Many chemotherapeutic agents, such as doxorubicin (DOX), interfere with DNA replication. Here, Dutta and colleagues show that DOX treatment produces clusters of stalled replication forks and transcriptional repression of neighboring genes. An ATR-dependent checkpoint pathway that down-regulates histone chaperone ASF1a is shown to repress genes overlapping with stalled replication forks. Furthermore, ASF1a-depleted cancer cells are more sensitive to DOX, suggesting that the loss of this histone chaperone, as seen in several cancers, could be a personalized tumor marker for sensitivity to DOX.
Keywords: doxorubicin, stalled forks, transcription repression, S-phase checkpoint, ASF1, ATR, CRL1βTRCP
Abstract
Many agents used for chemotherapy, such as doxorubicin, interfere with DNA replication, but the effect of this interference on transcription is largely unknown. Here we show that doxorubicin induces the firing of dense clusters of neoreplication origins that lead to clusters of stalled replication forks in gene-rich parts of the genome, particularly on expressed genes. Genes that overlap with these clusters of stalled forks are actively dechromatinized, unwound, and repressed by an ATR-dependent checkpoint pathway. The ATR checkpoint pathway causes a histone chaperone normally associated with the replication fork, ASF1a, to degrade through a CRL1βTRCP-dependent ubiquitination/proteasome pathway, leading to the localized dechromatinization and gene repression. Therefore, a globally active checkpoint pathway interacts with local clusters of stalled forks to specifically repress genes in the vicinity of the stalled forks, providing a new mechanism of action of chemotherapy drugs like doxorubicin. Finally, ASF1a-depleted cancer cells are more sensitive to doxorubicin, suggesting that the 7%–10% of prostate adenocarcinomas and adenoid cystic carcinomas reported to have homozygous deletion or significant underexpression of ASF1a should be tested for high sensitivity to doxorubicin.
Chromosomal DNA replication is a highly conserved process in all eukaryotes that starts at specific sites along DNA known as replication origins. The activation of DNA replication origins requires assembly of prereplicative complexes (pre-RCs) followed by recruitment of additional initiating factors and formation of replisomes (Bell and Dutta 2002; Sclafani and Holzen 2007). Assembly of the pre-RCs onto each replication origin occurs during the early G1 phase of the cell cycle through sequential recruitment of the origin recognition complex (ORC), CDC6, CDT1, and minichromosome maintenance 2–7 (MCM2–7 complex) proteins. However, excess MCM2–7 complexes are loaded, leading to the licensing of excess origins, most of which are dormant and passively replicated in the normal S phase. The excess licensed origins, however, serve as backup origins that can complete DNA replication when existing forks stall (Ibarra et al. 2008; Ge and Blow 2010). In the presence of DNA-damaging agents, when newly fired replication forks stall after replicating a few hundred bases, many additional (formerly dormant) origins begin firing in mammalian cells (Ge et al. 2007; Courbet et al. 2008; Karnani and Dutta 2011).
DNA replication is often the target of anti-cancer drugs, leading to the generation of stalled replication forks (Hoeijmakers 2001). ATR kinase recognizes these stalled replication forks and activates the intra-S-phase checkpoint signaling cascade. The activation of ATR requires the replication protein A (RPA) complex RPA70–RPA32–RPA14 and ATRIP (Cortez et al. 2001; Ball et al. 2005; Cimprich and Cortez 2008). The RPA complex coats ssDNA at stalled replication forks and recruits ATR and other checkpoint proteins (Zou and Elledge 2003). Once activated, ATR triggers the checkpoint by phosphorylating many downstream targets (including RPA, CHK1, and p53), ultimately leading to arrest of the cell cycle in S phase. Since transcription of neighboring genes is often repressed by even a single DNA double-strand break (DSB) in mammalian cells by active pathways using ATM and DNA-dependent protein kinases (Kruhlak et al. 2007; Shanbhag et al. 2010; Pankotai et al. 2012), we wondered whether and how stalled replication forks produced by anti-cancer drugs affect the transcription of neighboring genes.
Doxorubicin (DOX) is a widely used cancer chemotherapeutic drug that works by intercalating into dsDNA, inhibiting the activity of DNA topoisomerase II (Bodley et al. 1989; Capranico et al. 1990). Despite the extensive use and study of this drug, the locations of replication origins that fire in the presence of DOX, the locations of stalled forks, and the effects of the stalled forks on neighboring gene expression have not been studied. Previously, we found that hydroxyurea (HU), a ribonucleotide reductase inhibitor (Elford 1968), activates clusters of dormant replication origins that produce clusters of forks that stall after a few hundred bases of DNA synthesis (Karnani and Dutta 2011). Many of these neoreplication origin clusters overlap with coding genes. We therefore hypothesized that chemotherapy drugs like DOX could activate similar clusters of neoreplication origins and produce clusters of stalled replication forks so that the cis effects of these stalled forks on the local transcription machinery would be part of the gene expression changes seen in cells treated with DOX.
We show that DOX treatment produces clusters of stalled replication forks at specific sites in the genome and that the production of those clusters decreases the transcription of neighboring genes due to active changes in the chromatin and detachment of RNA polymerase II. Surprisingly, the transcriptional repression was not simply due to the mechanical effects of local clusters of stalled forks but an ATR-dependent checkpoint pathway that down-regulates ASF1a, a histone chaperone known to act in association with replication forks. The CRL1βTRCPE3 ligase complex polyubiquitinates ASF1a and targets it for degradation by proteasomes. This is a new mechanism by which a globally active checkpoint pathway interacts with local clusters of stalled forks to specifically repress genes in the vicinity of the stalled forks. These specific changes in gene expression produced by anti-S-phase chemotherapy drugs like DOX will contribute to the efficacy or toxicity of this class of drugs. Finally, decrease of ASF1a makes cancer cells more sensitive to DOX, suggesting that the homozygous deletion or underexpression of this histone chaperone, as seen in several cancers, could be a personalized tumor marker for sensitivity to DOX.
Results
DOX-induced DNA damage results in the firing of clusters of neoreplication origins
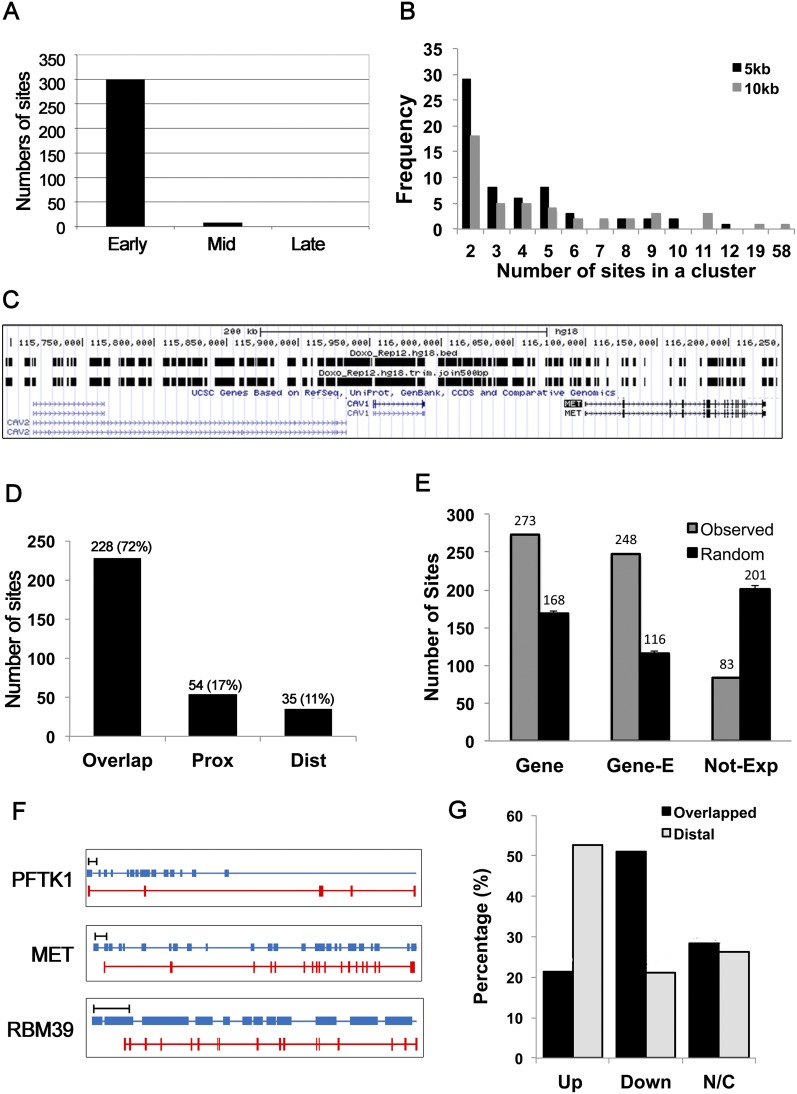
To characterize dormant origins that are fired in response to DOX-induced DNA damage, mitotically synchronized HeLa cells were released from a nocodazole block into medium containing DOX and 5-bromo-2′-deoxyuridine (BrdU). Newly replicated DNA labeled by BrdU was immunoprecipitated and hybridized to a high-resolution genome tiling array. The array covered the 1% of the human genome selected by the ENCODE pilot project to be representative of the entire genome so that it can be intensely investigated for biological questions (The ENCODE Project Consortium 2007). DOX-induced replication tracks were identified from two biological replicates. The hybridization data were analyzed by model-based analysis of tiling arrays (MAT) to identify genomic positions with a statistically significant enrichment (P ≤ 10−3) of BrdU labeling in DOX-treated cells compared with control cells (Supplemental Table S1A). A total of 433 DOX-BrIP (BrdU immunoprecipitation) sites were identified with a median length of 1355 base pairs (bp). When sites within 500 bp were considered to be part of the same replicon, there were 317 fired origins (Supplemental Tables S1A, S2). Of the DOX-BrIP sites, 94.6% were located within early-replicating regions of the genome, a few were in mid-replicating regions, and none were in late-replicating regions (Fig. 1A). Very few of these DOX-BrIP sites overlapped with the origins mapped in normally proliferating HeLa cells (Karnani et al. 2010), suggesting that these replication tracks arise from the firing of normally dormant origins (neo-origins). However, 60% of the DOX-BrIP sites overlapped with, and another 20% were within 10 kb of, sites detected when S phase was interrupted by another anti-S-phase agent, HU (Supplemental Fig. S1A). Therefore, certain sites in the genome preferentially fire neo-origins when replication is arrested by anti-S-phase agents.
Figure 1.
DOX induced firing of clusters of dormant replication origins and decreased the mRNA expression of overlapping genes. (A) Segregation of the DOX-BrIP sites (the joined sites in Supplemental Table S1A) by whether that region replicates early, mid, or late in a normal S phase. (B) Clusters defined as BrdU-labeled tracks within 5 kb or 10 kb of each other. Sixty-two or 46 clusters were seen, respectively. The plot shows the distribution of the number of initiation sites in a cluster. (C) Representative neoreplication origin clusters on a portion of chromosome 7. Vertical bars in the top row represent individual BrIP sites. The second row shows a track where BrIP sites within 500 bp of each other were joined. This figure was captured from the University of California at Santa Cruz genome browser (http://genome.ucsc.edu; Kent et al. 2002). (D) Genes overlapping with, proximal to (within 5 kb of the TSS or TES) or distal to (>5 kb away from the TSS and TES) DOX-induced BrIP sites. The bar graph shows the number and the percentage of genes in each class. (E) The overlap of DOX-induced BrIP sites, with the indicated parts of the genome compared with a random model, reveals a significant enrichment at genes (Z = 8.1) and “expressed” genes (Gene-E; Z = 12.2) and depletion (Z = −8.3) at “not expressed” genes (Not-Exp). The Z-score represents the observed random expected/standard deviation and is explained in the Supplemental Material. (F) Representative genes that overlap with neoreplication origin clusters. The bar at the top of the figures represents 10 kb. The middle blue bar and bottom red bar represent BrdU-labeled tracks and exons of the gene, respectively. These three genes will be studied in this study as examples of genes that overlap with clusters of stalled replication forks. (G) Compared with the DMSO-treated control, genes that were induced ≥1.3-fold in DOX were considered as up-regulated (Up), whereas genes ≤0.7 are considered down-regulated (Down). Genes expressing at 0.7∼1.3 times the level in DMSO were regarded as not changed (N/C).
Inhibition of the G1/S transition by roscovitine, a CDK inhibitor, suppressed the DOX-BrIP sites, consistent with the idea that DNA replication is involved in their generation (Supplemental Fig. S1B). Depletion of BRCA1, a key player in DNA damage repair, had no effect on the DOX-BrIP sites (Supplemental Fig. S1B). Inhibition of DNA damage-induced checkpoint pathways by caffeine also did not change the DOX-BrIP sites (Supplemental Fig. S1C). Multiple groups have shown by molecular combing that dormant origins fire to produce clusters of bidirectional replication forks when cells are exposed to agents that slow or stall replication forks (Ge et al. 2007; Courbet et al. 2008). Thus, although we cannot rule out the possibility that a few of the DOX-BrIP sites were generated by DNA repair, the bulk of the DOX-BrIP sites are generated by DNA replication, and their locations are not influenced by inhibiting the checkpoint pathways.
Previous studies by us and others have revealed extensive clustering of damage-induced replication origins (Karnani and Dutta 2011). Similarly, we found that DOX-induced neoreplication origins exhibit high levels of clustering. Two-hundred-thirty of the 317 DOX-BrIP sites (73%) were located within 5 kb of their neighbors and resolved into 62 DOX-BrIP clusters, defined as 5-kb stretches with more than two sites (Fig. 1B; Supplemental Table S1B). The largest cluster in this 1% of the genome contained 58 unique DOX-BrIP sites within a 10-kb stretch from chromosome 7, overlapping the FRA7G common fragile site (Huang et al. 1998) and multiple cancer-related genes: MET, CAV1, and CAV2 (Fig. 1C). Another cluster is seen on chromosome 20 overlapping with CPNE1 and RBM39. Additional examples are seen in Supplemental Table S2.
These results demonstrate that DOX treatment activates clusters of dormant replication origins within specific regions of the genome. The forks arising from these origins stall after replicating several hundred bases, yielding clusters of stalled replication forks in the same parts of the genome.
Neoreplication origin clusters overlap with transcriptionally expressed genes
Replication origins are located near transcriptional units in HeLa cells (Sequeira-Mendes et al. 2009; Karnani et al. 2010), and ORC-binding origin sites associate with RNA polymerase II-binding sites in Drosophila (MacAlpine et al. 2004). Therefore, we tested whether the DOX-induced neoreplication origins are at or near genes. DOX-induced neoreplication origin clusters were located within −5 to +5 kb relative to the transcribed regions of 47 genes (Supplemental Table S3). Of the total 317 DOX-BrIP sites, 228 (72%) overlapped with genes and 54 (17%) were located within 5 kb of a gene, leaving only 35 (11%) outside this range (Fig. 1D).
To address whether the DOX-BrIP sites preferentially arise in transcriptionally active genes, we compared their locations with the RNA sequencing (RNA-seq) data of HeLa cells generated by the ENCODE consortium (Derrien et al. 2012). Two-hundred-forty-eight BrIP sites intersected with expressed regions of the genome, a significant enrichment relative to the random expectation of overlap (Fig. 1E; Z-statistics are explained in the Supplemental Material). Conversely, the overlap of DOX-BrIP sites with nonexpressed genes was significantly lower than random expectation. PFTK1, MET, and RBM39 are examples of expressed genes that overlap with DOX-induced neoreplication origin clusters, genes that will be studied in detail later (Fig. 1F).
DOX-induced neoreplication origin clusters repress transcription of overlapping genes
We next tested in normally cycling (cell cycle-asynchronous) cells whether transcription of the 47 genes overlapping with the local clusters of fired origins and stalled forks is altered by DOX. We first confirmed that 12 representative DOX-BrIP clusters of neo-origins also fire in the presence of DOX in cell cycle-asynchronous HeLa cells (Supplemental Fig. S1D). Quantitative RT–PCR (qRT–PCR) of mRNA revealed that more (51%, 24 of 47) of the genes overlapping with the clusters of neo-origins were down-regulated by DOX than were up-regulated (21%, 10 of 47) or unchanged (28%, 13 of 47) (Fig. 1G; Supplemental Fig. S2A; Supplemental Table S3). In contrast, far fewer of the genes located ≥50 kb from DOX-BrIP sites (distal) were down-regulated by DOX: 21% (four of 19) (Fig. 1G; Supplemental Fig. S2B; Supplemental Table S3). Thus, the genes that overlap with the clusters of neoreplication origins were preferentially down-regulated by DOX in comparison with distal genes. The repression is not a direct effect of topoisomerase II inhibition on the transcription apparatus. First, genes overlapping with HU-induced neoreplication origin clusters (Karnani and Dutta 2011) were similarly repressed by HU, which is not a topoisomerase II inhibitor (Supplemental Fig. S3). Second, when roscovitine prevented the appearance of BrIP clusters in DOX-treated cells (Supplemental Fig. S1B), it also prevented the down-regulation of these overlapping genes (data not shown) even though topoisomerase II continued to be inhibited.
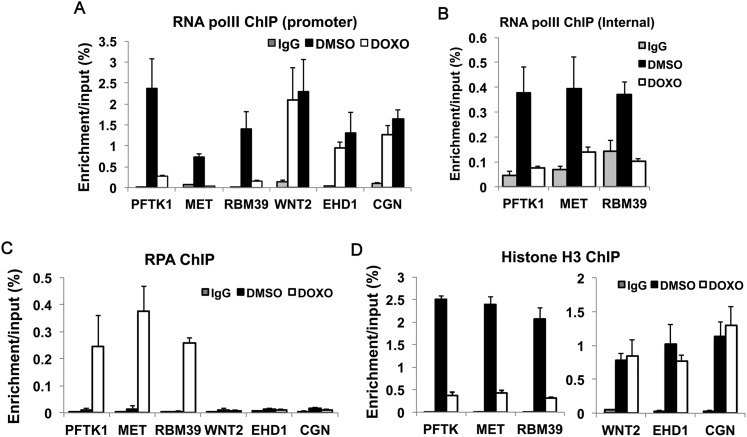
The mRNA decrease could be due to transcriptional or post-transcriptional repression. To distinguish between the two, we measured RNA polymerase II recruitment to three genes (PFTK1, MET, and RBM39) repressed in DOX and overlapping with clusters of neoreplication origins. Chromatin immunoprecipitation (ChIP) showed that DOX significantly decreased RNA polymerase II binding to the promoters (Fig. 2A) and bodies (Fig. 2B) of all three repressed genes. In contrast, RNA polymerase II recruitment to the promoter of a distally located gene, WNT2, was not decreased. Similarly, EHD1 and CGN genes overlapping with a single DOX-BrIP site, but not a cluster, and induced by DOX did not suffer a decrease in RNA polymerase II binding (Fig. 2A). Therefore, a local (geographical) effect of clusters of neoreplication origins may be involved in the specific transcriptional repression of genes overlapping with such clusters while sparing genes elsewhere in the genome.
Figure 2.
DOX produces excess RPA-bound, dechromatinized DNA near sites of stalled forks and disrupts the local recruitment of RNA polymerase II. (A,B) ChIP assay for RNA polymerase II (polII) at the promoter (A) and gene body (B) of three representative genes —(PFTK1, MET, and RBM39) overlapping with BrdU-labeled tracks and negative control genes (in A only). Each value represents a relative DNA concentration based on the standard curve of the input samples. (IgG) Normal IgG ChIP; (DMSO) anti-RNA polymerase II ChIP from DMSO-treated cells; (DOX) anti-RNA polymerase II ChIP from DOX-treated cells. Mean ± standard deviation (SD) of three measurements. (C,D) ChIP assay for RPA70 (C) and histone H3 (D) was conducted as for RNA polymerase II in A.
Stalled replication forks are formed at neoreplication origin cluster-coupled gene loci
We therefore hypothesized that neoreplication origin clusters suppress the transcriptional activity of overlapping genes because of the local accumulation of stalled replication forks. Stalled replication forks give rise to ssDNA that is coated by RPA (Zou and Elledge 2003; Minotti et al. 2004; Cimprich and Cortez 2008). Indeed, many foci of RPA were formed following DOX treatment (Supplemental Fig. S4A). ChIP assay for RPA70 showed increased loading of RPA at the promoters of the three genes overlapping with clusters of neo-origins and repressed in DOX (Fig. 2C). In contrast, no significant binding of RPA was observed at the WNT2, EHD1, and CGN promoters. RFC2, a subunit of the clamp loader present at replication forks, was also enriched at the same sites as RPA (Supplemental Fig. S4B). RPA and RFC2 recruitment indicates that ssDNA and stalled forks accumulate near the neoreplication origins in DOX-treated cells and overlap with the neighboring, transcriptionally repressed genes.
Stalled replication forks are known to evict nucleosomes locally (Groth et al. 2007; Jasencakova et al. 2010). There was a marked decrease in the association of histone H3 at the promoters of the repressed genes overlapping with stalled forks, PFTK, MET, and RBM39 (Fig. 2D), and this, too, was a local effect not seen at other sites, such as ENT2, EHD1, or CGN. The very localized RPA recruitment and histone eviction suggest that DOX-induced clusters of stalled forks are associated with abnormal chromatin structure and excessive ssDNA that may directly prevent RNA polymerase II recruitment. However, the stalled forks also have global effects on the cell, such as the activation of checkpoint enzymes, which will be shown below to also play a role in this repression.
The ATR-dependent checkpoint pathway degrades ASF1a via CRL1βTRCP E3 ligase
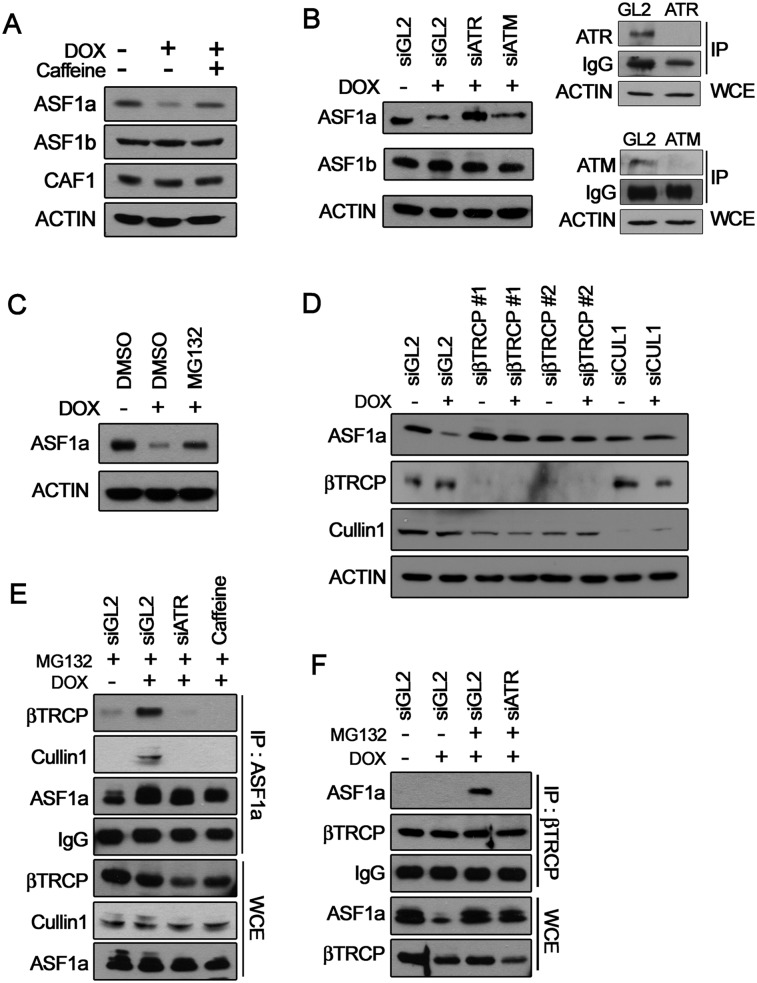
To understand the mechanism of the local dechromatinization associated with stalled replication forks, we examined ASF1, a histone H3/H4 chaperone that mediates nucleosomal chromatin assembly after DNA replication and repair and is involved in transcriptional regulation (Schulz and Tyler 2006; Goodfellow et al. 2007; Groth et al. 2007; Li et al. 2007; Moshkin et al. 2009; Takahata et al. 2009). There are two ASF1 isoforms in human cells: ASF1a and ASF1b. DOX specifically decreased ASF1a (Fig. 3A) but not ASF1b or CAF1, another histone chaperone that acts at replication forks (Hoek and Stillman 2003). Since the protein level of ASF1a increases in a normal S phase (Supplemental Fig. S5A), the decrease in ASF1a in DOX is not simply due to the accumulation of cells in S phase. As expected, the DOX-induced stalled replication forks activated the cell cycle checkpoint, as confirmed by CHK1 phosphorylation, and this activation was reversed by caffeine treatment (Supplemental Fig. S5B; Harrison and Haber 2006). Caffeine (Fig. 3A) and siRNA-mediated depletion of ATR, but not ATM (Fig. 3B), restored the level of ASF1a. Therefore, the decrease of ASF1a is downstream from the ATR-dependent checkpoint pathway.
Figure 3.
DOX induces ATR-CRL1βTRCP-dependent ASF1a degradation. (A) Immunoblots of cell lysates. ASF1a, but not ASF1b or CAF1, was decreased by DOX for 20 h, and this decrease was relieved by 5 mM caffeine added to cells from −4 h relative to DOX until the time of harvest. ACTIN was used as a loading control for the immunoblots. (B, left panel) ASF1a/b examined in cells transfected with siGL2, siATR, or siATM for 24 h and then treated with DOX for 20 h under continued exposure to siRNAs. (Right panel) Knockdown of endogenous ATR or ATM by siRNA was confirmed by immunoprecipitation (IP) and Western blot (WB). The IgG amount in each lane served as the loading control for the immunoprecipitates, and ACTIN shows equal amounts of whole -cell extracts (WCEs). (C) Cells treated with DOX for 20 h were exposed to 10 μM MG132 for 4 h before harvest. Endogenous ASF1a and ACTIN were immunoblotted. (D) Cells transfected with two different siRNAs to βTRCP for 24 h and a siRNA to Cullin1 for 48 h before DOX treatment for 20 h with continued incubation with siRNA and subjected to Western blotting for the indicated proteins and ACTIN (loading control). (E) After transfection of siGL2 or siATR (24 h) or exposure to 5 mM caffeine (4 h), cells were treated with DOX or DMSO for 20 h with continued incubation with siRNA or caffeine. Cells were harvested after exposure to 10 μM MG132 for 4 h to stabilize the ASF1a. Immunoprecipitates (top four panels) or input whole-cell extracts (bottom three panels) were immunoblotted for the indicated proteins. IgG served as a loading control for immunoprecipitates, and Cullin1 served as a loading control for whole-cell extracts. (F) An experiment similar to E, except immunoprecipitation done with anti-βTRCP antibody.
MG132, a proteasome inhibitor, prevented the decrease of ASF1a (Fig. 3C). Proteins targeted to the proteasome are usually polyubiquitinated by a substrate-specific E3 ligase. Since CRL1βTRCP activation has been shown in ATR-activated cells (Frescas and Pagano 2008), we examined whether this E3 ligase is involved in ASF1a degradation in response to DOX. Knockdown of βTRCP or Cullin1 (CRL1) restored ASF1a abundance in DOX (Fig. 3D), but knockdown of another E3 ligase, the CDT2 subunit of CRL4Cdt2, did not stabilize ASF1a (Supplemental Fig. S5C). When ASF1a degradation was prevented by MG132, it coimmunoprecipitated with βTRCP and Cullin1 only in DOX-treated cells (Fig. 3E). Conversely, immunoprecipitation of βTRCP coimmunoprecipitated MG132-stabilized ASF1a in DOX-treated cells (Fig. 3F). The interaction of ASF1a with CRL1βTRCP was disrupted by ATR knockdown or ATR inhibition with caffeine (Fig. 3E,F). These results suggest that the stalled replication forks induced by DOX activate the ATR-mediated checkpoint pathway to promote the interaction of ASF1a with CRL1βTRCP, setting in motion the polyubiquitination and proteasome-mediated global destruction of ASF1a.
Eviction of histone H3 and accumulation of RPA near DOX-induced clusters of stalled forks is dependent on ASF1a
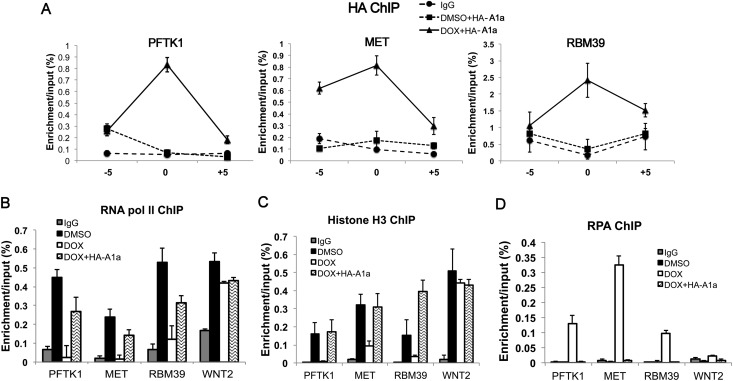
To verify whether the decrease of ASF1a is involved in the eviction of histones and loading of RPA at DOX-induced clusters of stalled forks, we overexpressed ectopic HA-tagged ASF1a (Supplemental Fig. S5D), which did not alter the appearance of BrIP sites in DOX (Supplemental Fig. S5E). The overexpression was sufficient to counter the decrease of endogenous ASF1a upon DOX treatment. The HA-ASF1a was specifically enriched upon DOX treatment in the neighborhood of the stalled forks, with significantly more enrichment at the transcription start site (TSS) than ±5 kb away from the TSS (Fig. 4A). In contrast, we did not see the enrichment of ectopic ASF1a at the negative control genes WNT2, EHD1, and CGN (Supplemental Fig. S5F), suggesting that the ASF1a, if present, is enriched specifically near the stalled replication forks. The ectopically overexpressed ASF1a restored RNA polymerase II and histone H3 to the promoters of the formerly repressed genes while inhibiting the accumulation of RPA (Fig. 4B–D). Therefore, the decrease of ASF1a in DOX-treated cells is responsible for the loss of histones, increase of RPA binding, and blocking of RNA polymerase II recruitment to the promoters of genes overlapping with the clusters of stalled replication forks.
Figure 4.
Ectopic ASF1a localizes near stalled forks, restoring RNA polymerase II and histone H3 while preventing RPA accumulation. (A) ChIP assay for 3HA-ASF1a at PFTK1, MET, and RBM39 was conducted after transfection with 3HA-ASF1a in the presence or absence of DOX. (−5) −5 kb from TSS; (0) TSS; (+5) +5 kb from TSS. The Y-axis indicates the amount of DNA in precipitate relative to input DNA. (B–D) RNA polymerase II (pol II) (B), histone H3 (C), or RPA (D) ChIP assay at the promoters of three genes overlapping with stalled forks and a control distal gene, WNT2.
The ATR-dependent checkpoint pathway is required for transcriptional repression of overlapping genes
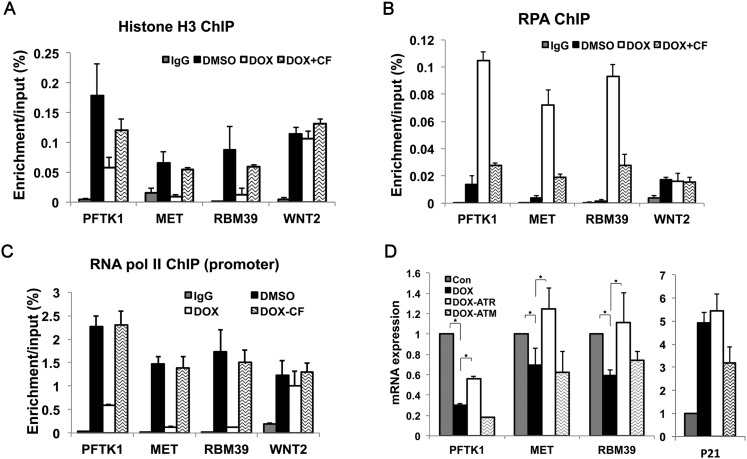
Since the ATR-dependent checkpoint pathway is required for the loss of ASF1a in DOX-treated cells (Fig. 3), we predicted that ATR will also be required for the loss of histones, accumulation of RPA, and loss of RNA polymerase II at DOX-induced clusters of stalled forks. Caffeine prevented the DOX-induced eviction of histone H3 at the promoters of the three genes overlapping with these clusters (Fig. 5A) and blocked the recruitment of RPA at these sites (Fig. 5B). The WNT gene locus, distal from the clusters, was not affected by these manipulations. Additionally, the overall number of cells positive for RPA foci following DOX treatment was decreased following ATR depletion by siRNA (Supplemental Fig. S6A). Caffeine also restored RNA polymerase II at the promoters (Fig. 5C) and bodies (Supplemental Fig. S6B) of the formerly repressed genes overlapping with the clusters of stalled forks with no effect at the promoter of a distal gene, WNT2 (Fig. 5C). Caffeine reversed the mRNA decrease of genes overlapping with the clusters of stalled forks (Supplemental Fig. S6C). Caffeine can inhibit both ATR and ATM. However, siRNA-mediated knockdown showed that ATR, but not ATM, is required for the suppression of mRNA levels following DOX treatment (Fig. 5D). UCN-01 specifically inhibits CHK1, an enzyme downstream from ATR (Graves et al. 2000). UCN-01, but not wortmannin or NU7441, also restored expression of the genes overlapping with stalled forks (Supplemental Fig. S6D–F). Wortmannin at the low doses used in this experiment inhibits DNA-PKcs and ATM but not ATR (Sarkaria et al. 1998), while NU7441 selectively inhibits DNA-PKcs and ATM (Leahy et al. 2004). These results indicate that the ATR–CHK1 checkpoint pathway, and not ATM or DNA-PK, is specifically required for (1) impairing chromatin assembly, (2) increasing ssDNA formation and RPA binding, and (3) preventing the association of RNA polymerase II and gene transcription near stalled replication forks.
Figure 5.
The ATR-dependent checkpoint pathway is required for the transcriptional repression of genes overlapping with clusters of stalled forks. (A,B) Histone H3 (A) and RPA (B) ChIP assay for three genes overlapping with stalled forks and a control distal gene, WNT2. The rest are as in Figure 2. (DOX-CF) DOX-treated cells exposed to caffeine for 24 h before harvesting. (C) RNA polymerase II (pol II) ChIP assay at the promoters of three genes overlapping with stalled forks and a control distal gene, WNT2. (DOX-CF) DOX-treated cells exposed to caffeine for 24 h before harvesting. (D) Cells depleted of ATR or ATM by siRNA were treated with 1.5 μM DOX or DMSO (Con). The relative mRNA expression of three overlapped genes and a control gene, p21, was analyzed by qRT–PCR as in the Materials and Methods. Student’s t-test analysis, (*) P < 0.05.
Interestingly, DOX-induced overexpression of p21 was significantly decreased by depletion of ATM, not ATR (Fig. 5D), suggesting that (1) DSBs also occur in DOX and activate ATM and (2) the DSB-dependent, ATM-mediated checkpoint pathway regulates the transcription of other genes in response to DNA damage. ATM inhibition did not reverse the inhibition of genes overlapping with stalled forks (Fig. 5D), suggesting that most of that inhibition in the vicinity of the stalled forks was not due to ATM or DSBs.
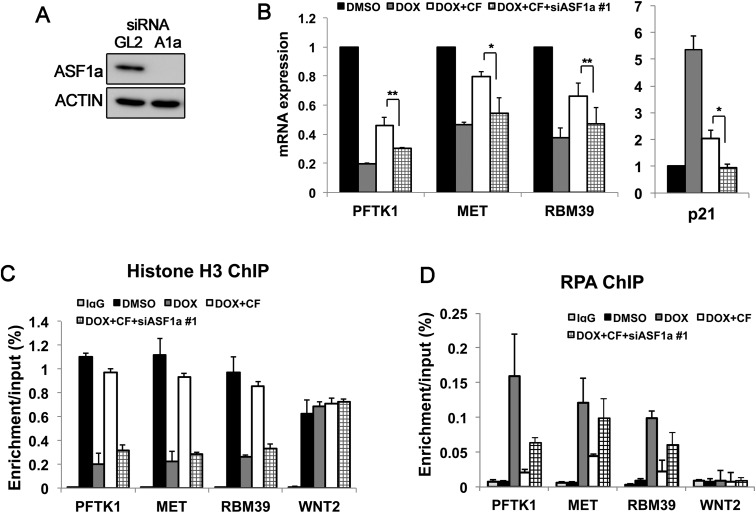
Knockdown of ASF1a antagonizes the recovery of transcription when the ATR-dependent checkpoint pathway is inhibited
If the ASF1a decrease in checkpoint-activated cells is responsible for the phenotypes observed, we reasoned that when the checkpoint is inhibited, restoration of transcription and histones and removal of RPA would be dependent on the restoration of ASF1a. To test this, DOX-treated cells were transfected with ASF1a siRNA for 24 h prior to addition of caffeine to block the checkpoint. mRNA levels were not rescued by caffeine when ASF1a was depleted (Fig. 6A,B). Identical results were obtained with a second siRNA to ASF1a (Supplemental Fig. S7A,B). Since knockdown of ASF1a by siRNA did not affect the locations of DOX-BrIP sites (Supplemental Fig. S7C), this result was not due to the disappearance of DOX-BrIP sites after ASF1a depletion. Histone H3 loading (Fig. 6C) and RPA removal (Fig. 6D) were also impaired when ASF1a was depleted. Thus, ASF1a decrease by itself can phenocopy the effects of checkpoint activation on transcription, chromatin, and ssDNA formation near stalled replication forks.
Figure 6.
The rescue of transcription upon inhibition of checkpoint kinase requires the restoration of ASF1a. (A) Knockdown of ASF1a by siRNA #1 for 24 h evaluated by immunoblotting. (B) mRNA levels of three genes that overlap with clusters of stalled forks analyzed by qRT–PCR. mRNA level normalized to GAPDH. Knockdown of ASF1a prevents the restoration of mRNA levels when the checkpoint is blunted with caffeine. (**) P < 0.01; (*) P < 0.05. (C,D) Chip assay for histone H3 (C) and RPA70 (D) was conducted after treatment of DOX and caffeine, as indicated, with or without knockdown of endogenous ASF1a. WNT2 was a negative control.
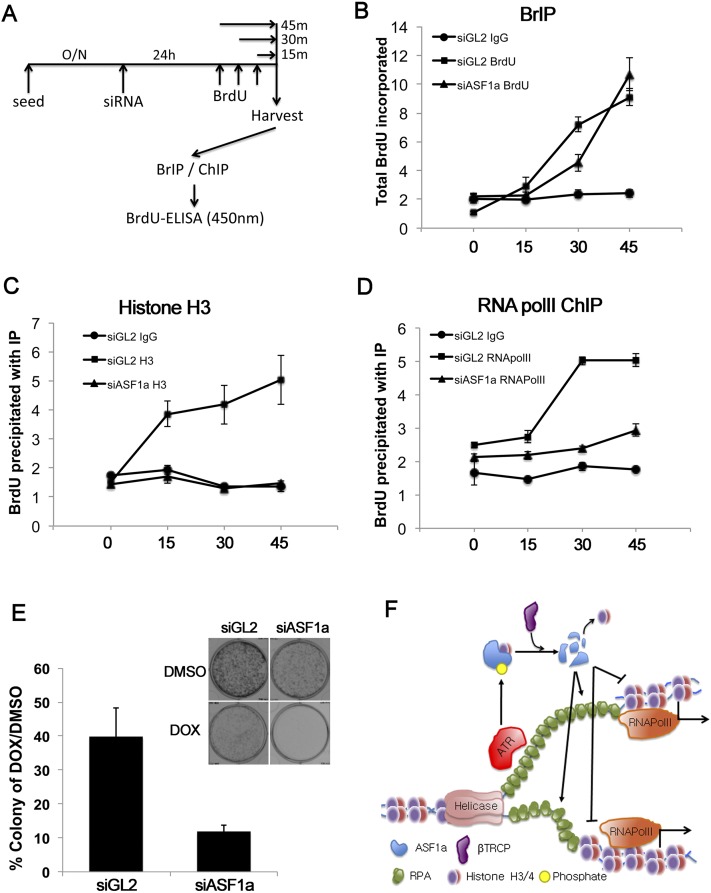
Depletion of ASF1a inhibits the loading of RNA polymerase II on newly replicated DNA
Finally, we asked whether ASF1a is required for the recruitment of RNA polymerase II on newly synthesized DNA even in an unperturbed S phase in the absence of DOX. After 24 h of exposure to siRNA for control GL2 and ASF1a, BrdU was added to the cells for variable periods of time before harvest, and BrdU-incorporated DNA was captured by BrIP or ChIP (Fig. 7A). The DNA eluted from the precipitates was assayed for newly synthesized DNA by ELISA with anti-BrdU antibodies. First, ELISA of BrIP precipitates showed that depletion of ASF1a did not decrease the rate of replication of DNA (Fig. 7B). Depletion of ASF1a decreased the amount of BrdU-incorporated DNA that was precipitated by ChIP with histone H3 (Fig. 7C), consistent with the suggestion that ASF1a is very important for rechromatinization of newly replicated DNA. Depletion of ASF1a also decreased the association of RNA polymerase II with the newly replicated DNA (Fig. 7D). Therefore, ASF1a is essential even in a normal S phase for the rapid rechromatinization and restoration of RNA polymerase II immediately after passage of a replication fork. It is intriguing that yeast Asf1 has a similar rechromatinization role immediately after passage of a transcription bubble (Schwabish and Struhl 2006).
Figure 7.
Depletion of ASF1a inhibits the loading of RNA polymerase II at newly synthesized DNA. (A–D) After 24 h of depletion of ASF1a, BrdU was added during the indicated time (shown in A). BrdU-incorporated DNA was captured by BrIP (B), histone H3 ChIP (C), or RNA polymerase II (polII) ChIP (D). BrdU-incorporated DNA eluted from BrIP or ChIP was analyzed by ELISA with anti-BrdU antibody. (E) The percentage of viable colonies after exposure of HeLa cells to DOX relative to DMSO with or without depletion of ASF1a (left) and representative examples of plates with colonies (top right). Mean ± SD from three independent experiments. (F) Schematic model of transcriptional repression of genes near DOX-induced clusters of stalled replication forks. ASF1a promotes nucleosome loading on the newly replicated DNA. This chromatinization promotes RNA polymerase II loading and suppresses unwinding or nuclease activity on the newly replicated dsDNA. ASF1a is itself degraded by ATR and CRL1βTRCP-dependent pathways activated by RPA-coated ssDNA that is present at stalled forks and that increases as more ssDNA is generated after dechromatinization of the newly replicated DNA.
Knockdown of ASF1a enhances the sensitivity of cancer cells to DOX
DOX is widely used for treating various cancers. Since our data indicated that ASF1a decrease is critical for the deleterious effects of DOX on the chromatin state and transcription in the vicinity of stalled replication forks, we wondered whether HeLa cells would be more sensitive to DOX upon predepletion of ASF1a. Clonogenic colony formation assays measure the long-term viability of cells after treatment with DOX. Depletion of ASF1a significantly sensitized the cells to DOX in this assay (Fig. 7E). While ASF1a alone is important for cell proliferation, we corrected for this effect by normalizing growth in DOX relative to growth in DMSO in the control and ASF1a-depleted cells. Therefore, ASF1a, which regulates chromatin structure soon after the passage of a replication fork, has an important role in protecting our cells from chemotherapeutic drugs like DOX.
Discussion
DOX is widely used in chemotherapy and is believed to act by DNA damage caused by the inhibition of topoisomerase II. We began with genomic studies to identify sites in the genome where DOX produces clusters of stalled replication forks, showed that the clusters tend to overlap on transcribed genes, and discovered that the very same genes are repressed by the clusters due to active dechromatinization and eviction of RNA polymerase II. The dechromatinization directed our attention to histone chaperones, and we discovered that one of them, ASF1a, is actively degraded by an ATR checkpoint-induced pathway that targets ASF1a to CRL1βTRCP E3 ubiquitin ligase. We then showed that restoring the ASF1a or suppressing the ATR–Chk1 checkpoint pathway prevents the dechromatinization and gene repression. Conversely, siRNA-mediated depletion of ASF1a phenocopied the effect of the checkpoint. Finally, we showed that even in a normal S phase, ASF1a is critical for the rechromatinization and restoration of RNA polymerase II on newly synthesized DNA soon after passage of the replication fork.
Thus, we present a novel mechanism in which DOX produces clusters of stalled replication forks at defined sites in the genome to activate checkpoint pathways that degrade ASF1a by CRL1βTRCP and proteasomes. Since ASF1a is present at replication forks and is required for the rapid chromatinization and RNA polymerase II re-loading after passage of a replication fork, the checkpoint-induced ASF1a degradation specifically leads to dechromatinization of and RNA polymerase II eviction from the newly replicated DNA. We suggest that the naked, newly replicated DNA is more susceptible to nucleases and helicases, leading to the production of more ssDNA in these same regions, perhaps contributing to the RNA polymerase II eviction (model in Fig. 7F). This is an interesting example of how a globally active checkpoint pathway is integrated by localized clusters of stalled replication forks to produce very local changes in chromatin and gene expression. It is also a novel mechanism of action of the commonly used anti-cancer drug DOX.
As explained above, the checkpoint-induced ASF1a degradation leads to the production of more ssDNA over the newly replicated DNA and thus contributes to more RPA binding. Since RPA-coated ssDNA is necessary to activate ATR, our results suggest a positive feedback loop by which activated ATR decreases ASF1a, leading to further loading of RPA and further activation of ATR. Our findings provide another mechanism by which ATR could, as reported, facilitate RPA loading at DNA damage foci and on chromatin in mammals and yeast (Barr et al. 2003; Cobb et al. 2005).
Previously, we characterized clusters of neoreplication origins in HU-treated HeLa cells in the 1% of the genome studied by the ENCODE pilot project. There was a strong concordance in the location of HU-induced and DOX-induced BrIP sites and clusters, suggesting that particular genomic regions are susceptible to firing normally dormant origins in response to a variety of DNA-damaging agents. The clusters of neo-origins fire preferentially in areas of the genome that are rich in transcriptionally active genes. Since transcriptional repression is a local phenomenon seen near the clusters of stalled forks, it will be interesting to assess whether the clusters appear at the same sites (genes) in cancer cells of different lineages with different gene expression patterns. If the sites (genes) affected are lineage-specific, DOX and other DNA-damaging chemotherapy agents could have lineage-specific toxicities due to differences in which genes are repressed by the mechanism described in this study.
Clusters of stalled forks are expected to not only produce ssDNA but also give rise to chromosome breaks or double-strand ends formed by fork regression. DSB-induced, ATM-dependent signaling is known to inhibit the promoter activity of RNA polymerase II reporter genes due to suppression of transcription elongation (Shanbhag et al. 2010). The kinase activity of ATM also inhibits RNA polymerase I following DSBs (Kruhlak et al. 2007). Inhibition of RNA polymerase II-dependent gene transcription following I-PpoI-induced DNA breakage is dependent on DNA-PKcs (Pankotai et al. 2012). Although we expected clusters of stalled forks to give rise to double-strand ends and activate ATM and DNA-PKcs, the checkpoint pathway reported here is exclusively mediated by ATR but not ATM and DNA-PKcs. Thus, the dechromatinization and RNA polymerase II eviction that are studied here are mostly due to excess ssDNA, leading to activation of ATR and CHK1 and decrease of ASF1a. However, our results do not rule out additional changes in the cell from ATR or even from the activation of ATM or DNA-PKcs upon DOX treatment.
ASF1 is a well-known histone chaperone that mediates chromatin assembly during DNA replication and repair (Tyler et al. 1999; Groth et al. 2005) and also modulates transcription (Schwabish and Struhl 2006; Minard et al. 2011). Yeast Asf1 is directly involved in the recruitment of RNA polymerase II to promoters and in the acetylation of Lys56 (K56) on newly synthesized H3 histone deposited on chromosomes during S phase (Recht et al. 2006; Schwabish and Struhl 2006). ASF1-mediated H3 acetylation is important for regulating gene transcription in yeast (Xu et al. 2005; Williams et al. 2008; Lin and Schultz 2011; Minard et al. 2011), Drosophila, and human cells (Das et al. 2009; Yuan et al. 2009; Battu et al. 2011). Our data demonstrate yet another context in which ASF1 regulates gene transcription. In the vicinity of replication forks, ASF1a, not ASF1b or CAF1, directly promotes chromatinization, RNA polymerase II binding, and transcription.
The two different isoforms of human ASF1—ASF1a (205a.a) and ASF1b (203a.a)—show a 71% identity with yeast ASF1 in the N-terminal region that associates with the H3–H4 histones but not in the C-terminal regions (Sillje and Nigg 2001; De Koning et al. 2007). Despite their highly conserved N-terminal regions and many similarities in cellular roles, ASF1a possesses functions different from ASF1b, as observed in this study. ASF1b, unlike ASF1a, was not decreased by the checkpoint and was not necessary for the restoration of transcription when the checkpoint was inhibited by caffeine (data not shown). Others have noted similar differences between ASF1a and ASF1b. S-phase-specific phosphorylation of ASF1a has been preferentially observed in HeLa cells compared with ASF1b, and this phosphorylation was regulated by DNA damage such as IR and UV (Sillje and Nigg 2001; Groth et al. 2003). ASF1a, but not ASF1b, regulates UV-induced checkpoint recovery through ATM and histone H3-K56 acetylation (Battu et al. 2011). On the other hand, only ASF1b can compensate for the growth defects of yeast lacking endogenous Asf1 (Tamburini et al. 2005); ASF1b also has a greater ability to increase breast cancer cell proliferation (Corpet et al. 2011).
The decrease of human ASF1a in response to stalled replication forks has some parallel to the inhibition of Asf1-controlled histone deposition after DNA damage in yeast, but the latter is achieved by phosphorylation of Rad53 (Chk2 equivalent) disrupting the interaction of Rad53 with Asf1 (Emili et al. 2001; Hu et al. 2001; Mousson et al. 2007; Jiao et al. 2012). In contrast, the ATR–CHK1 pathway activated by stalled replication forks in human cells appears to polyubiquitinate ASF1a by CRL1βTRCPE3 ligase, leading to degradation by proteasomes. Although DNA damage in mammals activates Tousled-like kinases (TLK1/2) through checkpoint kinases, and TLKs phosphorylate ASF1a (Sillje and Nigg 2001; Groth et al. 2003; Pilyugin et al. 2009), we failed to implicate TLKs in the decrease of ASF1a in DOX-treated cells (data not shown). CRL1βTRCP is well known to target several proteins, such as CDC25A/B and Claspin, to regulate the response to DNA damage and replication stress in mammalian cells (Busino et al. 2003; Jin et al. 2003; Frescas and Pagano 2008). ASF1a can now be added as an important target of the ATR–CHK1–βTRCP pathway in response to a high density of stalled replication forks.
DOX (Adriamycin), a commonly used chemotherapeutic agent, is an anthracycline antibiotic known to impede DNA replication by inhibiting DNA topoisomerase II (Bodley et al. 1989). An important result in this study is that knockdown of ASF1a enhances the sensitivity of cancer cell lines to DOX (Fig. 7E). While we cannot be certain that the additive toxicity of ASF1a depletion and DOX treatment is due to the role of ASF1a at the replication forks, the biochemical data provided in this study are supportive of such a hypothesis. Regardless of this caveat, the increased DOX sensitivity in ASF1a-depleted cells along with the reports of homozygous deletion of ASF1a in several human tumors (Supplemental Table S4) lead to an interesting possibility: As personalized therapy of cancers directed by sequencing of tumor DNA and RNA becomes a reality, it will be useful to evaluate whether loss of ASF1a predicts a superior responsiveness of these cancers to DOX.
Materials and methods
Cell culture, inhibitors, and siRNAs
HeLa cells, human cervical adenocarcinoma cells, were cultured in DMEM supplemented with 10% FBS and antibiotics at 37°C in a humidified incubator containing 5% CO2. Five millimolar caffeine (Sigma), 10 μM roscovitine (Sigma), 300 nM UCN-01 (Sigma), 1 μM wortmannin (Sigma), and 1 μM NU7441 (SelleckChem) were added in culture medium. All siRNAs were chemically synthesized from Invitrogen except for siATR and siATM (Dharmacon). Transfections of siRNAs were carried out with oligofectamine reagents following the manufacturer’s instruction. The sense strand sequences of siRNAs used in this study are listed in the Supplemental Material.
BrIP for hybridization to genome tiling microarray
The BrIP and hybridization to microarrays were performed as described in our previous reports (Karnani et al. 2010). For details, see the Supplemental Material.
Identification of clusters of BrIP sites and comparison of their locations with transcriptional units
These analyses were performed as previously described (Karnani and Dutta 2011). To identify the transcriptionally active regions in the HeLa cell genome, we used paired-end high-throughput whole-cell RNA-seq data from HeLaS3 cells. For details, see the Supplemental Material.
BrIP assay on asynchronous cells
We performed the BrIP assay as previously described with some modifications (Karnani et al. 2010). Asynchronous cells were incubated in 1.5 μM DOX for 20 h along with 100 μM BrdU for the last 14 h. The immunoprecipitated DNA–antibodies complex was washed once with 1× immunoprecipitation buffer and incubated in digestion buffer containing 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 0.5% SDS, and 250 μg/mL proteinase K for 2 h at 50°C. The eluted DNA was analyzed by qPCR using an ABI 7300 real-time PCR system (Applied Biosystems) or ELISA assay. The signal value in each experiment represents a relative DNA concentration based on the standard curve of the input samples. Error bars represent the standard deviation of the mean from triplicates. The result was confirmed by three independent experiments. Primer sequences for qPCR are provided in Supplemental Table S5.
qRT–PCR
To analyze the expression of target genes, total cellular RNA was purified from cells using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. cDNA was generated from 2 μg of RNA using the Superscript III first strand synthesis system (Invitrogen) and subjected to qRT–PCR using an ABI 7300 real-time PCR system (Applied Biosystems). The relative expression of each gene was normalized to a housekeeping gene, GAPDH, and analyzed by the ΔΔCT method (Schmittgen and Livak 2008). All bar graphs represent the average of three independent experiments. Error bars indicate the standard deviation of the mean, and statistical significance is shown using the Student’s t-test analysis: P < 0.001 (***), P < 0.01 (**),and P < 0.05 (*). Primer sequences are provided in Supplemental Table S5.
ChIP assay
The ChIP assay was performed as previously described with slight modifications (Negishi et al. 2010). Detailed methods are provided in the Supplemental Material.
Immunoprecipitation and immunoblotting
For immunoprecipitation, whole-cell extracts from cells were resuspended in buffer containing 20 mM Tris-HCl (pH 7.5), 100 mMNaCl, 1% Nonidet P-40, 5% glycerol, 1 mM EDTA, 1 mM MgCl2, 1 mM ATP, 1 mM DTT, 10 mM NaF, 1 mM sodium vanadate, and protease inhibitors. One milligram of total cell extract was incubated with the indicated antibodies and immunoprecipitated with protein G-conjugated agarose beads (GE Healthcare). For immunoblotting, ∼30–50 μg of protein was subjected to SDS-PAGE analysis. Antibodies against each protein were purchased from the following companies: CHK1, Cyclin A, Cyclin E, CUL1, and CAF1 from Santa Cruz Biotechnology; p-CHK1(Ser345), ASF1a, ASF1b, and βTRCP from Cell Signaling Technology; ATR from Affinity Bioreagent; ATM from Novus Biological; and BRCA1 from Milipore.
BrdU ELISA assay
BrdU-incorporated DNA was attached on a 96-well plate coated with poly-L-lysine (Becton Dickinson) and bound with POD-conjugated anti-BrdU antibody (Roche) for 30 min. After washing unbound antibodies with PBS, bound POD was activated by ultra-TMB-ELISA solution (Thermo Scientific), and the reaction was stopped after 5 min with 2 M H2SO4. The colorimetric signal was measured at a 450-nm wavelength in an ELISA plate reader (BioTek). The reading from a blank well was subtracted from all measurements, and error bars represent the standard deviation from three technical replicates. The result was confirmed by two independent measurements.
Clonogenic colony formation assay
After transfection of the indicated siRNA, HeLa cells were plated in six-well plates and treated with 1.5 μM DOX for 1 h. At 7 d after initial DNA damage, the HeLa cell colonies were stained with crystal violet and quantitated using GeneTools software (Syngene).
Acknowledgments
We thank Dr. Y.M. Moshkin for providing 3HA-ASF1a plasmid, and members of the Dutta laboratory for helpful discussions. We also thank Dr. Laura Dillon for critical editing of the manuscript. This work was supported by R01 CA166054 and CA60499 to A.D.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.239194.114.
References
- Ball HL, Myers JS, Cortez D 2005. ATRIP binding to replication protein A–single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell 16: 2372–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr SM, Leung CG, Chang EE, Cimprich KA 2003. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr Biol 13: 1047–1051 [DOI] [PubMed] [Google Scholar]
- Battu A, Ray A, Wani AA 2011. ASF1A and ATM regulate H3K56-mediated cell-cycle checkpoint recovery in response to UV irradiation. Nucleic Acids Res 39: 7931–7945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Bodley A, Liu LF, Israel M, Seshadri R, Koseki Y, Giuliani FC, Kirschenbaum S, Silber R, Potmesil M 1989. DNA topoisomerase II-mediated interaction of doxorubicin and daunorubicin congeners with DNA. Cancer Res 49: 5969–5978 [PubMed] [Google Scholar]
- Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF 2003. Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature 426: 87–91 [DOI] [PubMed] [Google Scholar]
- Capranico G, Kohn KW, Pommier Y 1990. Local sequence requirements for DNA cleavage by mammalian topoisomerase II in the presence of doxorubicin. Nucleic Acids Res 18: 6611–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D 2008. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM 2005. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet A, De Koning L, Toedling J, Savignoni A, Berger F, Lemaitre C, O’Sullivan RJ, Karlseder J, Barillot E, Asselain B, et al. 2011. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J 30: 480–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294: 1713–1716 [DOI] [PubMed] [Google Scholar]
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M 2008. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 455: 557–560 [DOI] [PubMed] [Google Scholar]
- Das C, Lucia MS, Hansen KC, Tyler JK 2009. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning L, Corpet A, Haber JE, Almouzni G 2007. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol 14: 997–1007 [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elford HL 1968. Effect of hydroxyurea on ribonucleotide reductase. Biochem Biophys Res Commun 33: 129–135 [DOI] [PubMed] [Google Scholar]
- Emili A, Schieltz DM, Yates JR 3rd, Hartwell LH 2001. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol Cell 7: 13–20 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D, Pagano M 2008. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat Rev Cancer 8: 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Blow JJ 2010. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J Cell Biol 191: 1285–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ 2007. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev 21: 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow H, Krejci A, Moshkin Y, Verrijzer CP, Karch F, Bray SJ 2007. Gene-specific targeting of the histone chaperone asf1 to mediate silencing. Dev Cell 13: 593–600 [DOI] [PubMed] [Google Scholar]
- Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O’Connor PM, Piwnica-Worms H 2000. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem 275: 5600–5605 [DOI] [PubMed] [Google Scholar]
- Groth A, Lukas J, Nigg EA, Sillje HH, Wernstedt C, Bartek J, Hansen K 2003. Human Tousled like kinases are targeted by an ATM- and Chk1-dependent DNA damage checkpoint. EMBO J 22: 1676–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G 2005. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol Cell 17: 301–311 [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G 2007. Chromatin challenges during DNA replication and repair. Cell 128: 721–733 [DOI] [PubMed] [Google Scholar]
- Harrison JC, Haber JE 2006. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40: 209–235 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH 2001. Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374 [DOI] [PubMed] [Google Scholar]
- Hoek M, Stillman B 2003. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci 100: 12183–12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Alcasabas AA, Elledge SJ 2001. Asf1 links Rad53 to control of chromatin assembly. Genes & Dev 15: 1061–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Qian C, Jenkins RB, Smith DI 1998. Fish mapping of YAC clones at human chromosomal band 7q31.2: identification of YACS spanning FRA7G within the common region of LOH in breast and prostate cancer. Genes Chromosomes Cancer 21: 152–159 [DOI] [PubMed] [Google Scholar]
- Ibarra A, Schwob E, Mendez J 2008. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci 105: 8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z, Scharf AN, Ask K, Corpet A, Imhof A, Almouzni G, Groth A 2010. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell 37: 736–743 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Seeger K, Lautrette A, Gaubert A, Mousson F, Guerois R, Mann C, Ochsenbein F 2012. Surprising complexity of the Asf1 histone chaperone-Rad53 kinase interaction. Proc Natl Acad Sci 109: 2866–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, Harper JW 2003. SCFβ-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes & Dev 17: 3062–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Dutta A 2011. The effect of the intra-S-phase checkpoint on origins of replication in human cells. Genes Dev 25: 621–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A 2010. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell 21: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R 2007. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 447: 730–734 [DOI] [PubMed] [Google Scholar]
- Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC 2004. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett 14: 6083–6087 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Lin LJ, Schultz MC 2011. Promoter regulation by distinct mechanisms of functional interplay between lysine acetylase Rtt109 and histone chaperone Asf1. Proc Natl Acad Sci 108: 19599–19604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP 2004. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev 18: 3094–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minard LV, Williams JS, Walker AC, Schultz MC 2011. Transcriptional regulation by Asf1: new mechanistic insights from studies of the DNA damage response to replication stress. J Biol Chem 286: 7082–7092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L 2004. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev 56: 185–229 [DOI] [PubMed] [Google Scholar]
- Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP 2009. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell 35: 782–793 [DOI] [PubMed] [Google Scholar]
- Mousson F, Ochsenbein F, Mann C 2007. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma 116: 79–93 [DOI] [PubMed] [Google Scholar]
- Negishi M, Saraya A, Mochizuki S, Helin K, Koseki H, Iwama A 2010. A novel zinc finger protein Zfp277 mediates transcriptional repression of the Ink4a/arf locus through polycomb repressive complex 1. PLoS ONE 5: e12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T, Bonhomme C, Chen D, Soutoglou E 2012. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol 19: 276–282 [DOI] [PubMed] [Google Scholar]
- Pilyugin M, Demmers J, Verrijzer CP, Karch F, Moshkin YM 2009. Phosphorylation-mediated control of histone chaperone ASF1 levels by Tousled-like kinases. PLoS ONE 4: e8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. 2006. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci 103: 6988–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT 1998. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res 58: 4375–4382 [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schulz LL, Tyler JK 2006. The histone chaperone ASF1 localizes to active DNA replication forks to mediate efficient DNA replication. FASEB J 20: 488–490 [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell 22: 415–422 [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM 2007. Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M 2009. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 5: e1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141: 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillje HH, Nigg EA 2001. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr Biol 11: 1068–1073 [DOI] [PubMed] [Google Scholar]
- Takahata S, Yu Y, Stillman DJ 2009. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell 34: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini BA, Carson JJ, Adkins MW, Tyler JK 2005. Functional conservation and specialization among eukaryotic anti-silencing function 1 histone chaperones. Eukaryot Cell 4: 1583–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402: 555–560 [DOI] [PubMed] [Google Scholar]
- Williams SK, Truong D, Tyler JK 2008. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci 105: 9000–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhang K, Grunstein M 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121: 375–385 [DOI] [PubMed] [Google Scholar]
- Yuan J, Pu M, Zhang Z, Lou Z 2009. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle 8: 1747–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]