Intracellular proteases combat proteotoxic stress by degrading damaged proteins, but their activity must be controlled to maintain cellular fitness. DegP is a conserved periplasmic protease essential for E. coli growth at high temperatures. Kim and Sauer investigate how allosteric activation and polyhedral cage formation contribute to DegP function and cellular fitness. The results suggest that allosteric control of active and inactive conformations is the primary mechanism regulating DegP proteolysis and cellular fitness, with cage assembly providing an additional layer of protection against excessive protease activity.
Keywords: protein quality control, proteolysis, cellular fitness, DegP, HtrA, Lpp
Abstract
Intracellular proteases combat proteotoxic stress by degrading damaged proteins, but their activity must be carefully controlled to maintain cellular fitness. The activity of Escherichia coli DegP, a highly conserved periplasmic protease, is regulated by substrate-dependent allosteric transformations between inactive and active trimer conformations and by the formation of polyhedral cages that confine the active sites within a proteolytic chamber. Here, we investigate how these distinct control mechanisms contribute to bacterial fitness under heat stress. We found that mutations that increase or decrease the equilibrium population of active DegP trimers reduce high-temperature fitness, that a mutation that blocks cage formation causes a mild fitness decrease, and that combining mutations that stabilize active DegP and block cage formation generates a lethal rogue protease. This lethality is suppressed by an extragenic mutation that prevents covalent attachment of an abundant outer-membrane lipoprotein to peptidoglycan and makes this protein an inhibitor of the rogue protease. Lethality is also suppressed by intragenic mutations that stabilize inactive DegP trimers. In combination, our results suggest that allosteric control of active and inactive conformations is the primary mechanism that regulates DegP proteolysis and fitness, with cage formation providing an additional layer of cellular protection against excessive protease activity.
Protein degradation is an essential cellular function. In the absence of degradation, toxic misfolded proteins or aggregates can accumulate after heat shock or other environmental stresses, resulting in reduced viability or cell death. Protein quality control (PQC) proteases play critical biological roles by degrading diverse misfolded proteins but also require careful regulation because proteolysis of functional proteins can be cytotoxic. Intracellular proteases whose activities are allosterically regulated have evolved to meet these conflicting needs.
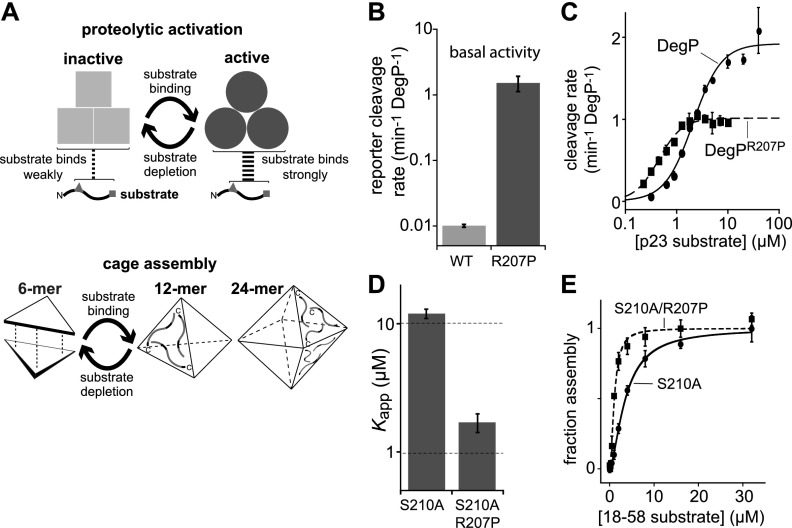
DegP is a member of the highly conserved HtrA family of proteases, is overexpressed at high temperatures as the major PQC protease in the periplasmic compartment of Escherichia coli and many other Gram-negative bacteria, and is implicated in the virulence of several pathogens (Lipinska et al. 1989; Strauch et al. 1989; Pallen and Wren 1997; Clausen et al. 2011). Each subunit of DegP contains a trypsin-like protease domain and two PDZ domains (Krojer et al. 2002, 2008b; Kim et al. 2011). The protease domains of three subunits pack together to form a stable trimer, which can switch between a proteolytically active conformation with high substrate affinity and a proteolytically inactive conformation with low substrate affinity (Fig. 1A; Kim et al. 2011). DegP trimers display positively cooperative substrate binding to the active enzyme, a hallmark of Monod-Wyman-Changeux (MWC) allostery, which allows proteolysis to be controlled. Substrate binding also triggers the assembly of DegP trimers and hexamers into large polyhedral cages that restrict the access of native proteins to the active sites within these cages (Fig. 1A; Jiang et al. 2008; Krojer et al. 2008b; Kim et al. 2011).
Figure 1.
The R207P mutation stabilizes the high-affinity active conformation of DegP. (A) Substrate-binding triggers proteolytic activation and cage assembly. The fundamental unit of DegP structure is a trimer, which reversibly transforms between an inactive conformation with low substrate affinity and an active conformation with high substrate affinity. DegP trimers and hexamers also assembled into cages with 12 or 24 subunits. (B) Basal cleavage of 100 µM Abz-KASPVSLGYNO2D ([Abz] 2-aminobenzoic acid; [YNO2] 3-nitrotyrosine), a poor substrate, by 10 µM DegP or 1 µM DegPR207P. Values are averages ± 1 SD (n = 3). (C) Cleavage rates of the p23 substrate by DegP or DegPR207P were measured at different substrate concentrations at room temperature (22°C). Values are averages ± 1 SD (n = 3). Lines are fits to the Hill equation: rate = Vmax/{1 + (KM/[p23])h}. For DegP, Vmax = 1.9 ± 0.1, KM = 2.1 ± 0.2, and h = 1.6 ± 0.2 (Kim and Sauer 2012). For DegPR207P, Vmax = 1.0 ± 0.03, KM = 0.45 ± 0.03, and h = 1.9 ± 0.2. (D) Apparent dissociation constants (Kapp) of a model substrate (flC18–58) for proteolytically inactive DegPS210A/R207P (1.7 µM ± 0.3 µM) or DegPS210A (12 µM ± 1 µM) (Kim et al. 2011) were measured by fluorescence anisotropy. Bars represent the nonlinear least squares fitting error. (E) Substrate-dependent assembly of DegPS210A or DegPS210A/R207P cages was monitored by Förster resonance energy transfer (FRET) between proteins labeled with acceptor and donor dyes (Kim et al. 2011). Values are averages ± 1 SD (n = 3). Lines are fits to a Hill equation. For DegPS210A, Kapp = 3.6 ± 0.2, and h = 1.6 ± 0.1. For DegPS210A/R207P, Kapp = 1.1 ± 0.1, and h = 1.9 ± 0.2.
Distinct modes of substrate binding control DegP activation and assembly. In unfolded or misfolded substrates, two degron sequences in a single polypeptide interact with DegP to promote allosteric activation (Kim et al. 2011). Substrate residues flanking scissile peptide bonds bind to the active site cleft and oxyanion hole of the active protease domain, whereas a hydrophobic residue at the substrate C terminus binds to a site in the PDZ1 domain. These linked interactions also result in the assembly of trimers into hollow polyhedral cages with 12, 18, or 24 subunits by stabilizing interactions between the PDZ1 domains of one trimer and the PDZ2′ domains of neighboring trimers (Kim and Sauer 2012). Substrate cleavage eventually generates peptide products that bind too weakly to maintain DegP in the active conformation, resulting in cage disassembly and a return to the inactive conformation. Thus, bivalent substrate binding helps ensure that DegP is activated and assembled only in the presence of appropriate substrates.
Several observations demonstrate the coupling of DegP activation and cage assembly. Crystal structures are known for a DegP hexamer in which trimers assume an inactive conformation with a malformed Ser–His–Asp catalytic triad and oxyanion hole and for substrate-bound cages with 12 or 24 subunits in which trimers assume an active conformation (Krojer et al. 2002, 2008b; Kim et al. 2011). In solution, proteolytic activation and cage assembly of DegP also occur synchronously (Krojer et al. 2008b; Kim et al. 2011), which led to the proposal that cage assembly was the molecular switch for activation (Krojer et al. 2008b). We found, however, that cage assembly was not required for either proteolytic activity in vitro or suppression of proteotoxic heat stress in vivo (Kim and Sauer 2012).
At present, it is unclear how allosteric activation and cage assembly contribute to DegP function in vivo and cellular fitness. Here, we show that mutations that increase or decrease the equilibrium population of active DegP trimers result in small decreases in high-temperature fitness. Combining a mutation that allosterically activates DegP with one that prevents cage formation converts the enzyme into a lethal rogue protease. This lethality appears to be caused by excessive proteolysis of proteins associated with cell envelope maintenance and can be relieved by an extragenic suppressor that alters the cell envelope and directly inhibits the rogue protease or by intragenic mutations that stabilize inactive DegP trimers. We propose that substrate-dependent allosteric control of the conformation and activity of DegP trimers represents the primary form of proteolytic regulation, whereas cage assembly protects cells from promiscuous proteolysis that would otherwise be triggered under heat stress and other conditions that result in protein misfolding and allosteric activation of DegP.
Results
A mutation that stabilizes active DegP
To test the consequences of increasing the equilibrium population of functional DegP enzymes, we sought to identify a mutation that preferentially stabilizes the active state. Because DegP mainly adopts an inactive conformation without substrate, mutational stabilization of the active conformation should result in higher basal proteolytic activity and tighter substrate binding (Fig. 1A). In DegS, a trimeric DegP paralog, the H198P mutation results in exactly this phenotype by stabilizing the active conformation of the oxyanion hole (Sohn and Sauer 2009; Sohn et al. 2010). Modeling suggested that the homologous R207P mutation in DegP could also stabilize the active conformation, and thus we constructed and purified DegPR207P for biochemical characterization.
Several results established that the R207P mutation increased the equilibrium fraction of active enzymes compared with wild-type DegP. First, DegPR207P had substantially higher basal activity in cleaving a poor peptide substrate (Fig. 1B). This reporter peptide binds weakly to active DegP and converts very little wild-type enzyme into the active conformation (Kim et al. 2011). Second, DegPR207P cleaved a good peptide substrate derived from lysozyme (p23) (Kim and Sauer 2012) with a lower KM (∼0.5 µM) than wild-type DegP (∼2 µM) (Fig. 1C), as expected if more DegPR207P trimers assume the active high-affinity conformation in the absence of substrate. At saturating concentrations of p23, DegPR207P had lower maximum cleavage activity than wild-type DegP (Fig. 1C), establishing that the higher basal cleavage activity of DegPR207P is not a result of intrinsically increased proteolytic activity. Third, DegPS210A/R207P bound a fluorescent peptide derived from residues 18–58 of lysozyme (flC18–58) more tightly (dissociation constant [Kapp] ∼1.7 µM) than DegPS210A (Kapp ∼12 µM) (Fig. 1D; Kim et al. 2011). In this assay, the S210A mutation was used to remove the nucleophilic serine from the catalytic triad and prevent degradation. Fourth, the R207P mutation resulted in cage formation at lower concentrations of substrate than required for cage assembly of otherwise identical proteins lacking R207P (Fig. 1E), as assayed in the S210A background by Förster resonance energy transfer (FRET) between trimers labeled with donor dyes and trimers labeled with acceptor dyes (Kim et al. 2011).
A dominant lethal DegP protease
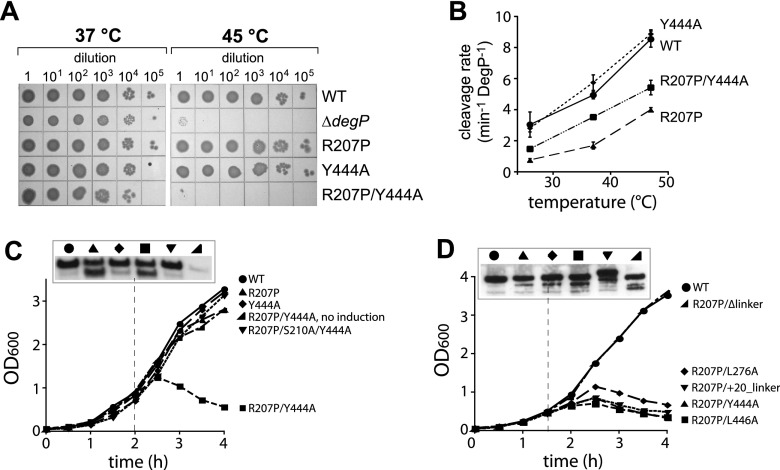
To test the cellular effects of R207P and DegP mutations that affect cage formation, we constructed E. coli strains with mutant alleles in chromosomal degP and assayed bacterial growth on Luria-Bertani (LB) agar plates at 37°C or 45°C. The latter temperature induces heat stress and high-level production of DegP. As expected from previous studies (Lipinska et al. 1989; Strauch et al. 1989), the ΔdegP strain grew at 37°C but not at 45°C (Fig. 2A). Both the degP+ strain and the otherwise isogenic degPR207P strain grew reasonably well at both temperatures (Fig. 2A), although the degPR207P strain showed reduced viability at 47°C (Supplemental Fig. S1A). DegPY444A, in which a mutation in the PDZ2 domain prevents cage assembly and makes the enzyme constitutively trimeric, supported growth at 37°C, 45°C, and 47°C (Fig. 2A; Supplemental Fig. S1A; Kim and Sauer 2012). Notably, however, the double-mutant degPR207P/Y444A strain was inviable at 45°C and 47°C but not at 37°C (Fig. 2A; Supplemental Fig. S1A). Purified DegPR207P/Y444A efficiently cleaved the p23 substrate at a rate faster than DegPR207P but slower than wild-type DegP or DegPY444A at 26°C, 37°C, and 47°C (Fig. 2B), suggesting that the high-temperature inviability of strains expressing DegPR207P/Y444A is not caused by the absence of proteolytic activity. Moreover, wild-type DegP, DegPR207P, DegPY444A, and DegPR207P/Y444A were expressed at similar levels at high temperature (Supplemental Fig. S1B), and thus the inability of the double mutant to support growth at 45°C is unlikely to be caused by a reduced concentration of enzyme.
Figure 2.
DegPR207P/Y444A is a rogue protease that dominantly kills cells. (A) Strains with different chromosomal degP alleles were grown to log phase in LB broth, serially diluted, spotted on LB agar plates, and grown overnight at 37°C or 45°C. (B) Cleavage rates of a saturating concentration of the p23 substrate (40 µM) by different DegP variants (0.5 µM) were determined at 26°C, 37°C, and 47°C. Values are averages ± 1 SD (n = 3). (C,D) A degP+ E. coli strain (W3110) was transformed with plasmids expressing wild-type DegP or variants under arabinose control. Overnight cultures grown in LB broth plus 10 µM/mL chloramphenicol were diluted 100-fold in the same medium plus or minus 0.2% arabinose, and growth at 37°C was monitored by OD600. The panels above the growth curves show DegP expression levels after 2 h (C) or 1.5 h (D), as determined by SDS-PAGE of equal quantities of cells and Western blotting using an anti-DegP antibody. The top band is full-length DegP; the bottom band arises from DegP autocleavage that does not affect activity.
To test for dominance, we cloned genes encoding DegP, DegPR207P, DegPY444A, DegPR207P/Y444A, and DegPR207P/S210A/Y444A into plasmids under transcriptional control of an arabinose-inducible PBAD promoter and transformed E. coli W3110, a strain that expresses wild-type DegP from the chromosome. Strains were grown overnight at 37°C without arabinose and diluted in medium containing 0.2% arabinose (except for one control culture), and growth at 37°C was monitored by OD600 (Fig. 2C). All strains grew at similar rates for the first ∼2.5 h, but the cell density of the strain expressing DegPR207P/Y444A then decreased, an indication of cell lysis. In contrast, no major growth defects were observed for strains expressing DegP, DegPR207P, or DegPY444A from the plasmid or for a strain in which expression of DegPR207P/Y444A was not induced (Fig. 2C). Expression of proteolytically inactive DegPR207P/S210A/Y444A also did not interfere with growth, indicating that proteolysis is required for the dominant lethality of DegPR207P/Y444A. Thus, combining a mutation that stabilizes the active conformation of DegP (R207P) with a mutation that prevents cage assembly (Y444A) results in dominant lethality, which appears to be caused by excessive proteolysis of one or more cellular proteins.
We also tested for toxic effects of the activating R207P mutation in combination with other cage-destabilizing mutations (L276A, L446A, and +20_linker) or a cage-stabilizing mutation (Δlinker) (Kim and Sauer 2012). Plasmid-mediated expression of DegPR207P/L276A, DegPR207P/L446A, or DegPR207P/+20_linker in E. coli W3110 resulted in dominant lethality at 37°C, whereas cells expressing DegPR207P/Δlinker grew normally (Fig. 2D; Supplemental Fig. S2). Thus, all cage-deficient mutants tested resulted in toxicity in the R207P background, supporting a model in which cage assembly prevents excessive proteolysis by activated DegP.
Excessive proteolysis results in cell envelope defects
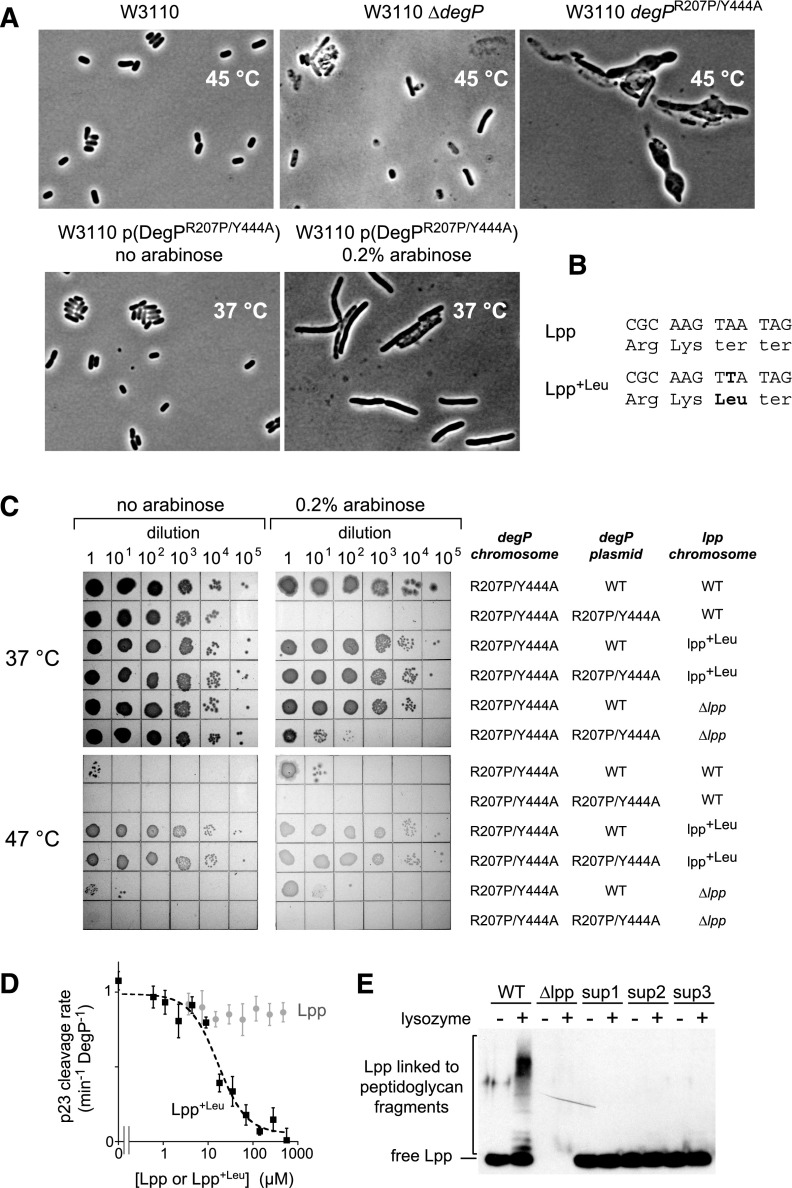
Periplasmic proteins are required for essential reactions in cell envelope maintenance and cell division. To test whether DegPR207P/Y444A expression interfered with either process, we examined cells expressing this double mutant for abnormal morphology. After a temperature shift to 45°C, chromosomal expression of DegPR207P/Y444A resulted in dramatically bloated, elongated, and irregular cells (Fig. 3A). This morphology suggests severe defects in cell wall biogenesis and was different from ΔdegP cells (Fig. 3A). In contrast, plasmid-mediated overproduction of DegPR207P/Y444A at 37°C resulted in elongated cells, as expected for cell division defects (Fig. 3A). It is possible that additional cell envelope proteins are degraded at the higher temperature or that degradation of one or more proteins is simply more extensive at the higher temperature.
Figure 3.
Proteolysis by DegPR207P/Y444A damages the cell envelope and is relieved by an lpp mutation. (A) Micrographs of E. coli W3110 strains with different degP alleles after 3–4 h of growth at 45°C or 37°C following dilution from overnight cultures. (B) Gene and C-terminal protein sequences for wild-type Lpp and Lpp+Leu from extragenic suppressor strains. (C) Strains SK339 (degPR207P/Y444A/lpp+), SK373 (degPR207P/Y444A/lpp+Leu), and SK367 (degPR207P/Y444A/Δlpp) expressing plasmid-borne DegP or DegPR207P/Y444A under arabinose control were serially diluted and grown on LB agar plates at 37°C or 47°C. The lethality of DegPR207P/Y444A expression was largely suppressed in the degPR207P/Y444A/lpp+Leu strain at both temperatures and partially suppressed in the degPR207P/Y444A/Δlpp strain at 37°C. (D) The rate of cleavage of 2 µM p23 substrate at 37°C by 0.2 µM DegPR207P/Y444A was assayed in the presence of increasing quantities of purified Lpp or Lpp+Leu. Values are averages ± 1 SD (n = 3). (E) SDS-PAGE and Western blots using anti-Lpp antibody were used to assay covalent attachment of Lpp to peptidoglycan. Following treatment of cells with lysozyme, a portion of wild-type Lpp electrophoreses as a ladder of higher-molecular-weight species, indicating covalent linkage to peptidoglycan. These larger species were not observed for Lpp+Leu from extragenic suppressor strains.
An extragenic suppressor of toxicity
To obtain additional information concerning the lethality of DegPR207P/Y444A, we isolated suppressor mutations that restored growth to cells producing this mutant from the chromosome and from a multicopy plasmid at 47°C (Supplemental Fig. S3A), a temperature even more restrictive than 45°C. After eliminating strains with additional mutations in degP, we obtained three isolates. In each isolate, whole-genome sequencing revealed a common mutation in the stop codon of the lpp gene that results in an Lpp variant with one additional leucine at the C terminus (Lpp+Leu) (Fig. 3B). Lpp, a trimeric coiled coil, is one of the most abundant E. coli proteins; a fatty acid attached to the N terminus of Lpp inserts into the outer membrane, whereas the ε-amino group of the C-terminal lysine is covalently attached to cell wall peptidoglycan, linking these structures and helping to maintain cell envelope integrity (Braun and Rehn 1969; Braun 1975; DiRienzo et al. 1978; McLachlan 1978). We reconstructed a strain with chromosomal lpp+Leu and degPR207P/Y444A mutations and transformed it with DegP or DegPR207P/Y444A plasmids. The lpp+Leu strain grew equally well when DegPR207P/Y444A or DegP was overproduced from the plasmid at 37°C and/or from the chromosome and plasmid at 47°C, whereas the lppWT strain grew poorly under all conditions where DegPR207P/Y444A was overproduced (Fig. 3C). The viability of the reconstructed strain was comparable with the three original suppressor strains (Supplemental Fig. S3B), indicating that the lpp+Leu allele is the major determinant of lethality suppression.
Because the PDZ1 domain of DegP preferentially binds C-terminal hydrophobic residues (Krojer et al. 2008a; Kim et al. 2011), we tested the possibility that the Lpp+Leu protein might be a substrate or inhibitor of DegPR207P/Y444A. We detected no cleavage of Lpp+Leu by DegPR207P/Y444A (Supplemental Fig. S3C). Notably, however, purified Lpp+Leu inhibited DegPR207P/Y444A cleavage of the p23 substrate, whereas purified Lpp did not inhibit cleavage (Fig. 3D). Lpp+Leu also inhibited wild-type DegP proteolysis, albeit substantially less efficiently than it inhibited DegPR207P/Y444A (Supplemental Fig. S3D). Thus, Lpp+Leu probably directly reduces the intracellular activity of the DegPR207P/Y444A protease. We also tested whether the C-terminal leucine of Lpp+Leu affected covalent attachment to peptidoglycan. Approximately one-third of Lpp in wild-type cells is linked to peptidoglycan (Inouye et al. 1972) and forms a ladder of higher-molecular-weight bands on SDS-PAGE after lysozyme treatment (Fig. 3E). In contrast, Lpp+Leu from each suppressor strain showed no evidence of peptidoglycan attachment (Fig. 3E), suggesting that lpp+Leu might be a biochemical loss-of-function allele as well as a gain-of-function allele. Indeed, we found that deletion of lpp partially suppressed DegPR207P/Y444A toxicity at 37°C (Fig. 3C). These results support a model in which Lpp+Leu relieves the toxic effects of DegPR207P/Y444A by direct inhibition of rogue proteolysis and by restructuring the cell envelope (see the Discussion).
Intragenic suppressors of toxicity
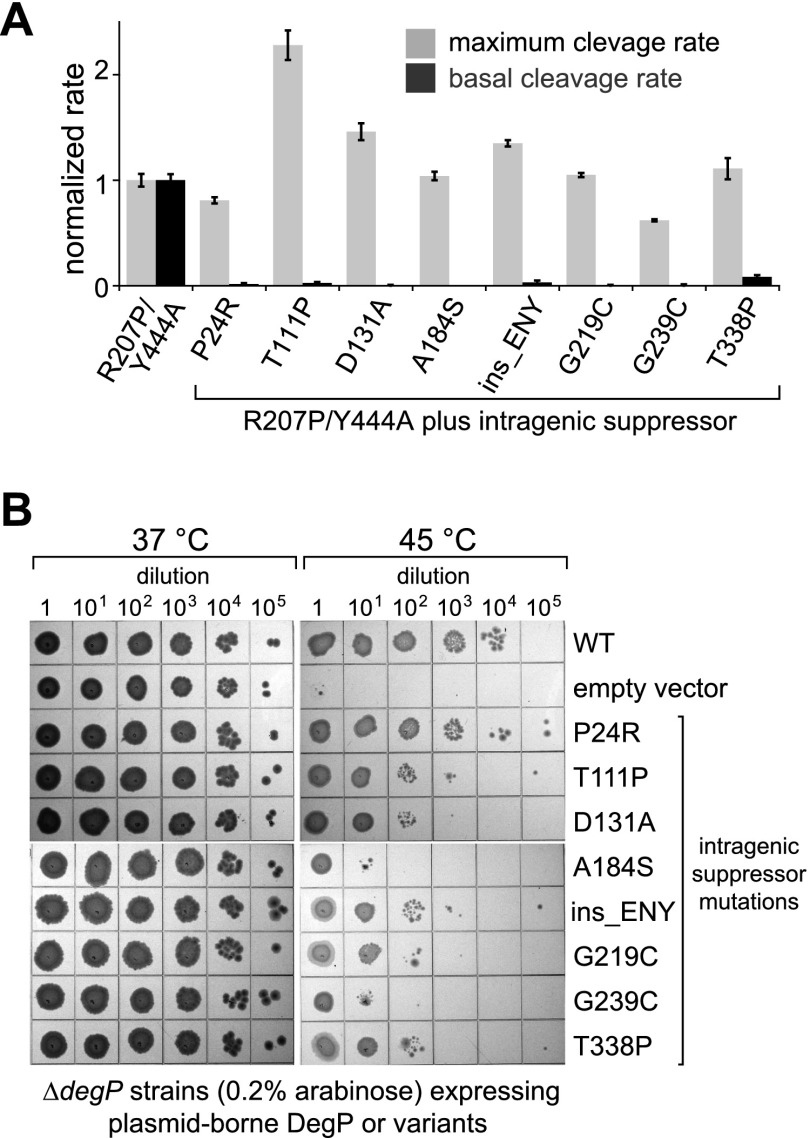
We also selected intragenic suppressor mutations that relieved R207P/Y444A toxicity and allowed growth at 47°C, a temperature at which DegP-mediated proteolysis is required for survival. Seven amino acid substitutions and a duplication of residues 193–195 (ENY) were obtained and subjected to further analysis. The positions of the suppressors, which were scattered throughout the protease domain and PDZ1 domain (Supplemental Fig. S4A), provided little indication of the suppression mechanism. Each suppressor mutation was reconstructed in the R207P/S210A/Y444A background, and the proteins were purified and tested for substrate-dependent cage assembly by gel filtration (Supplemental Fig. S4B). In no case were cages observed, indicating that these mutations do not restore cage assembly. Next, we purified enzymes containing individual suppressor mutations and the R207P/Y444A mutations and assayed basal cleavage of the reporter substrate. Strikingly, all of the enzymes with suppressor mutations displayed substantially lower levels of basal cleavage than the R207P/Y444A mutant (Fig. 4A, black bars). In contrast, when substrate-activated cleavage of a saturating concentration of p23 was assayed, most suppressor mutations supported rates similar to the parental R207P/Y444A mutant, and one mutation increased the rate (Fig. 4A, gray bars). DegP variants containing just the suppressor mutations also had lower levels of basal activity than wild-type DegP but mediated comparable or higher levels of saturated p23 cleavage (Supplemental Fig. S4C). In combination, these results suggest that the suppressor mutations modify DegP activity independently of the R207P and Y444A mutations and relieve R207P/Y444A toxicity by stabilizing the inactive conformation of DegP and reducing the excessive basal activity caused by the R207P mutation.
Figure 4.
Intragenic suppressors of DegPR207P/Y444A lethality. (A) DegPR207P/Y444A variants with or without mutations that suppress lethality were assayed for the maximum rate of cleavage of the p23 substrate by Michaelis-Menten analysis and fitting to the Hill equation (gray bars are the fitted Vmax normalized to the value for DegPR207P/Y444A ± the error of fitting) and were also assayed for basal cleavage of 100 µM Abz-KASPVSLGYNO2D peptide by 10 µM variant (black bars are averages [n = 3] normalized to the value for DegPR207P/Y444A ± 1 SD). (B) Plasmids expressing DegP or variants containing different intragenic suppressor mutations without the R207P/Y444A mutations were transformed into a ΔdegP strain, and growth of serial dilutions on LB agar plates was monitored at 37°C or 45°C in the presence of 0.2% arabinose.
To test whether DegP enzymes bearing just the suppressor mutations relieve high-temperature stress, we expressed variants harboring the single mutations from plasmids in ΔdegP cells and monitored growth on LB agar plates at 37°C or 45°C. Seven of the eight mutants resulted in reduced cell viability at 45°C (Fig. 4B). Thus, mutational stabilization of the inactive conformation reduces DegP’s ability to combat heat stress caused by protein unfolding or misfolding.
Single R207P or Y444A mutations reduce high-temperature cellular fitness
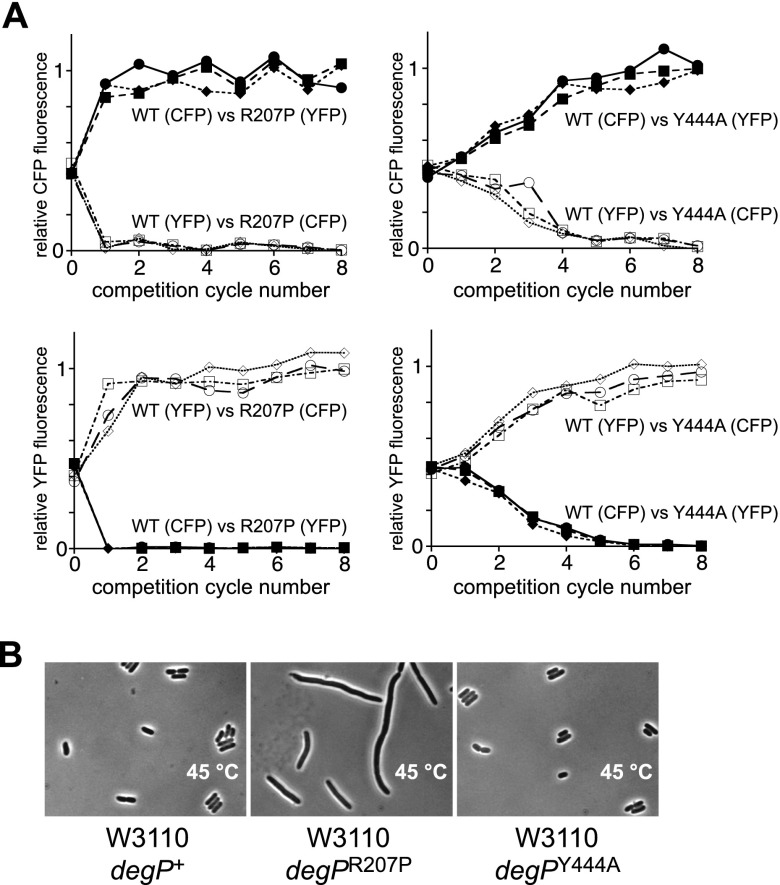
Although the individual R207P or Y444A mutations did not substantially alter cell viability in plating or growth assays, we suspected that they might reduce high-temperature cellular fitness. To test this idea, we constructed cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP) strains producing wild-type DegP, DegPR207P, or DegPY444A from the chromosome and performed growth competition experiments. Cells producing wild-type DegP and either DegPR207P or DegPY444A were initially mixed in a 1:1 ratio, diluted 104-fold in LB broth, and grown at 45°C for ∼12 h, and then additional cycles of dilution and growth were carried out. The relative population of each strain at the beginning of the experiment and after each cycle was quantified by CFP and YFP fluorescence. In one set of experiments, the strain expressing wild-type DegP was marked with YFP, and the mutant strain was marked with CFP. In a second set of experiments, the fluorescent markers were reversed to exclude possible effects of the markers on cell fitness. In competitions against DegPR207P, cells expressing wild-type DegP dominated the population after a single cycle of competition (Fig. 5A, left panels). Cells expressing wild-type DegP also competed effectively against DegPY444A but required five to six cycles to dominate the population (Fig. 5A, right panels). Examination of the morphology of degP+, degPR207P, and degPY444A cells at 45°C revealed that cells expressing DegPR207P were highly filamented compared with cells expressing wild-type DegP or DegPY444A (Fig. 5B), suggesting that DegPR207P degradation of one or more cell division factors contributes to the reduced fitness of cells expressing this mutant.
Figure 5.
Growth competitions between degP+ cells and degPR207P or degPY444A cells. (A) Three independent mixtures with equal initial quantities of a degP+ strain and a degPR207P or degPY444A strain were subjected to eight cycles of 104-fold dilution in LB broth and subsequent growth for ∼12 h at 45°C. At the beginning of the experiment and after each cycle, the fraction of each strain in the population was determined by relative CFP or YFP fluorescence. The degP+ cells dominated the population after one cycle of competition against degPR207P cells and after six cycles of competition against degPY444A cells when the wild-type strain was marked with CFP and the mutant strain was marked with YFP and vice versa. (B) Micrographs of strains expressing chromosomal DegP, DegPR207P, or DegPY444A at 45°C.
Discussion
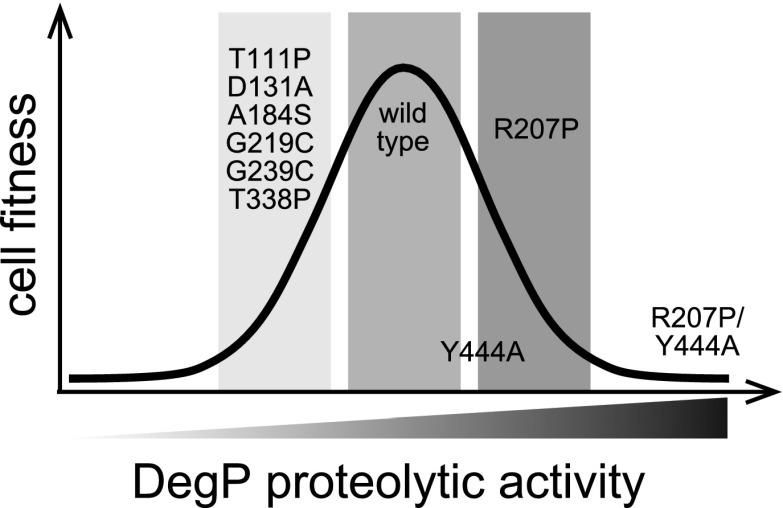
Our results establish that there is a delicate balance between the proteolytic activity of a PQC protease, like DegP, and the fitness of bacterial cells at temperatures that result in protein unfolding or misfolding (Fig. 6). For example, we found that mutations that alter the allosteric equilibrium between the active and inactive conformations of the DegP trimer reduce the fitness of E. coli at heat-shock temperatures. Moreover, combining a mutation that allosterically activates DegP with a mutation that prevents formation of DegP cages produces a toxic rogue protease. This toxicity can be relieved by intragenic mutations that restore allosteric balance or by an extragenic mutation that generates an inhibitor and alters the cell envelope. Thus, allosteric activation of DegP appears to be the major mechanism for regulating DegP proteolysis, whereas cage assembly provides a second level of protection by limiting the repertoire of substrates degraded by activated DegP.
Figure 6.
Relationship between proteolysis levels and cellular fitness. DegP proteolysis at a high enough level to protect cells from proteotoxic stress is required for fitness, but excessive degradation is harmful.
A critical allosteric balance
For any allosteric enzyme, the relative populations of inactive and active molecules are determined by a conformational equilibrium constant, the concentrations of substrates or allosteric effectors, and the relative affinities of these molecules for the two enzyme conformations. We found that most mutations that alter the conformational equilibrium constant for DegP reduce cellular fitness. For example, the R207P mutation stabilizes active DegP, increasing basal activity, and cells expressing DegPR207P compete poorly against wild-type cells and show morphologies consistent with defects in cell division. Moreover, in combination with cage-disrupting mutations, R207P results in proteolysis-dependent lethality, which can be suppressed by intragenic mutations that stabilize inactive DegP. In otherwise wild-type DegP, however, these intragenic suppressors reduce basal activity, and most also decrease cellular viability at 45°C. Thus, achieving a proper allosteric balance between active and inactive DegP is critical, as either too little or too much active DegP creates problems for the cell.
Mutations that stabilize inactive DegP are likely to reduce cell viability at 45°C because substrates present at subsaturating concentrations are not degraded sufficiently rapidly. Deleterious proteolysis by DegPR207P could, in principle, arise from a change in substrate specificity, a higher rate of degradation of saturating concentrations of normal substrates, or increased degradation of subsaturating levels of normal substrates. Compared with wild-type DegP, the R207P mutant had ∼100-fold higher basal activity but lower maximal activity in degrading model substrates. Moreover, the lethality of the R207P/Y444A variant could be suppressed by many different mutations that stabilize inactive DegP, suggesting that altered substrate specificity is unlikely to lead to degradation of “new” target proteins by DegPR207P. Hence, the deleterious effects of this mutant on cell fitness probably arise from increased levels of degradation of low concentrations of normal substrates.
At the position corresponding to residue 207 in E. coli DegP, many orthologs have a wild-type proline (Mohamedmohaideen et al. 2008; Cezairliyan and Sauer 2009). Based on our results, any potential deleterious effects of this “activating” proline are likely to be balanced by one or more “inactivating” residues elsewhere in the structure. Thus, the trimeric architecture of HtrA proteases allows the stabilities of the active and inactive conformations to be tuned to meet cellular requirements.
Protective effects of cage assembly
Assembly of DegP cages appears to protect cells from excessive or inappropriate proteolysis. For example, cells expressing DegPY444A, which cannot form cages, fared poorly in a competition with cells expressing wild-type DegP at 45°C, indicating that cage assembly provides a fitness advantage at this heat-shock temperature. Moreover, combining the activating R207P mutation with Y444A or other cage-destabilizing mutations resulted in dominant lethality. Why might cage-defective DegP variants mediate inappropriate degradation? Following cage assembly, the proteolytic active sites of DegP are accessible to unfolded substrates that can pass through small openings into the cage lumen, but misfolded substrates bound to chaperones or partially native substrates would be excluded. Once substrates are depleted, cages disassemble, and trimers revert to the inactive conformation. In contrast, the active sites of an activated cage-defective DegP trimer are exposed, and thus any polypeptide sequence with an appropriate P1 residue could be cleaved. Moreover, the binding of one or two good substrates to the active sites and PDZ1 domains of a trimer can transactivate cleavage of poor substrates by the remaining active sites. Because cage assembly should restrict the normal repertoire of DegP substrates, additional cellular proteins would be expected to be cleaved by cage-defective trimers, especially those that are easily activated, like DegPR207P/Y444A. Therefore, sequestering the active sites of DegP within cages allows modulation of proteolysis independently of the allosteric control of active and inactive states (Kim and Sauer 2012).
Based on the finding that outer-membrane proteins (OMPs) copurify with cages of DegPS210A, it was proposed that cages of wild-type DegP function as chaperones (Krojer et al. 2008b). However, OMP-associated DegPS210A cages assume a proteolytically active conformation (Krojer et al. 2008b; Kim et al. 2011), and thus any encapsulated partially folded protein in a wild-type DegP cage would have a high probability of being cleaved. Moreover, at 37°C, we found that the dominant lethality of DegPR207P/Y444A overexpression is suppressed by a mutation in the proteolytic catalytic triad, a result inconsistent with loss of chaperone activity being responsible for the deleterious effects of this cage assembly defect.
Toxicity, suppression of toxicity, and targets of rogue proteolysis
Combining the DegP R207P allosteric activation mutation with the Y444A cage-defective mutation results in rogue proteolysis and cellular toxicity. At heat-shock temperatures, where wild-type DegP is normally expressed at high levels to combat the stress associated with protein misfolding, high-level expression of DegPR207P/Y444A results in cell death and morphologies suggesting damage to proteins involved in cell wall maintenance and/or cell division. High-level expression of DegPR207P/Y444A at 37°C, a non-heat-shock temperature, also resulted in cell death.
Intriguingly, the lethality of DegPR207P/Y444A is suppressed by adding one additional leucine to the C terminus of Lpp, a lipoprotein that normally cross-links the bacterial outer membrane to the peptidoglycan lattice. Two distinct properties of the Lpp+Leu protein appear to mediate suppression of toxicity. First, this protein functions directly as a gain-of-function inhibitor of DegPR207P/Y444A proteolysis in vitro (Fig. 3D). Because DegP proteolysis is required for high-temperature survival (Lipinska et al. 1989; Strauch et al. 1989), we suspect that Lpp+Leu inhibition in the cell reduces but does not completely eliminate DegPR207P/Y444A activity, but the level of such inhibition is unclear. Second, Lpp+Leu did not cross-link to peptidoglycan (Fig. 3E), mimicking one cellular effect associated with loss-of-function Lpp mutations. Indeed, deletion of lpp partially suppressed toxicity associated with DegPR207P/Y444A overexpression at 37°C (Fig. 3C), suggesting that cross-linking of wild-type Lpp to peptidoglycan accounts for some lethal effects of rogue proteolysis. Importantly, previous studies show that depletion or inhibition of E. coli proteins needed for lipoprotein biogenesis causes lethality, which can be suppressed by deletion of lpp (Zwiebel et al. 1981; Matsuyama et al. 1997; Yakushi et al. 1997; Tajima et al. 1998; Kikuchi et al. 2000; Robichon et al. 2005; Pailler et al. 2012). Deleting lpp may suppress lethality by preventing toxic cross-linking of Lpp mislocalized to the inner membrane to peptidoglycan; by reducing the load on lipoprotein translocation machinery, thereby allowing better translocation of other essential lipoproteins; or by outer-membrane leakage that reduces toxic species in the periplasm. The toxicity of DegPR207P/Y444A overexpression may arise in part from excessive degradation of one or more proteins involved in lipoprotein biogenesis. Collectively, these results indicate that the toxicity of DegPR207P/Y444A is caused by excessive degradation of proteins that function in essential cellular processes in the bacterial periplasm. Thus, regulating wild-type DegP proteolysis by allosteric activation and cage formation limits damage to critical periplasmic proteins while allowing efficient degradation of proteotoxic proteins.
Protein unfolding and periplasmic degradation
The peptide bonds cleaved by proteases are inaccessible in native proteins, and unfolding or prevention of folding are therefore key steps in degradation. In the cytoplasm, AAA+ proteases execute most protein degradation (Sauer and Baker 2011). These enzymes use hexameric AAA+ rings to bind target proteins, unfold them, and translocate the polypeptide into a barrel-like proteolytic chamber in reactions fueled by ATP hydrolysis. Interestingly, the proteolytic barrel of the AAA+ ClpXP protease becomes a rogue enzyme when ADEP antibiotics bind and alter its conformation, allowing unfolded substrates to enter the chamber in an unregulated fashion (Brötz-Oesterhelt et al. 2005). Uncontrolled proteolysis by the ADEP•ClpP complex has recently been shown to kill persistent forms of pathogenic bacteria, which are extremely hard to eradicate with conventional antibiotics (Conlon et al. 2013). The periplasmic compartment of bacteria is devoid of ATP, and thus protein degradation largely depends on spontaneous unfolding or misfolding, which is enhanced at elevated temperatures. In E. coli, heat shock induces high-level expression of DegP, whose proteolytic activity is essential for cell survival and is regulated by allosteric activation and cage formation to avoid damaging important cellular processes in the periplasm. Thus, small molecule dysregulation of DegP proteolysis may also be a promising route to new antibiotics in the fight against pathogenic bacteria.
Materials and methods
Bacterial strains
Supplemental Table S1 lists strains, plasmids, and oligonucleotide primers used in this study. All plasmid constructions and cellular mutations introduced by site-directed mutagenesis were confirmed by DNA sequencing (Genewiz). Two-step recombineering (Davis et al. 2011) was used to replace the wild-type degP gene with mutant alleles as described (Kim and Sauer 2012). Briefly, in one recombination step, the degP gene in E. coli W3110 was replaced by an mPheS-kanR allele, which was subsequently replaced with a mutant degP gene in a second step. DNA fragments for the second recombination step were amplified with two primers, insP_ss_for1 and insP_rev2, from pSK617 (for SK341, which expresses DegPR207P) or insP_ss_for1 and insP_rev3 from pSK576 (for SK339, which expresses DegPR207P/Y444A).
The lpp deletion strain (JW1667-5 from the Keio collection; Coli Genetic Stock Center [CGSC] no. 9417) was obtained from the CGSC (Yale University, CT). The Δlpp-752∷kan allele in this strain was transferred to SK339 by P1 transduction (Thomason et al. 2007) to make strain SK367. Recombineering was also used to introduce lpp+Leu into W3110 to make strain SK371; the DNA fragment for the second recombination was amplified from genomic DNA of the suppressor strain with two primers, 5-lpp_up67 and 3-lpp_down75. Strain SK373, harboring both lpp+Leu and degPR207P/Y444A, was constructed by transduction of the mPheS-kanR allele into the degP locus and subsequent recombination with the degPR207P/Y444A allele.
IPTG-inducible genes encoding CFP (galK∷cfp-ampR) and YFP (galK∷yfp-ampR) were amplified with primers 5-galK_80up and 3-galK_80down from parental strains (Hegreness et al. 2006) and recombined into W3110 to generate SK352 and SK353, respectively. The CFP and YFP genes were then transferred to SK326 (degPR207P) and SK341 (degPY444A) by P1 transduction.
Plasmids
pBAD vectors were used for arabinose-induced expression of proteins (Guzman et al. 1995). Initially, the degP gene was amplified with primers 5-degP-NcoI and 3-degP-SalI and inserted between the NcoI and SalI sites of pBAD24, which contains a Shine-Dalgarno (SD) sequence upstream of the NcoI site. The SD_degP locus was amplified with primers 5-SacI_SD_degP and 3-SphI_degP from pBAD24_degP and inserted between the SacI and SphI sites in pBAD33 to generate pSK633 (pBAD33_SD_degP). Plasmids harboring degP mutations were constructed by mutagenesis of pBAD24_degP and subsequent transfer of alleles to pBAD33 or by direct mutagenesis of pSK633. Plasmids that overexpress DegP variants were constructed as reported (Kim et al. 2011).
To construct plasmids expressing His6-Lpp or His6-Lpp+Leu, a portion of the lpp gene missing the signal sequence and Cys21 was initially amplified from E. coli genomic DNA with primers 5-pET28_lpp-2 and 3-pET28_lpp (for pSK709) or 3-pET28_lpp_stop79L (for pSK710) and inserted between the NdeI and NotI sites of pET28b.
Proteins, peptides, and biochemical assays
DegP variants, 18–58 peptide, p23 peptide, reporter peptide, and flC18–58 (fluorescently labeled 18–58) were prepared as described (Kim et al. 2011; Kim and Sauer 2012). Activity assays, fluorescence anisotropy, FRET, and gel filtration chromatography were performed as described (Kim et al. 2011; Kim and Sauer 2012).
Bacterial growth and viability assays
To test strain viability, overnight cultures were diluted 1:100 in LB broth and grown to OD600 ∼0.2 at 37°C, and 5-µL aliquots from a set of 10-fold serial dilutions were spotted for growth on LB agar plates at 37°C. To test growth in liquid culture, overnight cultures were grown in LB broth with 10 µM/mL chloramphenicol at 37°C and diluted 100-fold in fresh medium with or without 0.2% arabinose, and continued growth at 37°C was monitored by OD600. To monitor the expression of DegP variants, cells were taken 1.5 h or 2 h after dilution, lysed by boiling in SDS loading buffer, and subjected to SDS-PAGE, and DegP was detected by Western blotting with anti-DegP antibody (a gift from T. Silhavy, Princeton University).
Microscopy
Cells harboring different chromosomal or plasmid degP alleles were grown in LB broth, diluted to OD600 0.05–0.2 with fresh LB, and placed on an LB 1.5% agarose pad. Phase microscopy images were taken using a Zeiss Axiovert 200 microscope with a 100×/1.45 oil immersion objective.
Isolation of suppressor mutations
SK339 cells encoding chromosomal degPR207P/Y444A were spread on LB agar plates and incubated overnight at 47°C, and colonies that grew were purified by restreaking and growth at 47°C. The degP genes of nine candidates contained mutations in addition to R207P/Y444A. Consistent with the absence of extragenic suppressor mutations in these strains, plasmid-mediated overexpression of DegPR207P/Y444A from pSK636 reduced viability at 37°C. Additional intragenic suppressor mutations were identified after selections at 47°C starting with strains SK345(ΔdegP)/pSK636 or SK339/pSK636 grown on LB agar plates containing 10 µg/mL chloramphenicol and 0.2% arabinose. Three survivors from the two SK339/pSK636 selection experiments contained no additional mutations in either the chromosomal or plasmid degP genes, and these strains overexpressed DegPR207P/Y444A at the expected high levels after 47°C heat shock or induction with arabinose at 37°C. To identify extragenic mutations, we isolated genomic DNA from the parental strain and the three suppressor strains and subjected them to Illumina whole-genome sequencing (BioMicro Center, Massachusetts Institute of Technology), which revealed one common suppressor mutation located in the stop codon of lpp.
Peptidoglycan attachment of Lpp
To test the attachment of Lpp or Lpp+Leu to peptidoglycan, overnight cultures were diluted 100-fold in LB broth and grown to log phase at 37°C, and the cells in 1 mL of culture were harvested by centrifugation and resuspended in 200 µL of 10 mM Tris (pH 7.9) and 5 mM EDTA. This material was divided into two 100-µL solutions, and 0.5 mg/mL lysozyme (final) was added to one solution. After 10 min at room temperature, 100 µL of SDS-PAGE sample buffer was added to both solutions, which were heated for 10 min at 100°C and centrifuged at 20,000g for 10 min. Aliquots of the supernatants were subjected to SDS-PAGE and Western blot analysis with anti-Lpp antibody (provided by H. Tokuda). The free form of Lpp electrophoreses at a position of the expected molecular weight, whereas peptidoglycan-bound Lpp electrophoreses as a ladder-like pattern of higher-molecular-weight fragments after lysozyme treatment.
Growth competition
Strains harboring chromosomal degP, degPR207P, or degPY444A and IPTG-inducible CFP or YFP were grown overnight at 37°C in LB broth. For competition experiments, equal amounts of a degP strain marked with one color and a degPR207P or degPY444A strain marked with the other color were mixed (cycle #0), diluted 104-fold in LB broth, and grown for ∼12 h at 45°C (cycle #1). Seven addition cycles of 104-fold dilution and growth were performed (cycles #2–#8). Cell mixtures were taken after each cycle, mixed with glycerol (15% final concentration), and stored at −80°C for further analysis. Three independent competition experiments were performed with each combination of strains. After all competition cycles were complete, frozen cell mixtures were thawed, diluted 100-fold in M9 minimal medium (1× M9 salts, 2 mM MgSO4, 0.1 mM CaCl2, 0.4% glucose) supplemented with 0.1% casamino acid and 1 mM IPTG, and grown for ∼3 h at 37°C. M9 supplemented medium was used to reduce background fluorescence, and ∼3 h of growth in this medium did not significantly change the initial cell ratios (Fig. 5A shows that the CFP/YFP cell ratios at cycle #0 were consistently 0.4–0.5). One-hundred microliters of cells was transferred to clear 96-well plates (Greiner, catalog no. 655096) to measure OD600, CFP fluorescence (excitation at 430 nm; emission at 480 nm; cutoff filter at 455 nm), and YFP fluorescence (excitation at 490 nm; emission at 530 nm; cutoff filter at 515 nm) in a SpectraMax M5 microplate reader (Molecular Devices). Blank signals from medium alone were subtracted from CFP and YFP fluorescence, which were normalized by OD600. Data were normalized by the average high fluorescence signals during the final cycles to obtain relative CFP/YFP fluorescence.
Acknowledgments
We thank A. Grossman, M. Laub, S. Levine, and C. Aakre for discussions and technical help. Anti-DegP and anti-Lpp antibodies were generous gifts from T. Silhavy and H. Tokuda, respectively. This work was supported by National Institutes of Health grant AI-16892 and by a Charles A. King Trust Post-doctoral Fellowship to S.K.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.238394.114.
References
- Braun V 1975. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim Biophys Acta 415: 335–377 [DOI] [PubMed] [Google Scholar]
- Braun V, Rehn K 1969. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem 10: 426–438 [DOI] [PubMed] [Google Scholar]
- Brötz-Oesterhelt H, Beyer D, Kroll H-P, Endermann R, Ladel C, Schroeder W, Hinzen B, Raddatz S, Paulsen H, Henninger K, et al. 2005. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat Med 11: 1082–1087 [DOI] [PubMed] [Google Scholar]
- Cezairliyan BO, Sauer RT 2009. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol 72: 368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T, Kaiser M, Huber R, Ehrmann M 2011. HTRA proteases: regulated proteolysis in protein quality control. Nat Rev Mol Cell Biol 12: 152–162 [DOI] [PubMed] [Google Scholar]
- Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JH, Baker TA, Sauer RT 2011. Small-molecule control of protein degradation using split adaptors. ACS Chem Biol 6: 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRienzo JM, Nakamura K, Inouye M 1978. The outer membrane proteins of Gram-negative bacteria: biosynthesis, assembly, and functions. Annu Rev Biochem 47: 481–532 [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegreness M, Shoresh N, Hartl D, Kishony R 2006. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311: 1615–1617 [DOI] [PubMed] [Google Scholar]
- Inouye M, Shaw J, Shen C 1972. The assembly of a structural lipoprotein in the envelope of Escherichia coli. J Biol Chem 247: 8154–8159 [PubMed] [Google Scholar]
- Jiang J, Zhang X, Chen Y, Wu Y, Zhou ZH, Chang Z, Sui S-F 2008. Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc Natl Acad Sci 105: 11939–11944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Shibuya I, Matsumoto K 2000. Viability of an Escherichia coli pgsA null mutant lacking detectable phosphatidylglycerol and cardiolipin. J Bacteriol 182: 371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Sauer RT 2012. Cage assembly of DegP protease is not required for substrate-dependent regulation of proteolytic activity or high-temperature cell survival. Proc Natl Acad Sci 109: 7263–7268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Grant RA, Sauer RT 2011. Covalent linkage of distinct substrate degrons controls assembly and disassembly of DegP proteolytic cages. Cell 145: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416: 455–459 [DOI] [PubMed] [Google Scholar]
- Krojer T, Pangerl K, Kurt J, Sawa J, Stingl C, Mechtler K, Huber R, Ehrmann M, Clausen T 2008a. Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc Natl Acad Sci 105: 7702–7707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krojer T, Sawa J, Schäfer E, Saibil HR, Ehrmann M, Clausen T 2008b. Structural basis for the regulated protease and chaperone function of DegP. Nature 453: 885–890 [DOI] [PubMed] [Google Scholar]
- Lipinska B, Fayet O, Baird L, Georgopoulos C 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J Bacteriol 171: 1574–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama Si, Yokota N, Tokuda H 1997. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J 16: 6947–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan AD 1978. The double helix coiled coil structure of murein lipoprotein from Escherichia coli. J Mol Biol 121: 493–506 [DOI] [PubMed] [Google Scholar]
- Mohamedmohaideen NN, Palaninathan SK, Morin PM, Williams BJ, Braunstein M, Tichy SE, Locker J, Russell DH, Jacobs WR Jr, Sacchettini JC 2008. Structure and function of the virulence-associated high-temperature requirement A of Mycobacterium tuberculosis. Biochemistry 47: 6092–6102 [DOI] [PubMed] [Google Scholar]
- Pailler J, Aucher W, Pires M, Buddelmeijer N 2012. Phosphatidylglycerol:prolipoprotein diacylglyceryl transferase (Lgt) of Escherichia coli has seven transmembrane segments, and its essential residues are embedded in the membrane. J Bacteriol 194: 2142–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen MJ, Wren BW 1997. The HtrA family of serine proteases. Mol Microbiol 26: 209–221 [DOI] [PubMed] [Google Scholar]
- Robichon C, Vidal-Ingigliardi D, Pugsley AP 2005. Depletion of apolipoprotein N-acyltransferase causes mislocalization of outer membrane lipoproteins in Escherichia coli. J Biol Chem 280: 974–983 [DOI] [PubMed] [Google Scholar]
- Sauer RT, Baker TA 2011. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem 80: 587–612 [DOI] [PubMed] [Google Scholar]
- Sohn J, Sauer RT 2009. OMP peptides modulate the activity of DegS protease by differential binding to active and inactive conformations. Mol Cell 33: 64–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J, Grant RA, Sauer RT 2010. Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution. J Biol Chem 285: 34039–34047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch KL, Johnson K, Beckwith J 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol 171: 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima T, Yokota N, Matsuyama S, Tokuda H 1998. Genetic analyses of the in vivo function of LolA, a periplasmic chaperone involved in the outer membrane localization of Escherichia coli lipoproteins. FEBS Lett 439: 51–54 [DOI] [PubMed] [Google Scholar]
- Thomason LC, Costantino N, Court DL 2007. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol 79: 1.17.1–1.17.8 [DOI] [PubMed] [Google Scholar]
- Yakushi T, Tajima T, Matsuyama S, Tokuda H 1997. Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J Bacteriol 179: 2857–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiebel LJ, Inukai M, Nakamura K, Inouye M 1981. Preferential selection of deletion mutations of the outer membrane lipoprotein gene of Escherichia coli by globomycin. J Bacteriol 145: 654–656 [DOI] [PMC free article] [PubMed] [Google Scholar]