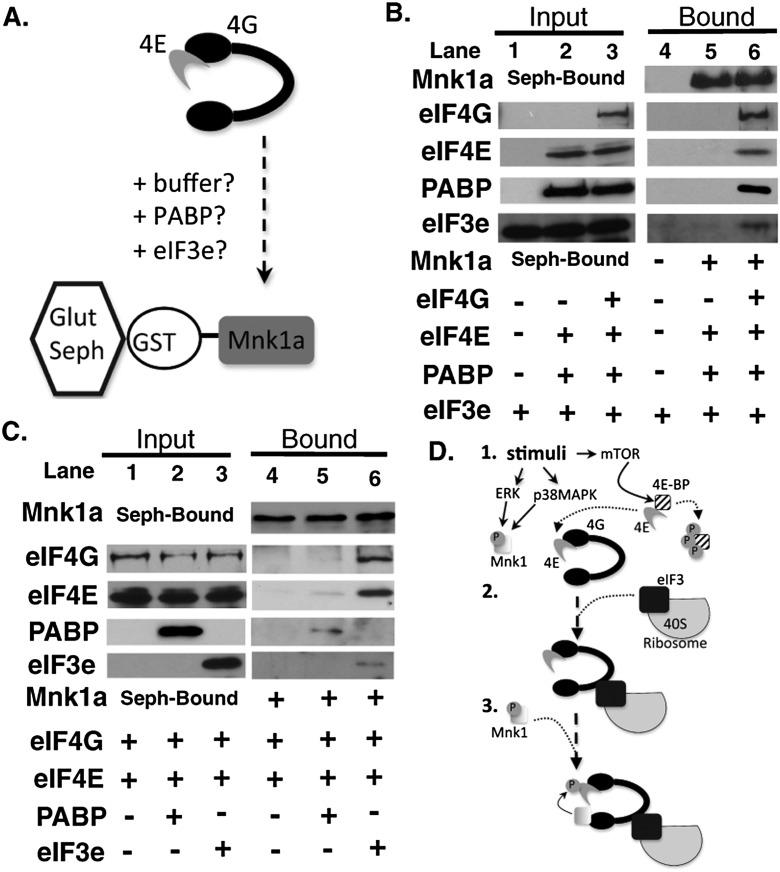
To test whether eIF3e was capable of directly modulating Mnk1 recruitment to eIF4F, the effects of purified proteins on Mnk1 binding to eIF4E–eIF4G complexes were assessed using an in vitro reconstituted system (Fig. 4A). Glutathione-conjugated beads were prebound with purified GST-tagged human Mnk1a (the longest Mnk1 isoform, containing all regulatory domains) (Buxade et al. 2008), and its capacity to retain exogenous purified eIF4G, eIF4E, PABP, and eIF3e was evaluated. Under these conditions, eIF4E, PABP, and eIF3e did not detectably bind Mnk1a, and their recovery on GST-Mnk1a-sepharose was dependent on the addition of eIF4G to reactions (Fig. 4B, cf. lanes 5 and 6). While eIF4E and eIF4G naturally associate and are recovered with GST-Mnk1a over time (Supplemental Fig. S3), suboptimal binding conditions were created by limiting both the amount of eIF4G (<1.5 μg/mL) and the reaction time such that minimal eIF4G and eIF4E were recovered bound to GST-Mnk1a (Fig. 4C, lane 4). Significantly, while retention of eIF4E and eIF4G increased at best only slightly upon addition of PABP, supplementing binding reactions with purified eIF3e dramatically stimulated binding of eIF4G and eIF4E to GST-Mnk1a (Fig. 4C, cf. lanes 4,5 and 6). This demonstrated that eIF3e was sufficient to stimulate recruitment of the kinase Mnk1a to eIF4G, which in turn is bound to the Mnk1a substrate eIF4E. Moreover, it suggests a model by which eIF3e couples 40S ribosome recruitment with phosphorylation of the cap-bound initiation factor eIF4F (Fig. 4D).
Figure 4.
eIF3e stimulates Mnk1a recruitment to eIF4G in vitro. (A) Reconstituted in vitro assay to measure Mnk1a binding to eIF4G. Retention of eIF4G:eIF4E complexes by GST-Mnk1a immobilized on glutathione sepharose (Glut Seph) was evaluated in the absence or presence of PABP and/or eIF3e. (B) eIF4E, eIF3e, and PABP were incubated for 1 h with GST-Mnk1a-bound beads ±300 ng of eIF4G. Input and bound samples were analyzed by immunoblotting with the indicated antibodies. (C) Purified eIF4G (150 ng) was incubated with the indicated proteins and GST-Mnk1a-bound beads for 35 min and then analyzed as in B. (D) Model for eIF3e-mediated control of eIF4E phosphorylation. (1) Signal activation; mTORC1 inactivates 4E-BP1, releasing eIF4E to bind eIF4G. ERK/p38MAPK are activated, but Mnk1 is not yet recruited to eIF4F. (2) An eIF3-associated ribosome engages eIF4F. (3) Interaction with eIF3e enables eIF4G to bind Mnk1 and facilitate eIF4E phosphorylation. This ensures eIF4E phosphorylation when an eIF3-bound 40S ribosome is engaged, functionally coupling eIF4E phosphorylation to 40S recruitment.
While its function in 40S recruitment has been studied, the potential for eIF3e to act in a regulatory capacity rather than solely as a tethering factor has not been explored. Here, we establish that eIF3e regulates recruitment of the eIF4E kinase Mnk1 to eIF4F to promote eIF4E phosphorylation, possibly by inducing changes in eIF4G conformation, which refolds in response to interactions with other proteins to initiate translation (Sonenberg and Hinnebusch 2009). Changes in eIF4G conformation could be triggered by either stoichiometric, stable association of eIF3e with eIF4G or a dynamic, transient association of eIF3e with eIF4G. In this latter scenario, substoichiometric eIF3e amounts could impact Mnk1 binding to eIF4G. At present, we are unable to distinguish between these two possibilities. Previous studies have reported that Mnk1 binding to eIF4G responds to a number of signal pathways (Orton et al. 2004; Parra et al. 2005; Shveygert et al. 2010; Dobrikov et al. 2011, 2013). Notably, phosphorylation of eIF4G on S1186 regulates its interaction with Mnk1 and lies near the minimal eIF3e-binding interface (residues 1015–1118) (LeFebvre et al. 2006) but far from the primary sequence determinants of Mnk1 binding (Dobrikov et al. 2011). In addition, a phylogenetic study linking eIF4G mutations to Parkinson’s disease identified a substitution at residue 1205 that disrupted eIF3e binding (Chartier-Harlin et al. 2011). Taken together, this suggests that eIF4G residues outside of the 1015–1118 minimal binding domain impact eIF3e binding. While phosphorylation has been proposed to induce eIF4G conformational changes that affect Mnk1 binding (Dobrikov et al. 2011), we demonstrated that eIF3e directly regulates Mnk1 recruitment to eIF4F both in cells and in vitro. This suggests an alternate explanation for why eIF4G phosphorylation within the eIF3-binding surface might affect Mnk1 recruitment. Indeed, phosphorylation regulates the eIF3–eIF4G interaction (Harris et al. 2006).
Recently, two distinct eIF3-binding domains in eIF4G were identified that interact with a larger eIF3 interface (Villa et al. 2013). While one eIF4G segment binds eIF3 subunits c and d, a second domain interacts with eIF3e. This potentially explains why eIF3d depletion did not detectably influence eIF4E phosphorylation and suggests that specific interaction of eIF3e with a discrete eIF4G region controls Mnk1 binding. Although eIF3c binds the same eIF4G segment as eIF3d, eIF3c recruits eIF3e into eIF3 complexes (Morris-Desbois et al. 1999; Zhou et al. 2008). This supports our finding that eIF3c depletion reduces eIF4E phosphorylation, most likely through effects on eIF3e levels or recruitment, although direct contributions by eIF3c cannot be excluded at this point.
Controlling eIF4E phosphorylation likely represents an important aspect of eIF3e’s role in regulating both global and specific mRNA translation (Asano et al. 1997; Rasmussen et al. 2001; Mayeur and Hershey 2002; Udagawa et al. 2008; Grzmil et al. 2010; Chiluiza et al. 2011; Suo et al. 2011; Neusiedler et al. 2012). Indeed, eIF4E phosphorylation exerts differential effects on mRNA translation and plays important roles in cell proliferation, transformation, immune responses, and viral infection (Walsh and Mohr 2004, 2011; Wendel et al. 2007; Furic et al. 2010; Ueda et al. 2010; Herdy et al. 2012). Notably, the gene encoding eIF3e is a frequent site of mouse mammary tumor virus integration, and the resulting truncated protein could influence eIF4E phosphorylation and possibly tumorigenesis in mice (Chiluiza et al. 2011). In addition, two distinct eIF3 complexes exist in cells defined by the presence or absence of eIF3e (Zhou et al. 2005; Sha et al. 2009). Instead of mRNAs differentially responding to eIF4E phosphorylation per se, eIF3e may dictate which eIF3-bound eIF4F complexes can recruit Mnk1 and could create distinct populations of Mnk1-responsive and Mnk1-unresponsive mRNA targets, potentially explaining why all capped mRNAs are not equally affected by eIF4E phosphorylation. Many eIF3 subunits have intrinsic RNA-binding activity (Hinnebusch 2006) that could discriminate among mRNAs and control their differential response to eIF4E phosphorylation.
A diverse array of cellular signaling pathways rapidly converges on the translation system, phosphorylating and activating a multitude of eIFs seemingly en masse (Sonenberg and Hinnebusch 2009). Although upstream mTOR, p38MAPK, and ERK signaling pathways are stimulated, these cues are not transmitted to eIF4E in eIF3e-depleted cells. Thus, additional regulation exists downstream from signal activation in which some eIFs ensure temporally correct assembly of the translation initiation machinery. In addition to tethering the 40S ribosome to eIF4F, eIF3e may also possess an intrinsic regulatory capacity to prevent premature eIF4E phosphorylation until an eIF3-bound ribosome has been engaged (Fig. 4D). This further illustrates how 40S binding recruits a kinase to impart a regulatory mark on cap-bound initiation factors. Moreover, it suggests how the deposition of coding marks in response to regulated interactions among cellular translation initiation factors might regulate selective mRNA translation (Xue and Barna 2012).
Materials and methods
Additional methods are described in the Supplemental Material.
m7-GTP chromatography and in vitro binding assays.
m7-GTP chromatography was described previously (Walsh and Mohr 2006). Proteins were purified from clarified, RNase A-treated cell extracts over NiNta (His-eIF3e or His-PABP) or glutathione (GST-Mnk1a) columns as described (Walsh and Mohr 2006). Purified Flag-tagged eIF4G was a gift of Dr. Simon Morley. For binding reactions, 10-μL packed bed volumes of glutathione sepharose 4B were washed three times in 500-μL reaction buffer (RB; 50 mM HEPES-KOH at pH 7.2, 1 mM EDTA, 20 mM NaF, 75 mM KCl, 0.66 mM Na3VO4, 25 mM β-glycerophosphate, 3 mM MgCl2, 5% glycerol [v/v], 1× complete mini-EDTA free protease inhibitor tablet [Roche]). Beads were blocked in RB+5% bovine serum albumin for 1 h at room temperature, incubated overnight at 4°C in RB containing 600 ng of purified GST-Mnk1a, and then washed three times in RB+0.125% NP40. Reactions were prepared in tubes containing 200 μL of RB+0.125% NP40. Flag-eIF4G (150 or 300 ng) was mixed with 5 μL of clarified protease inhibitor-treated and RNase A-treated bacterial extract containing human eIF4E (~50 ng/μL) (Walsh and Mohr 2006) to which 1 μg of either PABP or eIF3e was added. Where proteins were omitted from reactions, an equal volume of dialysis buffer was added. After mixing, input samples were taken, and binding reactions were initiated by adding 190 μL to washed glutathione sepharose preloaded with GST-Mnk1a and rocking at 4°C. Reactions were stopped by centrifugation, and the pelleted beads were washed four times in RB+0.125% NP40 and boiled in sample buffer.
Acknowledgments
We thank Simon Morley, Christopher Fraser, John Hershey, Robert Schneider, Christopher Proud, and Bernard Moss for reagents. This work was supported by National Institutes of Health grants R21AI105330 to D.W. and R01AI073898 and GM056927 to I.M.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.236752.113.
References
- Asano K, Merrick WC, Hershey JW 1997. The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J Biol Chem 272: 23477–23480 [DOI] [PubMed] [Google Scholar]
- Buxade M, Parra-Palau JL, Proud CG 2008. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases). Front Biosci 13: 5359–5373 [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Dachsel JC, Vilarino-Guell C, Lincoln SJ, Lepretre F, Hulihan MM, Kachergus J, Milnerwood AJ, Tapia L, Song MS, et al. 2011. Translation initiator EIF4G1 mutations in familial Parkinson disease. Am J Hum Genet 89: 398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiluiza D, Bargo S, Callahan R, Rhoads RE 2011. Expression of truncated eukaryotic initiation factor 3e (eIF3e) resulting from integration of mouse mammary tumor virus (MMTV) causes a shift from cap-dependent to cap-independent translation. J Biol Chem 286: 31288–31296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikov M, Dobrikova E, Shveygert M, Gromeier M 2011. Phosphorylation of eukaryotic translation initiation factor 4G1 (eIF4G1) by protein kinase Cα regulates eIF4G1 binding to Mnk1. Mol Cell Biol 31: 2947–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikov MI, Dobrikova EY, Gromeier M 2013. Dynamic regulation of the translation initiation helicase complex by mitogenic signal transduction to eukaryotic translation initiation factor 4G. Mol Cell Biol 33: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Berry KE, Hershey JW, Doudna JA 2007. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell 26: 811–819 [DOI] [PubMed] [Google Scholar]
- Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, et al. 2010. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci 107: 14134–14139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzmil M, Rzymski T, Milani M, Harris AL, Capper RG, Saunders NJ, Salhan A, Ragoussis J, Norbury CJ 2010. An oncogenic role of eIF3e/INT6 in human breast cancer. Oncogene 29: 4080–4089 [DOI] [PubMed] [Google Scholar]
- Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC Jr 2006. mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J 25: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdy B, Jaramillo M, Svitkin YV, Rosenfeld AB, Kobayashi M, Walsh D, Alain T, Sean P, Robichaud N, Topisirovic I, et al. 2012. Translational control of the activation of transcription factor NF-κB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat Immunol 13: 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG 2006. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 31: 553–562 [DOI] [PubMed] [Google Scholar]
- LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JW, Rhoads RE 2006. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem 281: 22917–22932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani M, Sonenberg N, Yokoyama S, Imataka H 2007. Reconstitution reveals the functional core of mammalian eIF3. EMBO J 26: 3373–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeur GL, Hershey JW 2002. Malignant transformation by the eukaryotic translation initiation factor 3 subunit p48 (eIF3e). FEBS Lett 514: 49–54 [DOI] [PubMed] [Google Scholar]
- Morris-Desbois C, Bochard V, Reynaud C, Jalinot P 1999. Interaction between the Ret finger protein and the Int-6 gene product and co-localisation into nuclear bodies. J Cell Sci 112: 3331–3342 [DOI] [PubMed] [Google Scholar]
- Neusiedler J, Mocquet V, Limousin T, Ohlmann T, Morris C, Jalinot P 2012. INT6 interacts with MIF4GD/SLIP1 and is necessary for efficient histone mRNA translation. RNA 18: 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orton KC, Ling J, Waskiewicz AJ, Cooper JA, Merrick WC, Korneeva NL, Rhoads RE, Sonenberg N, Traugh JA 2004. Phosphorylation of Mnk1 by caspase-activated Pak2/γ-PAK inhibits phosphorylation and interaction of eIF4G with Mnk. J Biol Chem 279: 38649–38657 [DOI] [PubMed] [Google Scholar]
- Parra JL, Buxade M, Proud CG 2005. Features of the catalytic domains and C termini of the MAPK signal-integrating kinases Mnk1 and Mnk2 determine their differing activities and regulatory properties. J Biol Chem 280: 37623–37633 [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J 18: 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querol-Audi J, Sun C, Vogan JM, Smith MD, Gu Y, Cate JH, Nogales E 2013. Architecture of human translation initiation factor 3. Structure 21: 920–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SB, Kordon E, Callahan R, Smith GH 2001. Evidence for the transforming activity of a truncated Int6 gene, in vitro. Oncogene 20: 5291–5301 [DOI] [PubMed] [Google Scholar]
- Scheper GC, Morrice NA, Kleijn M, Proud CG 2001. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol Cell Biol 21: 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Z, Brill LM, Cabrera R, Kleifeld O, Scheliga JS, Glickman MH, Chang EC, Wolf DA 2009. The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol Cell 36: 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shveygert M, Kaiser C, Bradrick SS, Gromeier M 2010. Regulation of eukaryotic initiation factor 4E (eIF4E) phosphorylation by mitogen-activated protein kinase occurs through modulation of Mnk1-eIF4G interaction. Mol Cell Biol 30: 5160–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Todorovic A, Querol-Audi J, Bai Y, Villa N, Snyder M, Ashchyan J, Lewis CS, Hartland A, Gradia S, et al. 2011. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Proc Natl Acad Sci 108: 20473–20478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo J, Snider SJ, Mills GB, Creighton CJ, Chen AC, Schiff R, Lloyd RE, Chang EC 2011. Int6 regulates both proteasomal degradation and translation initiation and is critical for proper formation of acini by human mammary epithelium. Oncogene 30: 724–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Nemoto N, Wilkinson CR, Narashimhan J, Jiang L, Watt S, Zook A, Jones N, Wek RC, Bahler J, et al. 2008. Int6/eIF3e promotes general translation and Atf1 abundance to modulate Sty1 MAPK-dependent stress response in fission yeast. J Biol Chem 283: 22063–22075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Sasaki M, Elia AJ, Chio II, Hamada K, Fukunaga R, Mak TW 2010. Combined deficiency for MAP kinase-interacting kinase 1 and 2 (Mnk1 and Mnk2) delays tumor development. Proc Natl Acad Sci 107: 13984–13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa N, Do A, Hershey JW, Fraser CS 2013. Human eukaryotic initiation factor 4G (eIF4G) binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem 288: 32932–32940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev 18: 660–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I 2006. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev 20: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I 2011. Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol 9: 860–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Arias C, Perez C, Halladin D, Escandon M, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Mohr I 2008. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol Cell Biol 28: 2648–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, et al. 2007. Dissecting eIF4E action in tumorigenesis. Genes Dev 21: 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Barna M 2012. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol 13: 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Arslan F, Wee S, Krishnan S, Ivanov AR, Oliva A, Leatherwood J, Wolf DA 2005. PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol 3: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW, et al. 2008. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci 105: 18139–18144 [DOI] [PMC free article] [PubMed] [Google Scholar]