miR-99a/100, let-7, and miR-125b paralogs are encoded in two tricistrons and are highly expressed in hematopoietic stem cells (HSCs). Emmrich et al. demonstrate that miR-99a/100∼125b tricistrons are transcribed as a polycistronic message and functionally converge at the combinatorial block of the TGFβ pathway. Down-regulating tumor suppressor genes APC/APC2 stabilized active β-catenin and enhanced Wnt signaling. These tricistronic miRNAs promoted sustained expansion of murine and human HSCs by switching the balance between Wnt and TGFβ signaling.
Keywords: AMKL, TGFβ, Wnt, hematopoiesis, let-7, miR-125
Abstract
Although regulation of stem cell homeostasis by microRNAs (miRNAs) is well studied, it is unclear how individual miRNAs genomically encoded within an organized polycistron can interact to induce an integrated phenotype. miR-99a/100, let-7, and miR-125b paralogs are encoded in two tricistrons on human chromosomes 11 and 21. They are highly expressed in hematopoietic stem cells (HSCs) and acute megakaryoblastic leukemia (AMKL), an aggressive form of leukemia with poor prognosis. Here, we show that miR-99a/100∼125b tricistrons are transcribed as a polycistronic message transactivated by the homeobox transcription factor HOXA10. Integrative analysis of global gene expression profiling, miRNA target prediction, and pathway architecture revealed that miR-99a/100, let-7, and miR-125b functionally converge at the combinatorial block of the transforming growth factor β (TGFβ) pathway by targeting four receptor subunits and two SMAD signaling transducers. In addition, down-regulation of tumor suppressor genes adenomatous polyposis coli (APC)/APC2 stabilizes active β-catenin and enhances Wnt signaling. By switching the balance between Wnt and TGFβ signaling, the concerted action of these tricistronic miRNAs promoted sustained expansion of murine and human HSCs in vitro or in vivo while favoring megakaryocytic differentiation. Hence, our study explains the high phylogenetic conservation of the miR-99a/100∼125b tricistrons controlling stem cell homeostasis, the deregulation of which contributes to the development of AMKL.
Self-renewal, quiescence, proliferation, and differentiation are tightly controlled during normal hematopoiesis to ensure lifelong hematopoietic stem cell (HSC) homeostasis and blood production. Deregulation of these processes results in hematologic dysplasia, deficiency, myeloproliferation, or leukemia. Therefore, a complex interplay of cytokines and signaling cascades, including their downstream transcriptional effectors, controls the balance between self-renewal and proliferation on the one hand and quiescence and differentiation on the other hand. The transforming growth factor β (TGFβ) and Wnt signaling pathways are major examples for two such opposing players.
Recently, microRNAs (miRNAs) have emerged as important regulators of hematopoiesis, which can fine-tune the activity of canonical signaling cascades. We and others have defined a central function of miR-125b in stem cell homeostasis and leukemogenesis (Klusmann et al. 2010b; O’Connell et al. 2010). In particular, we identified chromosome 21 (hsa21)-encoded miR-125b-2 to be highly up-regulated in acute megakaryoblastic leukemia (AMKL), especially in Down syndrome (DS) patients. AMKL blasts were sensitive to depletion of this miRNA (Klusmann et al. 2010b). However, miR-125b-2 is encoded within a phylogenetically highly conserved tricistron of miRNA genes on chromosome 21 that also contains miR-99a and let-7c (hsa21; miR-99a/let-7c/ miR-125b-2) (Fig. 1A). A highly similar homolog of this tricistron can be found in identical configuration on hsa11 (miR-100/let-7a-2/miR-125b-1). Another less conserved homolog resides on hsa19 (miR-99b/let-7e/miR-125a).
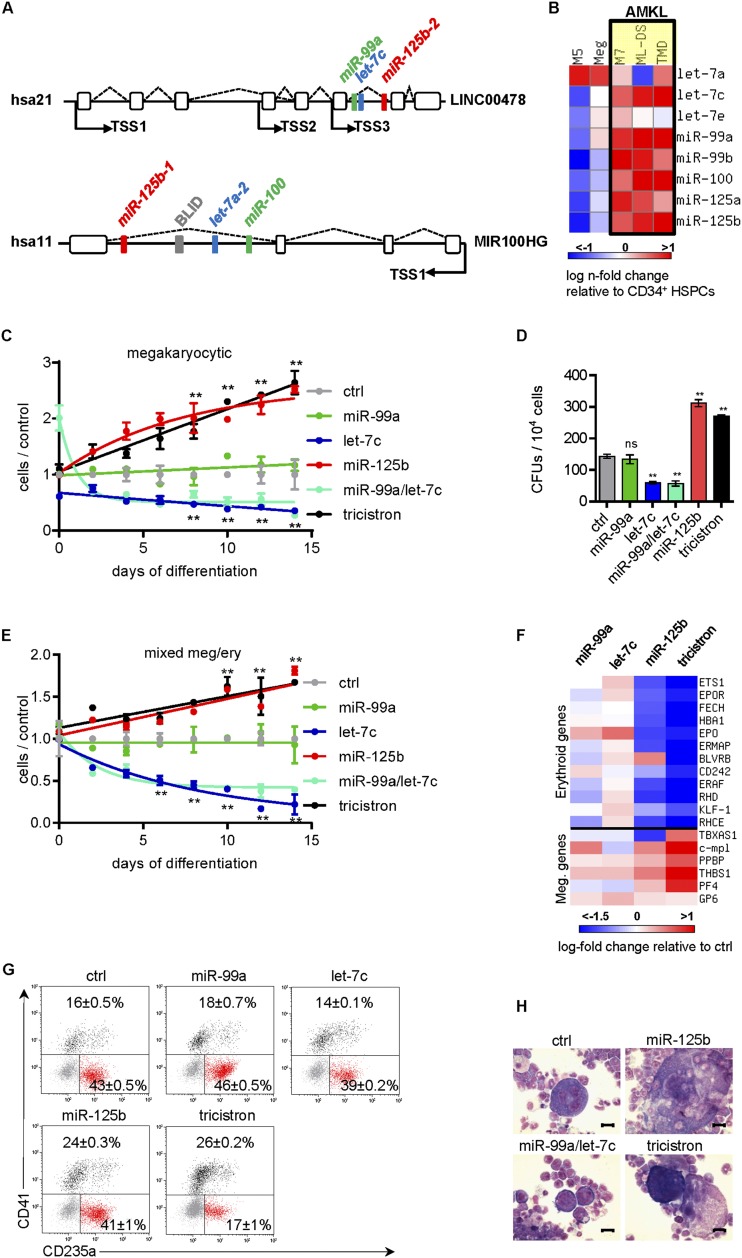
Figure 1.
miR-99/100∼125 clusters are highly expressed in HSPCs and AMKL and promote megakaryopoiesis. (A) Genomic organization of the miR-99a/let-7c/miR-125b-2 (hsa21) and miR-100/let-7a-2/miR-125b-1 (hsa11) tricistrons. LINC00478 and MIR100HG represent the lincRNA host genes of the tricistrons. Indicated TSSs were determined by 5′RACE-PCR (Supplemental Fig. S4A). (B) qRT-PCR quantification of the indicated mature miRNAs in CD34+ HSPCs (n = 2) and megakaryocytes (Meg; n = 1) from healthy donors as well as in sorted leukemic blasts from patients with DS-AMKL (ML-DS; n = 5), DS-transient leukemia (TMD; n = 4), non-DS-AMKL (M7; n = 3), and AML FAB M5 (M5; n = 2). Color bars depict log fold change over CD34+ HSPCs. (C) The number of miRNA-transduced CB CD34+ HSPCs (n = 3) during megakaryocytic in vitro differentiation relative to the control (nonsilencing miRNA [ctrl]). (D) Number of CD41+ CFU-MKs of miRNA-transduced CB CD34+ HSPCs (n = 3). (E) The number of miRNA-transduced CB CD34+ HSPCs (n = 2) during megakaryocytic/erythroid in vitro differentiation relative to the nonsilencing miRNA (ctrl). (F) Heat map of megakaryocytic and erythroid marker gene expression of miRNA-transduced CB CD34+ HSPCs, shown as log fold change over nonsilencing miRNA (ctrl). (G) Flow cytometric analysis on day 7 of megakaryocytic/erythroid in vitro differentiation of miRNA-transduced CB CD34+ HSPCs; representative dot plots from one of three independent experiments are shown. (H) Microscopic images of May-Grünwald-Giemsa (MGG)-stained cytospins of in vitro differentiated miRNA-transduced murine Lin− BM cells. Bars, 50 μm. (C–G) Data are presented as mean ± SD. (*) P < 0.05; (**) P < 0.01. (C,E) Two-way ANOVA was performed to compare the mean of each construct at each time point with control. (C–H) (tricistron) miR-99a∼125b-2; (ctrl) nonsilencing miRNA.
A significant number of miRNA genes are located closely adjacent to each other in miRNA polycistrons. The NCBI/hg18 human genome assembly features 1873 miRNA gene sequences, of which 42% are organized into polycistrons of two or more genes (Altuvia et al. 2005). Increasing evidence suggests that clustered miRNAs act together, achieving a regulatory net outcome on the cell (Chan et al. 2012). Accordingly, only the concerted action of two miRNA families within the miR-302/367 cluster enabled miRNA-mediated reprograming of somatic cells to induced pluripotent stem cells (Anokye-Danso et al. 2011). The miR-17∼92 cluster contributes to Myc-driven oncogenesis by forming a positive feedback loop (He et al. 2005). Dissecting the miR-17∼92 clusters revealed miR-19 as the crucial and dominant component of this effect (Mu et al. 2009).
Studies investigating the individual miRNAs of the miR-99/100∼125 tricistrons demonstrated conflicting functions. While let-7c was shown to stimulate granulocytic differentiation in AML cell lines and primary blasts, miR-100 inhibited it (Zheng et al. 2012; Pelosi et al. 2013). Also, in other cellular contexts, let-7 acts as a tumor suppressor, negatively regulating oncogenes such as MYC, RAS, and HMGA2 (Johnson et al. 2005; Lee and Dutta 2007). However, the high phylogenetic conservation as well as the genomic redundancy of the miR-99/100∼125 tricistrons implicate a common regulatory function of the three miRNAs.
Here, we deciphered the functional linkage of miR-99a/100, let-7, and miR-125b and demonstrate how polycistronic miRNAs cooperate to create a common phenotype. We show that the miRNAs are produced from a single primary transcript as a polycistronic message that is induced by the stem cell transcription factor HOXA10. The tricistron miRNAs form an interaction network in which the combined activity of all three miRNAs converged to block the TGFβ pathway and amplify Wnt signaling. A shift in the balance between these central pathways favors megakaryopoiesis and promotes expansion of HSCs. In AMKL, up-regulation of miR-99a/100∼125b represents a strategy to escape TGFβ-induced apoptosis and cell cycle arrest.
Results
miR-99a/100∼125b miRNAs are expressed together as polycistronic message in normal and leukemic hematopoietic cells
To test whether the miRNAs residing in the miR-99a/100∼125b tricistrons on hsa11 (miR-100, let-7a, and miR-125b-1) and hsa21 (miR-99a, let-7c, and miR-125b-2) are expressed as a polycistronic message (Fig. 1A), we quantified the expression of mature miRNAs and primary miRNAs in leukemic cell lines and primary cells. Hierarchical clustering indicated higher expression of all mature miR-99∼125 miRNAs in primary AMKL blasts and cell lines as compared with primary blasts or cell lines of other AML subtypes (Fig. 1B; Supplemental Fig. S1A). During normal hematopoiesis, miR-99a∼125b-2 (hsa21) and miR-100∼125b-1 (hsa11) miRNAs are primarily expressed together in CD34+ hematopoietic stem and progenitor cells (HSPCs), erythroid cells, and CD4+ T cells (Supplemental Fig. S1B). The expression of miR-99b∼125a miRNAs (hsa19) is higher in megakaryocytes, CD34+ HSPCs, and granulocytes as compared with the other blood lineages (Supplemental Fig. S1C). A previously published data set confirmed this expression pattern in murine cells (Petriv et al. 2010).
Both miR-99a/100∼125b tricistrons are embedded in the intron of long intervening noncoding RNA (lincRNA) host genes (hsa11, MIR100HG; hsa21, LINC00478) (Fig. 1A). Quantification of primary miRNA/lincRNA transcripts in AMKL cell line CMK showed the same expression level of the primary miRNAs of each tricistron and their lincRNA host gene (Supplemental Fig. S1D). Altogether, this suggests that miRNAs of the hsa11 and hsa21 tricistrons are produced from a single primary transcript, including the lincRNA host genes.
miR-99a∼125b-2 enhances megakaryopoiesis at the expense of erythropoiesis
The coordinated expression as a polycistronic message in HSPCs and megakaryoblasts suggests a common function of hsa11/hsa21 tricistronic miRNAs in these cells. To dissect the functional contribution of each miRNA within the hsa21 tricistron and their combination during hematopoiesis, we cloned lentiviral constructs containing single miRNAs (miR-99a, let-7c, and miR-125b-2), the miR-99a/let-7c bicistron, or a combined cassette of the whole hsa21 miR-99a∼125b-2 tricistron. In human cord blood (CB) CD34+ HSPCs or MV4:11 cells, our lentiviral constructs increased expression of these miRNAs within a physiological range (Supplemental Fig. S1E–H). During megakaryocytic in vitro differentiation, we observed a remarkable expansion of cells upon miR-125b-2 and miR-99a∼125b-2 overexpression (Fig. 1C). In contrast, let-7c or the miR-99a/let-7c bicistron had an opposite effect of impairing cell growth. miR-99a alone showed negligible effects. Similarly, megakaryocytic colony-forming unit (CFU-MK) assays indicated the expansion of CFU-MKs upon miR-125b-2 expression either alone or in the context of the hsa21 miR-99a∼125b-2 tricistron (Fig. 1D; Supplemental Fig. S1I).
In medium promoting megakaryocytic and erythroid differentiation, CD34+ HSPCs expressing miR-125b-2 or miR-99a∼125b-2 displayed a higher growth rate with increased differentiation into CD41+ megakaryocytes (Fig. 1E–G). miR-99a∼125b-2 decreased hemoglobinization and differentiation into CD235a+ erythroid cells (Fig. 1F–G). The block of erythroid development by the tricistron was even more aggravated in human fetal liver (FL) HSPCs (Supplemental Fig. S1J). This perturbed differentiation was preceded by increased megakaryocytic and decreased erythroid gene expression in CD34+ HSPCs after transduction (Fig. 1F). Moreover, when transduced murine lineage-depleted (Lin−) bone marrow (BM) cells (enriched for HSPCs) were cultured in stem cell expansion medium containing SCF, TPO, FGF, and IGF2, we observed spontaneous differentiation of miR-125b-2 or miR-99a∼125b tricistron-transduced cells into CD41+ megakaryocytic cells (Fig. 1H; Supplemental Fig. S1K).
Taken together, these data confirm previous observations that miR-125b-2 is a positive regulator of megakaryopoiesis (Klusmann et al. 2010b; Emmrich et al. 2012). Moreover, we show for the first time that when embedded in its tricistronic context, miR-125b-driven megakaryocytic differentiation is further intensified by a block of erythroid differentiation.
The miR-99a∼125b-2 phenotype is a cooperative effect of all three miRNAs
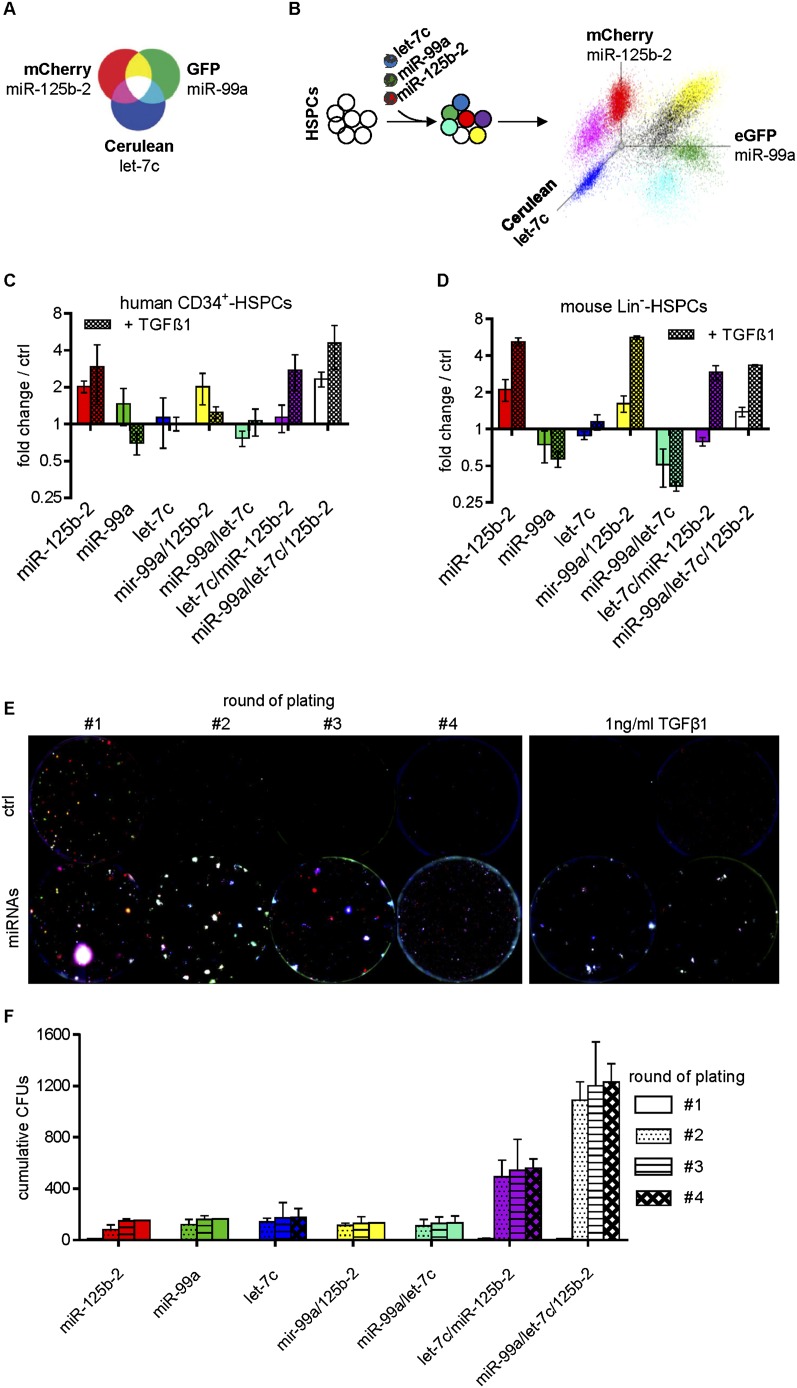
To test the genetic interaction of the miRNAs during megakaryopoiesis in an unbiased fashion, we applied red–green–blue (RGB) marking (Weber et al. 2011). By mixing red, green, and blue, four mixed colors can be obtained, each representing a distinct color combination. We assigned each miRNA to one color: miR-125b-2 to red, let-7c to blue, and miR-99a to green (Fig. 2A). Lentiviral constructs were pooled and transduced into human CD34+ HSPCs or murine FL Lin− HSPCs. Flow cytometric analysis demonstrated seven distinguishable populations resembling all possible miRNA combinations in equal starting amounts (Fig. 2B; Supplemental Fig. S2A). Monitoring the growth of the seven populations during megakaryocytic in vitro differentiation revealed that miR-125b-2, miR-99a/miR-125b-2, and miR-99a/let-7c/miR-125b-2 provided a growth advantage in both the human and murine cells (Fig. 2C,D). Correspondingly, in colony-forming assays, murine Lin− HSPCs containing miR-125b-2 developed a greater number of and larger-sized colonies (Fig. 2E). Strikingly, with each round of replating, white colonies overexpressing all three miRNAs became increasingly dominant (Fig. 2E,F).
Figure 2.
miR-99a/100∼125b miRNAs have a cooperative phenotype. (A) Schematic diagram showing the mixing spectra of RGB marking and the color assignments of miR-99a, let-7c, and miR-125b-2. Adapted with permission from Macmillan Publishers Ltd.: Nature Medicine (Weber et al. 2011), © 2011. (B) Experimental outline of RGB marking to quantify genetic interaction. (C,D) Diagrams showing the number of miRNA-transduced human (n = 3) (C) and murine (n = 4) (D) cells after megakaryocytic in vitro differentiation (day 8) with and without 1 ng/mL TGFβ1 in relation to the starting point (day 0) and the empty vector-transduced control cells (ctrl), measured by flow cytometry. (E, left panel) Merged microscopic fluorescence images of serial replatings of mouse FL cells. The right panel shows microscopic fluorescence images of the first replating of mouse FL cells treated with 1 ng/mL TGFβ1. Each fluorescent color was photo-acquired on day 11 separately using a BZ 9000 (Keyence) for subsequent image processing and merging with Biorevo software (Keyence). Colors represent distinct miRNA combinations: miR-125b-2 (red), miR-99a (green), let-7c (blue), miR-99a/miR-125b-2 (yellow), miR-99a/let-7c (turquoise), let-7c/miR-125b-2 (purple), and miR-99a/let-7c/miR-125b-2 (white). Representative images from n = 4 independent experiments are shown. (F) Cumulative number of colonies per round of plating. (C–F) Data are presented as mean ± SD.
Thus, RGB marking demonstrated that miR-125b-2 was mainly responsible for the growth advantage. However, only the combination of all three miRNAs could sustain this proliferative advantage over time.
The miR-99a∼125b-2 tricistron expands HSCs and HSPCs in vivo
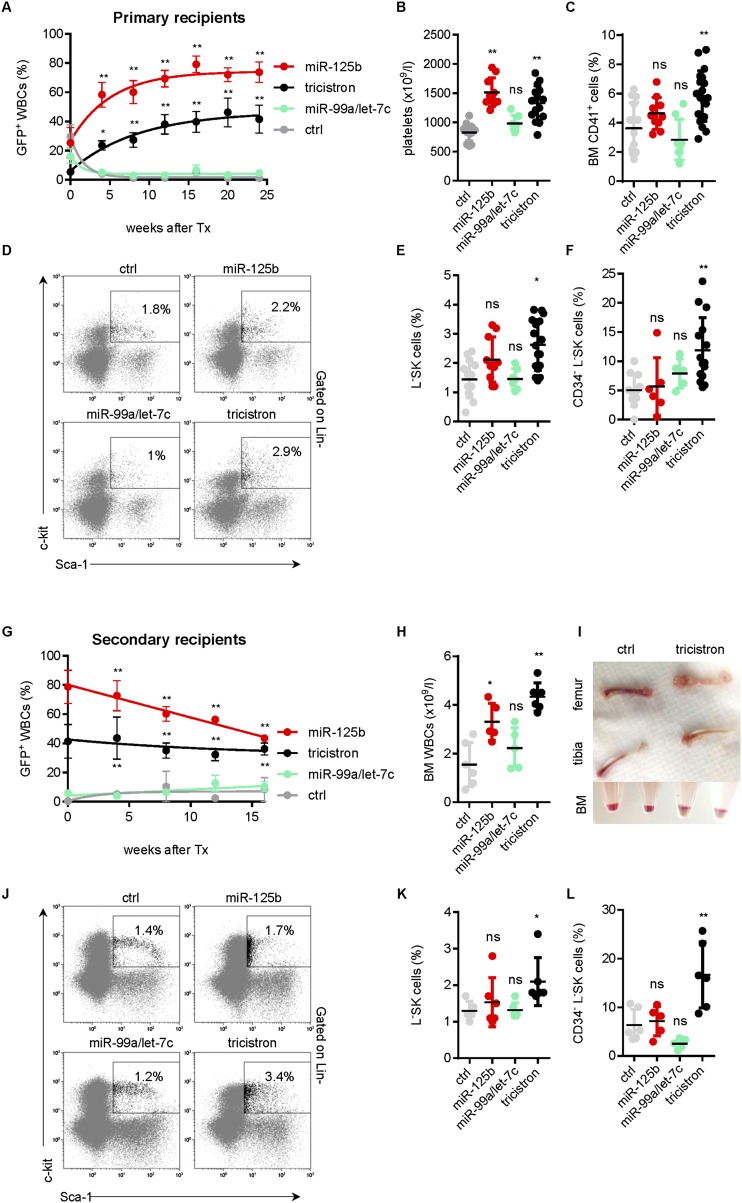
Next, we investigated the effect of the miRNA tricistron in competitive repopulation assays in vivo. We transplanted lentivirally transduced Lin− BM cells into lethally irradiated syngenic recipients. miR-125b-2 and miR-99a∼125b-2 overexpression conferred a competitive advantage to the cells, leading to a gradual increase of chimerism in the mononuclear cell fraction of the peripheral blood (PB). In contrast, nonsilencing shRNA control in the miR-30 context (ctrl) and the miR-99a/let-7c bicistron conferred a competitive disadvantage (Fig. 3A). Total platelet (Fig. 3B) and white blood cell (WBC) (data not shown) counts were significantly increased by miR-125b-2 and miR-99a∼125b-2. The fraction of GFP+ cells and the overall BM cellularity were similarly elevated (Supplemental Fig. S3A,B). Of note, we detected a significant expansion of the LSK cells and long-term (LT) HSCs (CD34− LSKs) as well as CD41+ megakaryocytic progenitors (MPs) exclusively in the marrow of miR-99a∼125b-2-transplanted mice (Fig. 3C–F). As previously described, we also observed a lineage skewing toward the myeloid lineage by miR-125b-2 (Bousquet et al. 2010; Guo et al. 2010; O’Connell et al. 2010) and miR-99a∼125b-2 (data not shown) in the BM.
Figure 3.
The miR-99a∼125b-2 cluster expands immature hematopoietic progenitors in vivo. (A) Percentage of GFP+ cells in the PB of recipient mice reconstituted with transduced Lin− HSPCs from syngenic donors at the indicated time points (ctrl, n = 14; miR-125b-2, n = 10; miR-99a/let-7c, n = 7; tricistron, n = 17). (B) PB platelets of the same mice as in A measured on week 24 after transplantation. (C) CD41+-expressing cells. (D) Representative flow cytometry plots. (E) Percentage of viable GFP+ LSK cells. (F) CD34− of LSK cells in the BM of the same mice as in A measured on week 24 after transplantation. (G) Percentage of GFP+ cells in the PB of secondary recipient mice reconstituted with transgenic whole BM of primary recipients from A (ctrl, n = 6; miR-125b-2, n = 5; miR-99a/let-7c, n = 5; tricistron, n = 6). (H) WBC counts. (I) Macroscopic images of bones (top panels) and of pellets from flushed BM samples before red blood cell lysis (bottom panel). (J) Representative LSK flow cytometry plots. (K) Percentage of viable GFP+ LSKs. (L) CD34− of LSK cells in the BM of the same mice as in E on week 16 after transplantation. (A,G) Data are presented as mean ± SEM. (B–F,H–L) Values for each mouse are shown. The black horizontal line indicates mean. (A–K) (*) P < 0.05; (**) P < 0.01. (A–L) (tricistron) miR-99a∼125b-2; (ctrl) nonsilencing miRNA.
Upon secondary transplantation of whole BM from primary recipients, miR-99a∼125b-2-reconstituted mice showed little change in their chimerism over time. In contrast miR-125b-2-transduced GFP+ cells were outcompeted (Fig. 3G). WBC, LSK, and LT-HSC expansion remained only in miR-99a∼125b-2 recipients, as evidenced by the paler femur and tibia and flow cytometric analyses (Fig. 3H–L). Thus, the competitive advantage conferred by miR-125b-2 is sustained in the tricistronic context of miR-99a∼125b-2.
HOXA10 binds and transactivates hsa21 and hsa11 miR-99a/100∼125b tricistrons
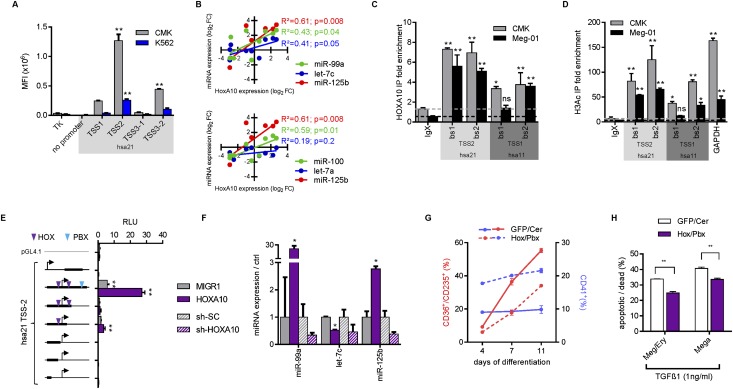
Quantification of primary miRNA/lincRNA transcripts suggested that miRNAs of hsa11 and hsa21 tricistrons are produced from a single primary transcript, including the lincRNA host genes (Supplemental Fig. S1D). Three common transcription start sites (TSSs) for the miR-99a∼125b-2 miRNAs were previously predicted (Fig. 1A; Marson et al. 2008). Similarly, the miR-100∼125b-1 tricistron harbors one common TSS for the host gene and all miRNAs. All predicted sites could be confirmed by 5′RACE-PCR (Supplemental Fig. S4A). In lentiviral reporter assays, the promoter adjacent to TSS2 among the hsa21 TSSs displayed the highest activity in CMK cells with high endogenous miRNA expression (Fig. 4A). This suggests that expression of the miR-99a∼125b-2 tricistron is mainly controlled by this promoter region.
Figure 4.
miR-99a/100∼125b clusters are transactivated by HOXA10. (A) Reporter assays in CMK and K562 cells. The empty TK-GFP and a vector without the TK promoter served as negative controls. The locations of TSS1, TSS2, and TSS3 are shown in Figure 1A. For each TSS, candidate promoters were amplified from genomic regions adjacent to the TSS (for TSS3, two candidate promoter fragments, TSS3-1 and TSS3-2, were amplified and tested). Mean fluorescence intensities (MFIs) were determined by flow cytometry for GFP (n = 2). (B) Correlation plots and statistics of HOXA10 and miRNA expression in NB4, NOMO-1, THP-1, Kasumi-1, Jurkat, K562, M-07e, Meg-01, CMK, and CMY cells measured by qRT–PCR. (C,D) ChIP analysis in CMK and Meg-01 cells for HOXA10 enrichment (C) or acetylated Histone H3 (H3Ac) (D) at predicted binding sites (bs) in experimentally validated hsa21 TSS2 (miR-99a∼125b-2) and hsa11 TSS1 (miR-100∼125b-1). Fold enrichment over background (IgG control) is shown (n = 2). (E) Luciferase reporter assays in 293T cells. The empty pGL4.1 and a 3000-bp fragment +1500 bp downstream from hsa21 TSS2 served as negative controls (n = 2). Relative luminescence units (RLUs) were determined by normalizing firefly to renilla luciferase activity. HOX-binding sites (purple) and PBX-binding sites (blue) are indicated. (Left) The bold black bar in the schemes indicates the size and location of the cloned promoter fragment in relation to the TSS (arrow). (F) qRT–PCR expression analysis of miR-99a∼125b-2 miRNAs in HOXA10-transduced MV4:11 cells or in sh-HOXA10-transduced CMK cells relative to empty vector-transduced (MIGR1) or control vector-transduced (sh-SC) cells. (G) Flow cytometric analysis of megakaryocytic in vitro differentiation of HOXA10/PBX1-transduced CB CD34+ HSPCs (n = 1). (H) Annexin V/7-AAD staining of HOXA10/PBX1-transduced CB CD34+ HSPCs on day 7 of mixed megakaryocytic/erythroid differentiation treated with 1 ng/mL TGFβ1. (A–H) Data are presented as mean ± SD. (*) P < 0.05; (**) P < 0.01.
Identification of the active TSSs for miR-99a/100∼125b tricistrons allowed mapping of transcription factor-binding sites (−5000 base pairs [bp] upstream of to +1000 bp downstream from hsa11 TSS1 and hsa21 TSS2). Mulan software predicted HOX-binding sites for both TSSs, phylogenetically conserved across seven species (Ovcharenko et al. 2005; data not shown). HoxA10 was previously shown to expand primitive HSCs and MPs, thereby acting as a leukemogenic transcription factor (Thorsteinsdottir et al. 1997; Bjornsson et al. 2001; Magnusson et al. 2007). We observed a significant positive correlation between the expression of HOXA10 and the mature miRNAs for each miR-99a/100∼125b miRNA except let-7a-2, which has two more isomers outside of the tricistrons (Fig. 4B). Particularly in AMKL blasts, HOXA10 is highly expressed (data not shown). Chromatin immunoprecipitation (ChIP) in two AMKL cell lines (CMK and Meg-01) with high miRNA and HOXA10 levels confirmed chromatin occupancy of HOXA10 at both promoter regions (Fig. 4C). The promoter regions occupied by HOXA10 were enriched for acetylated Histone H3, a typical epigenetic mark of transcriptional active promoters (Fig. 4D). Concordantly, ectopic expression of HOXA10 increased the activity of a hsa21 promoter fragment containing two HOX sites (−18 bp and +192 bp) and a more distal PBX site (+866 bp) in 293T cells (Fig. 4E). Fragment deletion analysis underscored the necessity of the PBX site and the HOX site +192 bp downstream from the TSS (Fig. 4E).
Ectopic HOXA10 expression in MV4:11 cells up-regulated the levels of mature miR-99a, miR-100, and miR-125b but not the let-7 members (Fig. 4F; Supplemental Fig. S4B). let-7 has several homologs, and its expression underlies post-transcriptional regulation, which might explain this finding (Hagan et al. 2009; Chen et al. 2011). Conversely, shRNA knockdown of HOXA10 in CMK cells led to decreased miR-99a, let-7c, and miR-125b expression (Fig. 4F; Supplemental Fig. S4C). As previously reported, ectopic expression of HOXA10 and its cofactor, PBX1, promoted the expansion of CD41+ MPs while inhibiting erythroid development (Fig. 4G; Magnusson et al. 2007). These data provide an explanation for the physiologic expression of miR-99a/100∼125b miRNAs in HSPCs and aberrant expression in malignant megakaryoblasts.
miR-99a/100∼125b target TGFβ pathway activators and Wnt repressors
To identify the downstream effectors targeted by the miR-99a/100∼125b tricistrons, we performed global gene expression profiling of human CD34+ HSPCs transduced with single miRNAs or the miR-99a∼125b-2 tricistron. Target transcripts (predicted by at least three independent prediction programs in miRecords) (Xiao et al. 2009) of each miRNA were strongly repressed by the miR-99a∼125b-2 tricistron (Fig. 5A; Supplemental Table S1). We hypothesized that the combinatorial action of the three miRNAs does not converge on individual targets but rather on gene expression programs or signaling cascades. Therefore, we analyzed by integrated pathway architecture which signaling pathways are enriched for down-regulated targets of miR-99a∼125b-2 (Supplemental Table S2). Positive effectors of the TGFβ pathway were targeted on all layers of the signaling cascade: receptors (TGFβ receptor I [ALK5], activin A receptor ACVR1C [ALK7], and BMP receptors BMPR1A [ALK3] and BMPR2), transmitters, and/or transcription factors (SMAD2 and SMAD4) (Fig. 5B; for quantitative RT–PCR [qRT–PCR] validation, see Supplemental Fig. S5A). To promote their effects, TGFβ superfamily ligands (TGFβ, Activin, Nodal, and BMP) bind to type I (e.g., TGFβR1) and type II (e.g., TGFβR2) receptors, which together form an activated complex and phosphorylate regulatory SMADs (R-SMADs: SMAD1–3, SMAD5, and SMAD8). R-SMADs complex with common mediator SMAD (co-SMAD: SMAD4) and several lineage-specific cofactors to induce transcription in a cell context-dependent manner (Fig. 5B; Moustakas and Heldin 2009). TGFβ, Activins, and Nodals signal through SMAD2/3, and BMP signals through SMAD1/5/8. In the hematopoietic system, TGFβ1 ensures stem cell homeostasis by keeping HSCs in quiescence and blocking their transition into faster-cycling short-term (ST) HSCs (Pietras et al. 2011). TGFβ1 stimulates differentiation of late erythroid and myeloid progenitors but blocks megakaryopoiesis (Jacobsen et al. 1991; Fortunel et al. 1998). This constitutes a negative feedback loop, as megakaryoblasts and megakaryocytes secrete active TGFβ1, while platelets’ α granules are the major source of TGFβ1 (Fava et al. 1990; Fortunel et al. 2000). Thus, we reasoned that the observed expansion of HSPCs, stimulation of megakaryopoiesis, and blocking of erythropoiesis by miR-99a∼125b-2 could be mediated by inhibition of TGFβ1 signaling. Deregulation of miR-99∼125 tricistrons could provide AMKL blasts a mechanism to evade the growth inhibitory function of TGFβ.
Figure 5.
miR-99/100∼125 miRNAs target the TGFβ and Wnt pathways. (A) Hierarchical clustering of miR-99/100 targets (green box), let-7 targets (blue box), and miR-125 targets (red box) from miRNA-transduced human CB CD34+ HSPCs on day 7 of megakaryocytic differentiation (n = 2). The normalized enrichment score (NES) of each target gene set compared with the control calculated by GSEA is indicated. (*) P < 0.05; (**) P < 0.01. (B) Schematic diagram of the TGFβ and Wnt pathways. miR-99∼125 putative target genes are framed in red. (C) Western blots of the indicated proteins in miRNA-transduced MV4:11 or THP-1 cells and SP-transduced Meg-01 cells. (D) Luciferase reporter assays in 293T cells (n = 3) with 3′ UTR fragments harboring color-indicated wild-type miRNA-binding sites in relation to the nonsilencing miRNA (ctrl). The presence or absence of binding sites corresponded with the regulatory effect. (Green) miR-99/100; (blue) let-7; (red) miR-125. The black line indicates the full-length 3′ UTR. (Left) The bold black bar in the schemes indicates the size and location of the cloned UTR fragment. (D) Data are presented as mean ± SD. (A,C,D) (tricistron) miR-99a∼125b-2; (ctrl) nonsilencing miRNA.
Gene set enrichment analysis (GSEA) confirmed the inverse correlation of miR-99∼125 with TGFβ pathway genes (Supplemental Fig. S5B). Down-regulation of the predicted targets was validated at the protein level in transduced MV4:11 and THP-1 cells (Fig. 5C). Moreover, stable inhibition of all three tricistron miRNAs alone (SP-99, SP-let7, and SP-125) or the tricistron (SP-TRI) using lentiviral sponge (SP) constructs (Emmrich et al. 2013) in Meg-01 cells up-regulated the target proteins (Fig. 5C; Supplemental Fig. S5C,D). Direct miRNA regulation of putative 3′ untranslated region (UTR) target sites was validated by luciferase reporter assays (Fig. 5D). Of note, luciferase reporter assays and Western blot analyses confirmed that targets of only one miRNA showed at least equal regulation by the tricistron compared with one miRNA alone. Conversely, targets with binding sites for two miRNAs, such as ACVR1C or SMAD2, were targeted more efficiently by the tricistron. Thus, miR-99a/100, let-7, and miR-125b converge in repressing targets of the TGFβ pathway.
In contrast to TGFβ signaling, canonical Wnt signaling induces self-renewal and proliferation of HSCs in a dose-dependent manner (Luis et al. 2011). Bioinformatics predicted targeting of the core component of the destruction complex adenomatous polyposis coli (APC) and its homolog, APC2, by miR-99a∼125b-2 miRNAs. When Wnt signaling is inactive, β-catenin is bound and phosphorylated by the destruction complex composed of AXIN1, GSK3β, and APC, leading to its degradation. Inhibition of the destruction complex upon binding of Wnt ligands to its membrane-bound receptor results in the accumulation and nuclear translocation of β-catenin, where it forms a complex with TCF/LEF to induce transcription (Logan and Nusse 2004). In tricistron-transduced cells, we detected decreased protein levels of APC and APC2 (Fig. 5C). APC2 is targeted by all three miRNAs. APC is targeted by let-7 and miR-125. Consistent with this, reporter assays for APC demonstrated binding of let-7c and miR-125b-2 alone or with stronger effect in the context of the tricistron (Fig. 5D).
Wnt pathway modulation correlates with the miR-99a∼125b-2 megakaryocytic phenotypes
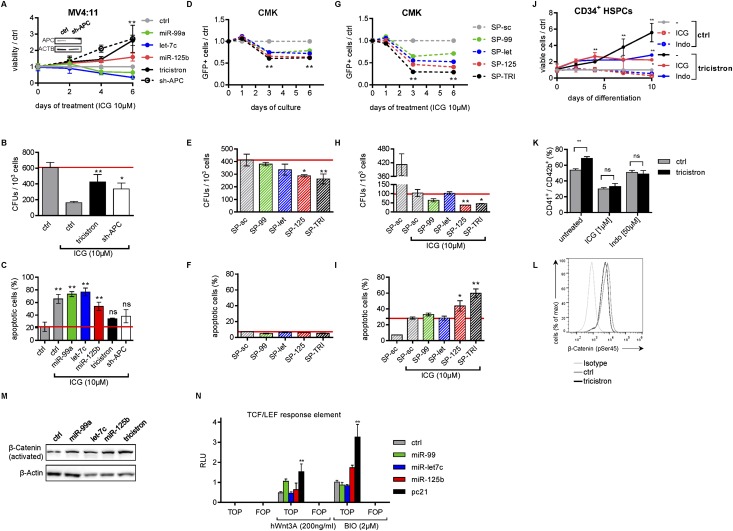
We sought to examine whether Wnt activation is a central function of miR-99a∼125b-2, conferring a growth advantage to normal HSPCs and malignant AML blasts. Wnt activation is a common feature of several forms of cancer, including leukemia, and targeting Wnt signaling was shown to be an attractive treatment strategy in AML (Klaus and Birchmeier 2008; Wang et al. 2010). We first tested whether genetically enhancing Wnt signaling by miR-99a∼125b-2 could ameliorate pharmacologic Wnt inhibition. Treatment of MV4:11 cells with Wnt inhibitor ICG-001, which disrupts the β-catenin/CBP interaction (Emami et al. 2004), or the reversible COX inhibitor indomethacin (Indo), which suppresses β-catenin expression (Hawcroft et al. 2002; Goessling et al. 2009), reduced proliferation and colony-forming capacity while inducing apoptosis (Fig. 6A–C; Supplemental Fig. S6A–C). These effects were rescued by enforced expression of miR-99a∼125b-2 or shRNA-mediated knockdown of the miR-125/let-7 common target APC (Fig. 6A–C; Supplemental Fig. S6A–C). Inversely, we asked whether pharmacologic Wnt inhibition would cooperate with miR-99∼125 inhibition in reducing leukemic growth in AMKL. Stable inhibition of miR-125 or miR-99∼125 in CMK cells using SP constructs reduced proliferation and colony formation but did not induce apoptosis (Fig. 6D–F). K562 cells with low miR-99a/100∼125b expression (Supplemental Fig. S7B) were not affected by the sponge constructs (Supplemental Fig. S6D–F). Treatment of nonsilencing sponge (SP-sc) control-transduced CMK and Meg-01 cells with ICG-001 reduced their proliferation and colony-forming capacity while inducing apoptosis. In all assays, miR-99∼125 knockdown by SP-TRI synergized with ICG-001 and aggravated its effects (Fig. 6G–I; Supplemental Fig. S6G–I).
Figure 6.
miR-99a/100∼125b miRNAs augment Wnt signaling. (A) Viability of miRNA- or shRNA-transduced MV4:11 cells treated with 10 μM ICG-001, relative to nonsilencing miRNA (ctrl; n = 2). (Inset) shRNA knockdown validation by Western blotting. (B) Methylcellulose colony assay of transduced MV4:11 cells (n = 2). (C) Annexin V/7-AAD staining after 4 d of ICG-001 treatment (n = 2). (D) Fraction of SP-transduced GFP+ cells competing with untransduced cells relative to the control (SP-sc). (E) Methylcellulose colony-forming assays of SP-transduced CMK cells. (F) Annexin V/7-AAD staining of SP-transduced CMK cells 5 d after selection. (G) Fraction of SP-transduced GFP+ cells competing with untransduced cells relative to the control (SP-sc) upon ICG-001 treatment. (H) Methylcellulose colony-forming assays of ICG-001-treated or untreated SP-transduced CMK cells. (I) Annexin V/7-AAD staining of SP-transduced CMK cells after 5 d of treatment with 10 μM ICG-001 (n = 3). (J) Cell number in relation to nonsilencing miRNA (ctrl). (K) Percentage of CD41+/ CD42b+ megakaryocytes of tricistron-transduced CB CD34+ HSPCs treated with either 1 μM ICG or 50 μM Indo (n = 2). (L) Flow cytometric analysis of Ser45 phosphorylation of β-catenin during combined megakaryocytic/erythroid in vitro differentiation of miRNA-transduced CB CD34+ HSPCs 1 h after stimulation with 200 ng/mL Wnt3a. Isotype-stained nonsilencing miRNA (ctrl)-transduced cells are shown. (M) Western blots of activated (unphosphorylated) β-catenin in miRNA-transduced MV4:11 cells. (N) Luciferase reporter assay using a TCF/LEF response element (TOPFlash) and a mutated version (FOPFlash) in untreated, Wnt3A-treated, and BIO-treated 293T cells, respectively, cotransfected with the indicated miRNAs (n = 4). RLUs are presented normalized to Renilla luciferase intensities. (A–C,J–N) (tricistron) miR-99a∼125b-2; (ctrl) nonsilencing miRNA. (A–N) Data are presented as mean ± SD. (*) P < 0.05; (**) P < 0.01; (ns) not significant.
Next, we aimed to modulate Wnt activity to recapitulate or mitigate miR-99a∼125b-2-mediated expansion of MPs. Wnt3a, one of 19 distinct Wnt ligands, has been reported to induce Wnt signaling in the hematopoietic system (Luis et al. 2012). Similar to miR-99a∼125b-2, supplementation with Wnt3a during megakaryocytic differentiation increased the total number of cells (Supplemental Fig. S6J). These observations are in accordance with a previous report (Macaulay et al. 2013). Inversely, Wnt inhibition using ICG-001 or Indo reduced the stimulatory effects of miR-99a∼125b-2 on in vitro megakaryopoiesis (Fig. 6J–K).
To study Wnt activation by miR-99a∼125b-2 on the molecular level, we quantified phosphorylated β-catenin in transduced CD34+ HSPCs. We found that degradation-primed phosphorylated β-catenin (Ser45) was decreased upon ectopic miR-99a∼125b-2 expression (Fig. 6L). In contrast, levels of β-catenin in its active (unphosphorylated) form were elevated in tricistron-transduced cells, as confirmed by Western blotting (Fig. 6M; Supplemental Fig. S6K). Up-regulation of Wnt downstream targets in miR-99a∼125b-2-transduced MV4:11 cells, such as cyclins, BTRC, BCL-9, and LEF-1, was confirmed by qRT–PCR (Supplemental Fig. S6L). Moreover, in 293T cells, miR-99a∼125b-2 augmented Wnt activation by GSK3β inhibitor BIO and Wnt3A, as evidenced by the increased luciferase activity of a TCF4 consensus binding site element (TOPFlash) but not of a mutated version (FOPFlash) (Fig. 6N).
In conclusion, miR-99/100∼125b tricistrons activated Wnt signaling by inhibiting APC/APC2, stabilizing cytoplasmic β-catenin, and increasing activated β-catenin. Pharmacologic or genetic Wnt activation could partially mimic, whereas Wnt inhibition could mitigate, miR-99a∼125b-2-mediated expansion of MPs. Accordingly, miR-99∼125 knockdown and Wnt inhibition synergized to induce growth arrest and apoptosis in leukemic cell lines.
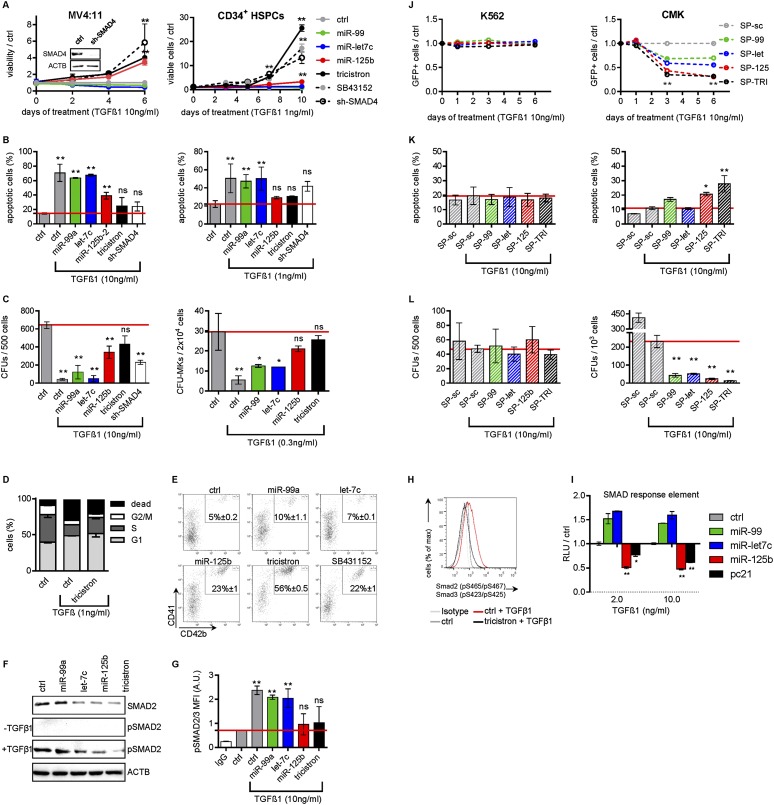
miR-99a∼125b-2 rescues TGFβ-mediated apoptosis, cell cycle arrest, and blocking of megakaryopoiesis
Our data suggest that up-regulation of miR-99a∼125b-2 might represent a strategy of leukemic blasts to escape TGFβ1-induced apoptosis and cell cycle arrest, as it has been demonstrated for other oncogenes (Massague 2008). Ectopic miR-125b-2 and miR-99a∼125b-2 expression or shRNA-mediated knockdown of miR-125/let-7 common target SMAD4 in TGFβ1-sensitive MV4:11 and THP-1 cells with low endogenous miR-99/100∼125b expression (Supplemental Fig. S7A,B) conferred resistance against the growth-suppressing and proapoptotic effects of TGFβ1 (Fig. 7A,B; Supplemental Fig. S7C). In methylcellulose CFU assays, reduction of colony formation upon TGFβ1 treatment was almost neutralized by miR-99a∼125b-2 (Fig. 7C; Supplemental Fig. S7D).
Figure 7.
miR-99a/100∼125b miRNAs antagonize TGFβ1 function. (A) The number of miRNA-transduced or sh-SMAD4-transduced MV4:11 cells (left) or human CB CD34+ HSPCs (right) during megakaryocytic in vitro differentiation, treated with the indicated concentrations of TGFβ1, normalized to nonsilencing miRNA (ctrl)-transduced cells (n = 3). (Inset) shRNA knockdown validation by Western blotting. Nonsilencing miRNA (ctrl)-transduced CD34+ HSPCs treated with 10 μM TGFβ inhibitor SB43152 are included. (B) Annexin V/7-AAD staining of miRNA-transduced or sh-SMAD4-transduced MV4:11 cells on day 4 (left) or miRNA-transduced or sh-SMAD4-transduced CB CD34+ HSPCs (right) on day 7 of combined megakaryocytic/erythroid differentiation, treated with the indicated concentrations of TGFβ1 in comparison with untreated controls (ctrl; n = 3). (C) Methylcellulose colony-forming assays of miRNA-transduced or sh-SMAD4-transduced MV4:11 cells (left) or the number of CD41+ CFU-MKs of miRNA-transduced CB CD34+ HSPCs (right) treated with the indicated concentrations of TGFβ1 in comparison with untreated nonsilencing miRNA (ctrl)-transduced cells (n = 3). (D) BrdU incorporation of miRNA-transduced CB CD34+ HSPCs on day 5 of combined megakaryocytic/erythroid differentiation (n = 3) in comparison with nonsilencing miRNA (ctrl). (E) Dot plots of flow cytometric analysis of miRNA-transduced CB CD34+ HSPCs on day 11 of combined megakaryocytic/erythroid in vitro differentiation treated with 1 ng/mL TGFβ1 for the last 72 h (n = 2). (F) Western blots of the indicated proteins in miRNA-transduced MV4:11 cells with our without 10 ng/mL TGFβ1. (G) Phosflow analysis of Smad2/3 phosphorylation in miRNA-transduced MV4:11 cells stimulated with TGFβ1, shown as MFI. (H) Phosflow analysis of Smad2/3 phosphorylation during megakaryocytic/erythroid in vitro differentiation of miRNA-transduced CB CD34+ HSPCs. Cells were stimulated for 1 h with TGFβ1. Isotype-stained and untreated nonsilencing miRNA (ctrl)-transduced cells are shown as controls. (I) Luciferase reporter assay using a SMAD response element (PAI-I promoter) in TGFβ1-treated 293T cells cotransfected with the indicated miRNAs (n = 3). RLUs are presented in relation to the nonsilencing miRNA (ctrl). (J) Fraction of SP-transduced GFP+ K562 (left) and CMK (right) cells competing with untransduced cells relative to the control (SP-sc) treated with 10 ng/mL TGFβ1 (n = 3). (K) Annexin V/7-AAD staining of SP-transduced K562 (left) and CMK (right) cells after 5 d of treatment with 10 ng/mL TGFβ1 (n = 3). (L) Methylcellulose colony-forming assays of SP-transduced K562 (left) and CMK (right) cells treated with 10 ng/mL TGFβ1 (n = 3). (A–D,F–I) (tricistron) miR-99a∼125b-2; (ctrl) nonsilencing miRNA. (A–L) Data are presented as mean ± SD. (*) P < 0.05; (**) P < 0.01.
Next, we analyzed the effects of miR-99a∼125b-2 on TGFβ signaling in primary CD34+ HSPCs. TGFβ1 is released by platelets (Fava et al. 1990; Fortunel et al. 2000) and blocks megakaryopoiesis while stimulating differentiation of late erythroid and myeloid progenitors (Jacobsen et al. 1991; Fortunel et al. 2000). As expected, TGFβ1 treatment strongly reduced the number of viable cells during megakaryocytic in vitro differentiation. However, miR-99a∼125b-2-transduced or sh-SMAD4-transduced HSPCs expanded 25-fold and 13-fold, respectively, as compared with controls (Fig. 7A). In comparison, cells treated with specific TGFβ receptor kinase inhibitor SB43152 (ALK4/5/7 inhibitor) expanded 17-fold. The cytoprotectivity of miR-125b-2 and miR-99a∼125b-2 was associated with an abrogation of TGFβ1-induced apoptosis and a partial rescue of TGFβ1-induced cell cycle arrest during both megakaryocytic and combined megakaryocytic/erythroid differentiation (Fig. 7B,D; Supplemental Fig. S7E). This was also observed upon activation of the miR-99/100∼125b tricistrons by ectopic expression of HOXA10 and its cofactor, PBX1 (Fig. 4H). The megakaryocytic differentiation block of TGFβ1 during combined erythroid/megakaryocytic in vitro differentiation was released by the miR-99a∼125b-2 tricistron, as assessed by the percentage of CD41+ (Fig. 7E) and CD36+ (marker for erythroid progenitors and MPs) (Supplemental Fig S7F) cells. The rescue by miR-99a∼125b-2 expression was stronger than by ALK inhibitor SB43152 (Fig. 7E). Similarly, TGFβ1-mediated suppression of CFU-MK formation was almost neutralized by miR-99a∼125b-2 expression (Fig. 7C).
RGB marking indicated that the resistance to TGFβ1 can be largely attributed to miR-125b-2. Each combination containing miR-125b-2 led to expansion of cells when either human or murine HSPCs were treated with TGFβ1 (Fig. 2C,D). However, upon replating of TGFβ1-treated cells, white colonies (i.e., miR-99a/let-7c/miR-125b-2-transduced) became prevalent (Fig. 2E; Supplemental Fig. S2B).
At the molecular level, let-7c, miR-125b, and the tricistron reduced total SMAD2 protein and inhibited phosphorylation of SMAD2 following TGFβ1 stimulation (Fig. 7F). Intracellular flow cytometry staining confirmed that miR-125b and the tricistron reduced TGFβ1-mediated phosphorylation (activation) of SMAD2/3 (Fig. 7G,H). In concordance with this observation, miR-125b and miR-99a∼125b-2 repressed transcriptional activity of TGFβ pathway downstream effector SMAD. Treatment with TGFβ1 at intermediate and high levels resulted in significantly lower activity of a reporter plasmid (PAI-1 promoter) containing 12 repeats of the SMAD-responsive element CAGA (Dennler et al. 1998) in miR-125b and miR-99a∼125b-2 transfected 293T cells (Fig. 7I). A different reporter with four repeats of SMAD-binding elements (SBE4x) yielded similar results (data not shown). In addition, downstream target genes of the TGFβ pathway, such as FOS, ID1, and ID2, were repressed in miR-99a∼125b-2-transduced MV4:11 cells treated with TGFβ1 (Supplemental Fig. S6L).
Last, we wanted to determine whether the tricistron’s effect might also be attributed to the other branches of the TGFβ superfamily. However, leukemic growth of miRNA- or control-transduced MV4:11 cells was not affected by BMP4 or Activin A (Supplemental Fig. S7G–J). Activin A reduced the expansion of MPs during in vitro culture, while high levels of BMP4 slightly increased their proliferation (Maguer-Satta et al. 2003; Jeanpierre et al. 2008). Tricistron-transduced CD34+ HSPCs were resistant to both effects (Supplemental Fig. S7O). In concordance with this observation, miR-99a∼125b-2 reduced BMP4-mediated phosphorylation of SMAD1/8 (Supplemental Fig. S7L).
We conclude that miR-99/100∼125b tricistrons block the TGFβ pathway, thereby abrogating its effect on differentiation, apoptosis, and cell cycle arrest during megakaryopoiesis.
Inhibition of miR-99∼125 miRNAs restores TGFβ sensitivity in AMKL cell lines
If ectopic miR-99a/100∼125b expression blocks the TGFβ pathway, then inhibition of tricistron function should enhance the TGFβ pathway and abrogate TGFβ1 resistance in AMKL. CMK and Meg-01 cells have high endogenous miR-99a/100∼125b expression and are mainly TGFβ-resistant. K562 cells served as negative controls, as they are also TGFβ-resistant but have low miR-99a/100∼125b expression (Supplemental Fig. S7A,B). CFU, proliferation, and apoptosis assays showed no response of SP-sc-transduced K562 cells to TGFβ1. As expected, SP-mediated inhibition of miR-99/100, let-7, or miR-125 did not sensitize K562 cells to TGFβ1 (Fig. 7J–L). In contrast, blocking miR-99/100, let-7, or miR-125 in CMK and Meg-01 cells significantly reduced in vitro growth, proliferation, cell cycle arrest, and colony formation in synergy with TGFβ1 (Fig. 7J–L; Supplemental Fig. S7K–N; for untreated SP-transduced CMK and K562 cells, see Fig. 6D–F; Supplemental Fig. S6D–F). The growth inhibitory effect, measured by GFP competition assays, was most pronounced in SP-125- and SP-TRI-transduced CMK and Meg-01 cells. A significant increase of apoptosis was observed only in SP-125-transduced and SP-TRI-transduced cells (Fig. 7K; Supplemental Fig. S7L). Thus, combined inhibition of miR-99∼125 miRNAs could sensitize TGFβ-resistant CMK and Meg-01 cells to TGFβ-mediated cell cycle arrest and apoptosis.
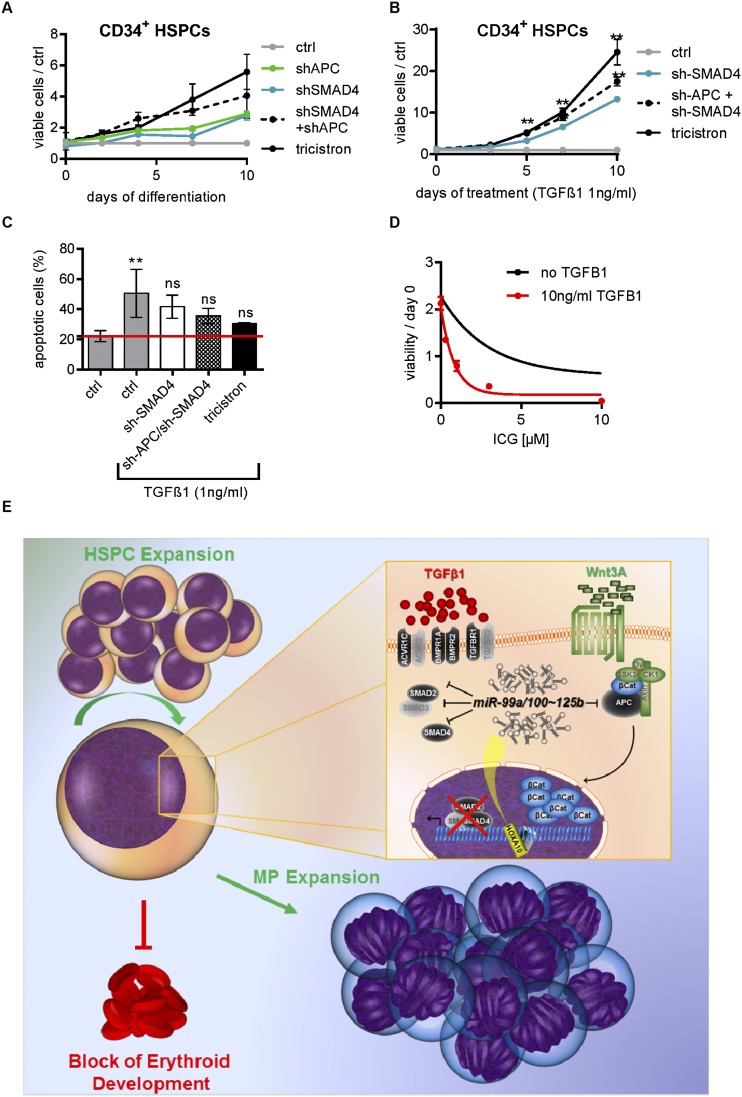
Inhibition of APC and SMAD4 efficiently mimics miR-99a∼125b-2 in HSPCs
If the concerted activity of miR-99a/100∼125b miRNAs on two pathways underlies the observed phenotype, then inhibition of single targets in one pathway may not fully recapitulate the miR-99a/100∼125b tricistron’s phenotype. To this end, we used superinfection of shRNA constructs directed to APC (marked with Venus) and SMAD4 (marked with mCherry) to transduce and sort double-positive HSPCs. During in vitro megakaryopoiesis, simultaneous knockdown of APC and SMAD4 more closely resembled the effect of the tricistron as compared with knockdown of either gene alone (Fig. 8A). The same applied for abrogation of TGFβ1-induced cell cycle arrest and apoptosis during megakaryopoiesis (Fig. 8B,C). Finally, we tested the synergy of activating the TGFβ pathway and inhibiting Wnt signaling to antagonize the tricistron in AMKL cells. Increasing doses of Wnt inhibitors ICG-001 or Indo greatly sensitized TGFβ1-resistant CMK and Meg-01 cells to TGFβ1 (Fig. 8D; Supplemental Fig. S8A,B), providing a novel option for combination treatment.
Figure 8.
miR-99a/100∼125b tricistrons regulate HSPC homeostasis by shifting the balance between TGFβ and Wnt signaling. (A,B) The number of miRNA- or shRNA-transduced CB CD34+ HSPCs (n = 2) during megakaryocytic in vitro differentiation relative to the control (nonsilencing miRNA [ctrl]). (B) TGFβ1 (1 ng/mL) was added to the culture. (C) AnnexinV/7-AAD staining on day 4 of megakaryocytic in vitro differentiation after treatment with 1 ng/mL TGFβ1. (D) The number of viable CMK cells upon treatment with increasing concentrations of ICG-001 in the presence or absence of 10 ng/mL TGFβ1 (n = 4). (E) Graphical model summarizing our study. By switching the balance between Wnt and TGFβ signaling, the concerted action of miR-99/100, let-7, and miR-125b promotes sustained expansion of murine and human HSPCs in vitro or in vivo while favoring megakaryocytic differentiation. (A–D) Data are presented as mean ± SD. (*) P < 0.05; (**) P < 0.01.
Discussion
Here, we unraveled a common regulatory function of miR-99a/100, let-7, and miR-125b homologs in the hematopoietic system. The miRNAs are encoded in two highly conserved miR-99a/100∼125b tricistrons on hsa11 and hsa21. They are highly expressed in HSCs/HSPCs and AMKL, a very aggressive form of leukemia with poor prognosis (Creutzig et al. 2012). We demonstrate that the tricistrons are produced from one primary transcript transactivated by the stem cell regulator HOXA10. It is the concerted activity of all three tricistron miRNAs that leads to the expansion of HSCs and MPs. This is achieved by jointly blocking the TGFβ pathway via repression of receptors and SMADs while elevating Wnt signaling via inhibition of the destruction complex (Fig. 8E). The TGFβ and Wnt pathways are two major regulatory signaling cascades that inversely ascertain the balance between HSC self-renewal and proliferation on the one hand and quiescence and differentiation on the other hand.
TGFβ ensures stem cell homeostasis by keeping HSCs in quiescence (Pietras et al. 2011) while inducing growth arrest and apoptosis in early HSPCs (Rorby et al. 2012). Thus, insensitivity to this inhibitory growth factor would render cells resistant to normal homeostatic mechanisms. This is a frequently observed mechanism in transformed hematopoietic cells to gain a growth advantage over their normal counterparts; e.g., the AML1/ETO fusion protein physically interacts with SMADs to disrupt SMAD-mediated transcription (Jakubowiak et al. 2000). Thus, overexpression of miR-99∼125 tricistrons constitutes a novel mechanism to evade the growth inhibitory function of TGFβ, which can be exploited by leukemic cells. Megakaryoblasts secrete active TGFβ1, determining the requirement to disrupt this autoregulatory feedback loop during malignant transformation in AMKL. Strikingly, when miR-99∼125 were inhibited in AMKL cells, TGFβ sensitivity was restored, opening the avenue for future therapeutic intervention.
However, the miR-99a/100∼125b tricistrons not only render hematopoietic cells insensitive to inhibitory growth factors but also increase their sensitivity to Wnt signaling. This is achieved by down-regulation of key members in the destruction complex (APC/APC2). In contrast to TGFβ, Wnt signaling is a positive regulator of self-renewal in HSCs and for megakaryopoiesis (Luis et al. 2012; Macaulay et al. 2013). When Wnt signaling is modestly enhanced, HSCs reconstitute better. However, a higher level of Wnt signaling results in failure to reconstitute irradiated recipient mice and multilineage defects (Kirstetter et al. 2006). The deregulation of Wnt signaling as a causative factor in leukemogenesis has become more and more apparent (Klaus and Birchmeier 2008). We hypothesize that this careful titration of the optimal activation level of signaling cascades determines the necessity for three miRNAs within the tricistron to coevolve.
Thus, our study shifts the focus from proliferation- and apoptosis-related miR-125 targets such as Bak1, Puma, p53, Trp53inp1, and ST18 (for review, see Shaham et al. 2012) to a tricistron target gene set affecting crucial upstream signaling pathways. Our study is in accordance with previous studies on individual miR-125 and let-7 family members in other cellular systems that demonstrated targeting of TGFβ or Wnt pathway genes. In embryonic stem cells (ESCs), miR-125a targets Dies1 to maintain strong BMP signaling and low to absent Activin/Nodal messaging. This keeps ESCs in an undifferentiated state (Parisi et al. 2012). miR-125 isoforms can also repress SMAD4 in human ESCs to favor neural commitment during neural conversion (Boissart et al. 2012). let-7 family members inhibit TGFBR1 to prevent endothelial-to-mesenchymal transition, while, in chicken embryos, down-regulation of let-7 permits active ACVR2 to contribute to gastrulation (Bobbs et al. 2012; Chen et al. 2012).
We showed that miR-125b was responsible for the majority of the tricistron effects on the hematopoietic system; e.g., the tricistron recapitulated the miR-125b phenotype during human in vitro megakaryopoiesis. Thus, despite targeting several common targets in the TGFβ and Wnt pathway, miR99/100 and let-7 exert different effects on HSPCs than miR-125b. These observations are consistent with previous reports on the miR-99b∼125a tricistron in the murine system (Guo et al. 2010; Gerrits et al. 2012). It was previously reported that miR-125a or miR-125b overexpression confers a competitive advantage to HSCs, leading to a gradual increase of chimerism in murine transplantation models (Guo et al. 2010; Ooi et al. 2010; Enomoto et al. 2011). When γ-retroviral vectors were used, which increased miR-125 expression level by 500-fold to several thousand-fold, development of myeloproliferative disease (MPD) and leukemia was observed (Gerrits et al. 2012; for discussion, see O’Connell et al. 2010). Using a lentiviral system with a SFFV promoter-driven miRNA cassette, we achieved expression levels comparable with those seen in AMKL. The miR-99a, let-7c, and miR-125b expression level in tricistron-transduced cells was equal to or lower than that in single miRNA-transduced cells. This excludes the possibility that the observed tricistron phenotype reflects a miR-125 dose-dependent phenomenon. Similar to other recent studies using lentiviral systems, we observed a competitive repopulation advantage and myeloid lineage bias but no MPD or leukemia from miR-125b-2- and miR-99a∼125b-2-transduced Lin− BM cells in vivo (Surdziel et al. 2011). Interestingly, the competitive advantage conferred by miR-125b-2 was only sustained in the tricistronic context of miR-99a∼125b-2. Only the tricistron was able to expand the LSK and LT-HSC compartment in primary and secondary recipients. The same applies to RGB marking, where only miR-99a, let-7c, and miR-125b-2 together allowed colony formation for more than two rounds of replating. Thus, miR-125b enhances proliferation of HSCs and MPs by negatively regulating anti-proliferative and proapoptotic genes; many of these genes are either part of the TGFβ or Wnt pathway or downstream effectors of this pathway. If no leukemic lesions are acquired, the miR-125b-transduced HSC pool will exhaust. Tumor suppressor let-7c or miR-99a/100 lacks those anti-apoptotic and proproliferative target genes of miR-125b. In contrast, both miRNAs negatively regulate proproliferative genes such as oncogenic MYC, MYCN, MYCL1, RAS, and HMGA2 and TCF-responsive genes CCND1 and CCND2 (Johnson et al. 2005; Lee and Dutta 2007). These differences in the targetome might explain distinctive hematopoietic phenotypes of let-7c, miR-99a/100, and miR-125b. Our results indicate that let-7 and miR-99/100 are not sufficient to fully block TGFβ or activate Wnt signaling. We hypothesize that miR-99/100 and let-7 assist miR-125b in suppressing TGFβ pathway genes or the β-catenin destruction complex. By repressing additional proproliferative targets, miR-99/100 and let-7 partially counteract miR-125b-induced hyperproliferation of HSCs, which confers a steady-state growth advantage and prevents exhaustion of miR-99∼125-transduced HSCs.
Thus, miR-99a/100∼125b tricistronic miRNAs converge in a combined phenotype in which miR-99/100 and let-7 add to or subtract from the hematologically dominant miR-125 phenotype. In addition, the interplay between let-7 and miR-125 via LIN28 proteins in HSPCs should not be neglected. While LIN28B represses let-7 to maintain stemness and block erythroid development (Lee et al. 2013), high miR-125 levels repress LIN28A (Chaudhuri et al. 2012). These are expected to add another layer of cross-talk between miRNAs in the miR-99a/100∼125b tricistrons.
With a plethora of miRNAs organized in polycistrons and regulatory versatility by genetic interactions, we believe our study paves the way to integration of combined gene regulation by miRNA clusters in stem cell biology. Furthermore, Wnt and TGFβ signaling also plays crucial roles in other developmental and cellular systems (Oshimori and Fuchs 2012), thus potentially extending the regulatory repertoire of miR-99a/100∼125b tricistrons beyond hematopoiesis.
Overall, we explored a HOXA10–miR-99a/100∼125b–TGFβ/Wnt axis responsible for hematopoietic stemness, megakaryocytic development, and leukemogenesis (Fig. 8E). Our study holds promising potential for future treatment rationales of hematologic malignancies.
Materials and methods
Patient samples and cell lines
The AML-Berlin–Frankfurt–Münster study group (AML-BFM-SG; Hannover, Germany) provided all patient samples. CB HSPCs from donors were positively selected by labeling CD34-expressing cells with magnetic cell-sorting beads (Miltenyi Biotec). FL CD34+ HSPCs were purchased from Novogenic Laboratories. Culture conditions for maintenance, megakaryocytic, or megakaryocytic/erythroid in vitro differentiation of CD34+ HSPCs were described elsewhere (Klusmann et al. 2010a; Kumar et al. 2011; Emmrich et al. 2012; Stankov et al. 2014). Cell lines (CMK, K562, MV4:11, THP-1, and NB4) were purchased from the German National Resource Center for Biological Material (DSMZ) and maintained under recommended conditions. The ALK inhibitor SB43152 (SelleckChem), Indo (SelleckChem), BIO (Sigma), ICG-001 (SelleckChem), human recombinant TGFβ1 (Peprotech), and human Wnt3A (R&D Systems) were applied as indicated. All investigations were approved by the local ethics committee. Fluorescence microscopy was carried out with a BZ9000 (Keyence) microscope.
Mice
All studies involving mice were approved by the Hannover Medical School Institute for Laboratory Animal Science and local authorities and performed in accordance with the relevant protocol. BM from C57BL/6J mice was lineage-depleted using mouse hematopoietic progenitor cell enrichment kit (Stem Cell Technologies) and used for transduction. FL cells were depleted using biotinylated TER119 and Ly-6G/C antibodies (Stem Cell Technologies). Recipients were lethally irradiated (10 Gy) and transplanted with 3 × 105 transduced cells. CBC and WBC counts were obtained from the scil Vet ABC blood counter (Horiba Medical). Flow cytometry of total BM and PB was performed after red blood cell lysis (Pharm Lyse reagent, Becton Dickinson). For quantification of cells, stained samples were analyzed with TRUcount tubes (Becton Dickinson) according to the manufacturer’s instructions. For secondary transplantation, one-third of the whole BM of each mouse per construct was pooled and injected per lethally irradiated secondary recipient.
Constructs and lentivirus
Cloning of miR-99a, let-7c, and miR-125b-2 (alone and in combination) as well as the miR-100/let-7a-2/miR-125b-1 tricistron from genomic DNA into a modified LeGO vector was performed as previously described (Weber et al. 2008; Emmrich et al. 2012). A nonsilencing shMiR in the miR-30 backbone (Open Biosystems) or miR-E backbone (Fellmann et al. 2013) was subcloned to the LeGO construct and used as control (referred to as nonsilencing miRNA). shRNAs directed to human HOXA10 were obtained from Open Biosystems (clone IDs: V2LHS_192251, and V2LHS_244712) and subcloned into the LeGO miR-30 backbone construct. shRNAs directed to human APC (ID: APC.3311) and SMAD4 (ID: SMAD4.2580) were cloned into the miR-E backbone. For the superinfection experiments, we sorted single-positive (Venus- or mCherry-positive) and double-positive cells, resulting in either sh-APC-expressing, sh-SMAD4-expressing, or sh-APC/sh-SMAD4-expressing cells. Endogenous promoter activity assays in cell lines were performed with a TK-GFP lentiviral reporter construct (kind gift of Dr. Orkin’s laboratory, Boston Children’s Hospital), where the TK promoter was replaced by the putative promoter fragments. Stable miRNA knockdown was achieved using the sponge technology (Loya et al. 2009). Briefly, we inserted eight to 16 specific target sites of each miRNA as an artificial 3′ UTR behind the sense SFFV-driven mCherry cassette, while an antisense CMV-driven GFP-IRES-Puro served as selection marker (pLBid-GIP/C vector) (Supplemental Fig. S5C,D; Maetzig et al. 2010). The SP-sc control sequence was adapted from Loya et al. (2009). The MSCV:HoxA10 vector was obtained as a kind gift from Dr. Heuser (Hannover Medical School). For the HSPC studies, human HOXA10 and PBX1 were cloned in a modified LeGO vector with a cerulean or GFP expression cassette, respectively. Lentiviral supernatant was generated and collected using standard protocols and as described (Emmrich et al. 2012; Maroz et al. 2013).
Microarray data collection and analysis
Microarray expression profiles were collected using human GE 4x44K v2 chips for mRNA and human miRNA microarray release 14.0, 8x15K chips for miRNA expression (both Agilent) and analyzed using GeneSpringXII (Agilent) and GSEA (Subramanian et al. 2005). For miRNA target gene prediction, we used the miRecords database (Xiao et al. 2009). All microarray data have been deposited in NCBI’s Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) with GEO series accession number GSE56335.
Statistics
Statistical evaluation between the two groups was carried out using Student’s t-test and for more than two groups by one-way ANOVA with Tukey’s or Sidak’s post-hoc analysis. The level of significance was set at P < 0.05. All data are presented as mean ± SD or SEM, as indicated. Calculations were performed using GraphPad Prism 6.
Acknowledgments
We thank Dr. S.H. Orkin and Dr. S. Meyer for critically reading the manuscript and helpful discussions; M. Ballmaier for cell sorting; M. El-Khatib for help with experimental validation; E. Pittermann, R. Jammal, R.-M. Struß, D. Heckl, and L. Queisser for technical assistance; and J. Zuber, K. Weber, and B. Fehse for providing plasmids. This work was supported by a grant to J.-H.K. from the German Research Foundation (DFG) within the Emmy Noether-Programme (KL-2374/2-1). S.E., M.R., S.K., and A.M. were supported by the Hannover Biomedical Research School. S.K. is supported by the DAAD. Z.L. was supported by the American Association for Cancer Research-Aflac, Incorporated Career Development Award for Pediatric Cancer Research (10-20-10-LI) and National Institutes of Health grant R01 HL107663.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.233791.113.
References
- Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H 2005. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res 33: 2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. 2011. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8: 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson JM, Andersson E, Lundstrom P, Larsson N, Xu X, Repetowska E, Humphries RK, Karlsson S 2001. Proliferation of primitive myeloid progenitors can be reversibly induced by HOXA10. Blood 98: 3301–3308 [DOI] [PubMed] [Google Scholar]
- Bobbs AS, Saarela AV, Yatskievych TA, Antin PB 2012. Fibroblast growth factor (FGF) signaling during gastrulation negatively modulates the abundance of microRNAs that regulate proteins required for cell migration and embryo patterning. J Biol Chem 287: 38505–38514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissart C, Nissan X, Giraud-Triboult K, Peschanski M, Benchoua A 2012. miR-125 potentiates early neural specification of human embryonic stem cells. Development 139: 1247–1257 [DOI] [PubMed] [Google Scholar]
- Bousquet M, Harris MH, Zhou B, Lodish HF 2010. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci 107: 21558–21563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WC, Ho MR, Li SC, Tsai KW, Lai CH, Hsu CN, Lin WC 2012. MetaMirClust: discovery of miRNA cluster patterns using a data-mining approach. Genomics 100: 141–148 [DOI] [PubMed] [Google Scholar]
- Chaudhuri AA, So AY, Mehta A, Minisandram A, Sinha N, Jonsson VD, Rao DS, O’Connell RM, Baltimore D 2012. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proc Natl Acad Sci 109: 4233–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PS, Su JL, Cha ST, Tarn WY, Wang MY, Hsu HC, Lin MT, Chu CY, Hua KT, Chen CN, et al. 2011. miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans. J Clin Invest 121: 3442–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Qin L, Barnes C, Charisse K, Yi T, Zhang X, Ali R, Medina PP, Yu J, Slack FJ, et al. 2012. FGF regulates TGF-β signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Rep. 2: 1684–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutzig U, van den Heuvel-Eibrink MM, Gibson B, Dworzak MN, Adachi S, de Bont E, Harbott J, Hasle H, Johnston D, Kinoshita A, et al. 2012. Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood 120: 3187–3205 [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM 1998. Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J 17: 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, Moon RT, Teo JL, Kim HY, Moon SH, et al. 2004. A small molecule inhibitor of β-catenin/CREB-binding protein transcription [corrected] Proc Natl Acad Sci 101: 12682–12687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmrich S, Henke K, Hegermann J, Ochs M, Reinhardt D, Klusmann JH 2012. miRNAs can increase the efficiency of ex vivo platelet generation. Ann Hematol 91: 1673–1684 [DOI] [PubMed] [Google Scholar]
- Emmrich S, Katsman-Kuipers JE, Henke K, Khatib ME, Jammal R, Engeland F, Dasci F, Zwaan CM, den Boer ML, Verboon L, et al. 2013. miR-9 is a tumor suppressor in pediatric AML with t(8;21). Leukemia doi: 10.1038/leu.2013.357 [DOI] [PubMed] [Google Scholar]
- Enomoto Y, Kitaura J, Hatakeyama K, Watanuki J, Akasaka T, Kato N, Shimanuki M, Nishimura K, Takahashi M, Taniwaki M, et al. 2011. Emu/miR-125b transgenic mice develop lethal B-cell malignancies. Leukemia 25: 1849–1856 [DOI] [PubMed] [Google Scholar]
- Fava RA, Casey TT, Wilcox J, Pelton RW, Moses HL, Nanney LB 1990. Synthesis of transforming growth factor-β1 by megakaryocytes and its localization to megakaryocyte and platelet α-granules. Blood 76: 1946–1955 [PubMed] [Google Scholar]
- Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, et al. 2013. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 5: 1704–1713 [DOI] [PubMed] [Google Scholar]
- Fortunel N, Batard P, Hatzfeld A, Monier MN, Panterne B, Lebkowski J, Hatzfeld J 1998. High proliferative potential-quiescent cells: a working model to study primitive quiescent hematopoietic cells. J Cell Sci 111: 1867–1875 [DOI] [PubMed] [Google Scholar]
- Fortunel NO, Hatzfeld A, Hatzfeld JA 2000. Transforming growth factor-β: pleiotropic role in the regulation of hematopoiesis. Blood 96: 2022–2036 [PubMed] [Google Scholar]
- Gerrits A, Walasek MA, Olthof S, Weersing E, Ritsema M, Zwart E, van Os R, Bystrykh LV, de Haan G 2012. Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood 119: 377–387 [DOI] [PubMed] [Google Scholar]
- Goessling W, North TE, Loewer S, Lord AM, Lee S, Stoick-Cooper CL, Weidinger G, Puder M, Daley GQ, Moon RT, et al. 2009. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell 136: 1136–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Lu J, Schlanger R, Zhang H, Wang JY, Fox MC, Purton LE, Fleming HH, Cobb B, Merkenschlager M, et al. 2010. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci 107: 14229–14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16: 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawcroft G, D’Amico M, Albanese C, Markham AF, Pestell RG, Hull MA 2002. Indomethacin induces differential expression of β-catenin, γ-catenin and T-cell factor target genes in human colorectal cancer cells. Carcinogenesis 23: 107–114 [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. 2005. A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Keller JR, Ruscetti FW, Kondaiah P, Roberts AB, Falk LA 1991. Bidirectional effects of transforming growth factor β (TGF-β) on colony-stimulating factor-induced human myelopoiesis in vitro: differential effects of distinct TGF-β isoforms. Blood 78: 2239–2247 [PubMed] [Google Scholar]
- Jakubowiak A, Pouponnot C, Berguido F, Frank R, Mao S, Massague J, Nimer SD 2000. Inhibition of the transforming growth factor β1 signaling pathway by the AML1/ETO leukemia-associated fusion protein. J Biol Chem 275: 40282–40287 [DOI] [PubMed] [Google Scholar]
- Jeanpierre S, Nicolini FE, Kaniewski B, Dumontet C, Rimokh R, Puisieux A, Maguer-Satta V 2008. BMP4 regulation of human megakaryocytic differentiation is involved in thrombopoietin signaling. Blood 112: 3154–3163 [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ 2005. RAS is regulated by the let-7 microRNA family. Cell 120: 635–647 [DOI] [PubMed] [Google Scholar]
- Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C 2006. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol 7: 1048–1056 [DOI] [PubMed] [Google Scholar]
- Klaus A, Birchmeier W 2008. Wnt signalling and its impact on development and cancer. Nat Rev Cancer 8: 387–398 [DOI] [PubMed] [Google Scholar]
- Klusmann JH, Godinho FJ, Heitmann K, Maroz A, Koch ML, Reinhardt D, Orkin SH, Li Z 2010a. Developmental stage-specific interplay of GATA1 and IGF signaling in fetal megakaryopoiesis and leukemogenesis. Genes Dev 24: 1659–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klusmann JH, Li Z, Bohmer K, Maroz A, Koch ML, Emmrich S, Godinho FJ, Orkin SH, Reinhardt D 2010b. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev 24: 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Narla A, Nonami A, Mullally A, Dimitrova N, Ball B, McAuley JR, Poveromo L, Kutok JL, Galili N, et al. 2011. Coordinate loss of a microRNA and protein-coding gene cooperate in the pathogenesis of 5q− syndrome. Blood 118: 4666–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Dutta A 2007. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21: 1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YT, de Vasconcellos JF, Yuan J, Byrnes C, Noh SJ, Meier ER, Kim KS, Rabel A, Kaushal M, Muljo SA, et al. 2013. LIN28B-mediated expression of fetal hemoglobin and production of fetal-like erythrocytes from adult human erythroblasts ex vivo. Blood 122: 1034–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R 2004. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20: 781–810 [DOI] [PubMed] [Google Scholar]
- Loya CM, Lu CS, Van Vactor D, Fulga TA 2009. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods 6: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis TC, Naber BA, Roozen PP, Brugman MH, de Haas EF, Ghazvini M, Fibbe WE, van Dongen JJ, Fodde R, Staal FJ 2011. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell 9: 345–356 [DOI] [PubMed] [Google Scholar]
- Luis TC, Ichii M, Brugman MH, Kincade P, Staal FJ 2012. Wnt signaling strength regulates normal hematopoiesis and its deregulation is involved in leukemia development. Leukemia 26: 414–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Thon JN, Tijssen MR, Steele BM, MacDonald BT, Meade G, Burns P, Rendon A, Salunkhe V, Murphy RP, et al. 2013. Canonical Wnt signaling in megakaryocytes regulates proplatelet formation. Blood 121: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetzig T, Galla M, Brugman MH, Loew R, Baum C, Schambach A 2010. Mechanisms controlling titer and expression of bidirectional lentiviral and γretroviral vectors. Gene Ther 17: 400–411 [DOI] [PubMed] [Google Scholar]
- Magnusson M, Brun AC, Miyake N, Larsson J, Ehinger M, Bjornsson JM, Wutz A, Sigvardsson M, Karlsson S 2007. HOXA10 is a critical regulator for hematopoietic stem cells and erythroid/megakaryocyte development. Blood 109: 3687–3696 [DOI] [PubMed] [Google Scholar]
- Maguer-Satta V, Bartholin L, Jeanpierre S, Ffrench M, Martel S, Magaud JP, Rimokh R 2003. Regulation of human erythropoiesis by activin A, BMP2, and BMP4, members of the TGFβ family. Exp Cell Res 282: 110–120 [DOI] [PubMed] [Google Scholar]
- Maroz A, Stachorski L, Emmrich S, Reinhardt K, Xu J, Shao Z, Kabler S, Dertmann T, Hitzler J, Roberts I, et al. 2013. GATA1s induces hyperproliferation of eosinophil precursors in Down syndrome transient leukemia. Leukemia doi: 10.1038/leu.2013.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J 2008. TGFβ in cancer. Cell 134: 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH 2009. The regulation of TGFβ signal transduction. Development 136: 3699–3714 [DOI] [PubMed] [Google Scholar]
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D’Andrea A, Sander C, Ventura A 2009. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev 23: 2806–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D 2010. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci 107: 14235–14240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi AG, Sahoo D, Adorno M, Wang Y, Weissman IL, Park CY 2010. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci 107: 21505–21510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N, Fuchs E 2012. The harmonies played by TGF-β in stem cell biology. Cell Stem Cell 11: 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Giardine BM, Hou M, Ma J, Hardison RC, Stubbs L, Miller W 2005. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res 15: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi S, Battista M, Musto A, Navarra A, Tarantino C, Russo T 2012. A regulatory loop involving Dies1 and miR-125a controls BMP4 signaling in mouse embryonic stem cells. FASEB J 26: 3957–3968 [DOI] [PubMed] [Google Scholar]
- Pelosi A, Careccia S, Lulli V, Romania P, Marziali G, Testa U, Lavorgna S, Lo-Coco F, Petti MC, Calabretta B, et al. 2013. miRNA let-7c promotes granulocytic differentiation in acute myeloid leukemia. Oncogene 32: 3648–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petriv OI, Kuchenbauer F, Delaney AD, Lecault V, White A, Kent D, Marmolejo L, Heuser M, Berg T, Copley M, et al. 2010. Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc Natl Acad Sci 107: 15443–15448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras EM, Warr MR, Passegue E 2011. Cell cycle regulation in hematopoietic stem cells. J Cell Biol 195: 709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorby E, Hagerstrom MN, Blank U, Karlsson G, Karlsson S 2012. Human hematopoietic stem/progenitor cells overexpressing Smad4 exhibit impaired reconstitution potential in vivo. Blood 120: 4343–4351 [DOI] [PubMed] [Google Scholar]
- Shaham L, Binder V, Gefen N, Borkhardt A, Izraeli S 2012. miR-125 in normal and malignant hematopoiesis. Leukemia 26: 2011–2018 [DOI] [PubMed] [Google Scholar]
- Stankov MV, El Khatib M, Kumar TB, Heitmann K, Panayotova-Dimitrova D, Schoening J, Bourquin JP, Schweitzer N, Leverkus M, Welte K, et al. 2014. Histone deacetylase inhibitors induce apoptosis in myeloid leukemia by suppressing autophagy. Leukemia 28: 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surdziel E, Cabanski M, Dallmann I, Lyszkiewicz M, Krueger A, Ganser A, Scherr M, Eder M 2011. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood 117: 4338–4348 [DOI] [PubMed] [Google Scholar]
- Thorsteinsdottir U, Sauvageau G, Hough MR, Dragowska W, Lansdorp PM, Lawrence HJ, Largman C, Humphries RK 1997. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol 17: 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA 2010. The Wnt/β-catenin pathway is required for the development of leukemia stem cells in AML. Science 327: 1650–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Bartsch U, Stocking C, Fehse B 2008. A multicolor panel of novel lentiviral ‘gene ontology’ (LeGO) vectors for functional gene analysis. Mol Ther 16: 698–706 [DOI] [PubMed] [Google Scholar]
- Weber K, Thomaschewski M, Warlich M, Volz T, Cornils K, Niebuhr B, Tager M, Lutgehetmann M, Pollok JM, Stocking C, et al. 2011. RGB marking facilitates multicolor clonal cell tracking. Nat Med 17: 504–509 [DOI] [PubMed] [Google Scholar]
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T 2009. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 37: D105–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YS, Zhang H, Zhang XJ, Feng DD, Luo XQ, Zeng CW, Lin KY, Zhou H, Qu LH, Zhang P, et al. 2012. MiR-100 regulates cell differentiation and survival by targeting RBSP3, a phosphatase-like tumor suppressor in acute myeloid leukemia. Oncogene 31: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]