A key question in biology is how the final size of an organism is set. Coherent plant growth requires spatial integration of hormonal signaling and cell wall remodeling activities, yet little is known about how these pathways are coordinated. Fridman et al. uncover the mechanism by which hormonal perturbation in two types of epidermal root cells, hair and nonhair cells, impacts whole root growth. The results suggest that the relative spatial distribution of brassinosteroid hormone receptor BRI1, and not its absolute level, fine-tunes growth, thus providing a new concept in hormone-mediated growth control.
Keywords: brassinosteroids, cell size determination, cell wall, hormone signaling, intercellular communication, root development
Abstract
Coherent plant growth requires spatial integration of hormonal pathways and cell wall remodeling activities. However, the mechanisms governing sensitivity to hormones and how cell wall structure integrates with hormonal effects are poorly understood. We found that coordination between two types of epidermal root cells, hair and nonhair cells, establishes root sensitivity to the plant hormones brassinosteroids (BRs). While expression of the BR receptor BRASSINOSTEROID-INSENSITIVE1 (BRI1) in hair cells promotes cell elongation in all tissues, its high relative expression in nonhair cells is inhibitory. Elevated ethylene and deposition of crystalline cellulose underlie the inhibitory effect of BRI1. We propose that the relative spatial distribution of BRI1, and not its absolute level, fine-tunes growth.
A fundamental question in developmental biology relates to the mode through which the final size of a cell, organ, and whole organism is set. Plant growth involves consecutive stages of cell proliferation and post-mitotic cell enlargement. In roots, these stages form a developmental gradient along the apical–basal axis that ultimately determines their length (Petricka et al. 2012). As the constituent root cells are linked through their rigid walls and do not migrate, intercellular coordination of growth processes is critical to maintain root integrity. Such coordination is reflected in the ability of different hormonal signal transduction pathways to control whole-organ growth via distinct cell types (Ubeda-Tomas et al. 2012). Moreover, a given hormone can confer opposing (promoting and restricting) effects on growth, traditionally attributed to the quantity and intensity of the stimulus. The mode by which hormonal sensitivity is integrated in space has been a long-standing open question that is critical in understanding plant growth (Bradford and Trewavas 1994).
The cell enlargement stage in plants is an important contributor to organ size. It is driven by turgor pressure and regulated by the extensibility of the cell’s surrounding wall (Cosgrove 2005; Wolf et al. 2012a). Cellulose microfibrils are the major load-bearing components of the cell wall and feature regions of high crystallinity alongside zones with disorganized glucan chain arrangements. A relative reduction in the crystalline to amorphous cellulose ratio is thought to allow access to microfibril tethering-related modifications that maintain anisotropic (unidirectional) expansion during rapid growth, whereas sustained high crystallinity limits unidirectional cell expansion (Baskin 2005; Fujita et al. 2011). It is largely unknown whether localized cell wall modifications, including modulation of crystalline cellulose, occur during primary root growth and whether they integrate with specific hormonal effects.
The promoting activity of the steroid group of hormones, brassinosteroids (BRs), and the inhibitory effect of the gaseous hormone ethylene (Le et al. 2001; Mussig et al. 2003; Hacham et al. 2011; Fridman and Savaldi-Goldstein 2013) are among the hormonal pathways regulating cell elongation in roots. Ethylene inhibits unidirectional cell expansion via its well-established and complex interactions with auxin, which involve mutual elevation of their corresponding biosynthesis genes and auxin transport from the root tip to elongating cells (Ruzicka et al. 2007; Stepanova et al. 2007; Swarup et al. 2007; Robles et al. 2013). Previous studies have shown that exogenous application of BRs triggers ethylene production in Arabidopsis seedlings (Woeste et al. 1999), but the significance of this effect during root growth remained unclear (Mussig et al. 2003). Hence, while moderate BR levels promote root growth, their high levels are inhibitory (Mussig et al. 2003). This inhibitory effect has been recently explained by a premature cell exit from mitosis (Gonzalez-Garcia et al. 2011).
BRs bind the BRASSINOSTEROID-INSENSITIVE1 (BRI1) cell surface receptor, consequentially initiating a sequence of events that activates the receptor complex (Clouse 2011). The signal is then transmitted to the nucleus in a multistep process that enables the activation of downstream homologous transcription factors BRASSINAZOLE-RESISTANT1 (BZR1) and BRI1–EMS SUPPRESSOR1 (BES1)/BZR2, which regulate gene expression, including that of a prominent group of cell wall biosynthesis and remodeling genes (Sun et al. 2010; Yu et al. 2011).
In Arabidopsis, the root epidermis is organized into two types of cells whose fates are determined by a positional effect at the embryonic stage and that differentiate to root hair cells and nonhair cells upon completion of elongation (Fig. 1A; Dolan et al. 1993, 1994). The two cell types differ in their final cell length and cellular organization, as illustrated by the stained cytoplasm feature of the hair cell type when in their growing stages (Fig. 1A; Dolan et al. 1994; Masucci et al. 1996). We previously showed that restriction of the otherwise ubiquitous expression of BRI1 to nonhair cells in its corresponding bri1 mutant background is sufficient to drive the cell proliferation stage of all cells in the primary root (Hacham et al. 2011).
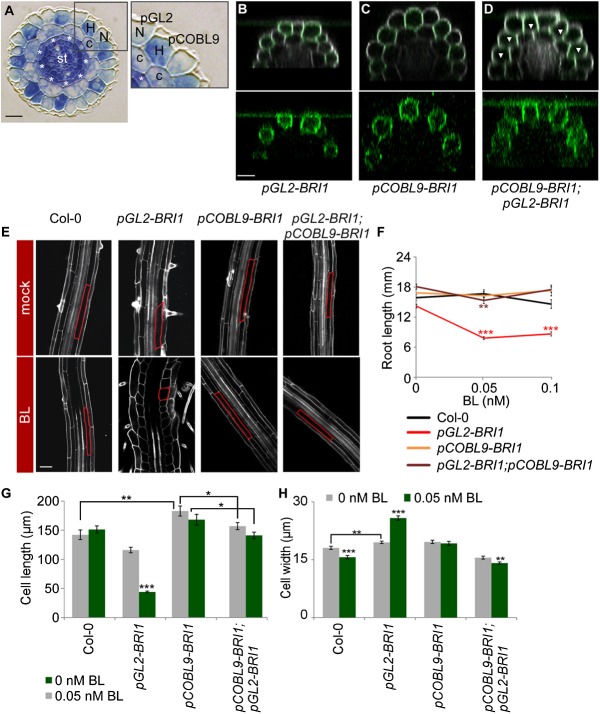
Figure 1.
The impact of BRs on root cell elongation is determined by the relative expression of BRI1 in neighboring epidermal cells. (A) Cross-section of the Arabidopsis primary root showing radial organization of its constituent tissues. (N) Nonhair cells; (H) hair cells; (c) cortex; (st) stele. Asterisks mark the endodermis. pGL2 and pCOBL9 promoter fragments mark nonhair and hair cells, respectively. Bar, 10 µm. (B–D) Expression patterns of BRI1-GFP in the different transgenic lines, all in the bri1 mutant background. Note the GFP signal (green, with intensified contrast in the bottom panels) in nonhair cells in pGL2-BRI1 (B), hair cells in pCOBL9-BRI1 (C), and throughout the epidermis of a cross between pCOBL9-BRI1 and pGL2-BRI1 (D). Arrowheads mark hair cells. Cells were stained with propidium iodide (gray). Bar, 20 µm. (E) Confocal microscopy image of these same lines and wild type (Col-0), untreated or treated with BL, with the cortical cell highlighted in red. Bar, 50 µm. (F) pGL2-BRI1 root length is shorter when exposed to low BL concentrations. In contrast, the root length of lines with BRI1 expression and overexpression throughout the epidermal tissue (as in wild type [Col-0] and pGL2-BRI1;pCOBL9-BRI1, respectively) remained similar (mean ± SE; 17 < n < 30). (G,H) Average mature cortical cell length (G) and width (H) in roots of wild-type and transgenic lines untreated or treated with BL (mean ± SE; 26 < n < 95 [G]; 32 < n < 45 [H]). Note the opposing effect of BRI1 on cell elongation upon its high relative expression in hair (pCOBL9-BRI1) versus nonhair (pGL2-BRI1) cells. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 with two tailed t-test.
Here, we studied the cell expansion stage and demonstrate that high BRI1 expression in hair cells drives cell elongation in all tissues, whereas its higher relative expression in nonhair cells inhibits root cell elongation. We reveal that the inhibition of cell elongation by nonhair cell BRI1 activity is due to enhanced sensitivity to the hormone and elevated downstream BR response. This response triggers activation of ethylene biosynthesis genes in a pathway involving BES1/BZR2; we show that ethylene activity is both necessary and sufficient in nonhair cells to inhibit cell elongation and, consequentially, whole-root growth. The local rise in BR and ethylene activities brings on enhanced accumulation of crystalline cellulose in the wall of nonhair cells, which impairs unidirectional cell expansion, cell elongation, and overall root length. Based on our results, we propose that BRI1 activity in hair cells restrains sensitivity to brassinolide (BL) imposed by nonhair cells expressing BRI1. Thus, the spatial rather than absolute density of BRI1 is an important determinant in coordination of organ growth.
Results
Differential density of BRI1 in hair and nonhair cells imposes opposing effects on root cell elongation
Our studies of the cell elongation stage in the Arabidopsis primary root revealed that bri1;pGL2-BRI1-GFP lines, in which BRI1 is targeted to nonhair cells of bri1 (hereafter referred to as pGL2-BRI1), exhibit reduced final cell length as compared with wild type, as also noted in our previous study (Hacham et al. 2011). This is exemplified by analysis of cortical cells that were also slightly but significantly wider as compared with wild-type roots, indicating reduced unidirectional cell expansion (Fig. 1B,E–H; Supplemental Fig. S1A). Similar analysis of epidermal cells also revealed wider nonhair cells in this background (Supplemental Fig. S1A). Intriguingly, unlike wild type and lines with ubiquitous overexpression of the receptor (pUBQ10-BRI1), all tested independent pGL2-BRI1 lines with varied BRI1 expression levels featured moderate reduction in root length that was dramatically enhanced in response to low concentrations of exogenously applied BL (the most active BR) (Fig. 1F; Supplemental Fig. S1C,D,F). Cellular analysis revealed that root length inhibition in BL-treated pGL2-BRI1 lines was the result of impaired unidirectional cell expansion, as implicated by swelled nonhair cells, a decrease in cell length, and an increase in the width of the two epidermal cell types and cortical cells (Fig. 1E,G,H; Supplemental Fig. S1A), while the number of meristematic cells remained unaffected (Supplemental Fig. S1G). In addition, root length and the short cortical cells of pGL2-BRI1 were suppressed in response to low concentrations of the BR biosynthesis inhibitor BRZ (Supplemental Fig. S1H). Thus, restriction of BRI1 activity to nonhair cells limits cell elongation and hence root length in a BR-dependent manner.
BRI1 promotes growth when expressed throughout the shoot epidermis (Savaldi-Goldstein et al. 2007; Savaldi-Goldstein and Chory 2008). In addition, roots expressing BRI1-GFP under the BRI1 endogenous promoter (Geldner et al. 2007) had similar receptor density in hair and nonhair cells (quantification of BRI1 along the anticlinal cell walls of the first elongating cells is shown in Supplemental Fig. S2A, left panel). We therefore reasoned that BRI1’s inhibitory effect in nonhair cells results from its uncoupled expression in neighboring epidermal cells. To explore this possibility, we established bri1 mutant lines with BRI1 expression targeted to elongating hair cells using the pCOBL9 promoter (pCOBL9-BRI1) (Fig. 1A,C; Brady et al. 2007). Remarkably, cellular analysis of pCOBL9-BRI1 lines revealed somewhat longer hair and cortical cells, which were unresponsive to the applied BL and, in agreement, had root length similar to that of wild type (Fig. 1E–H; Supplemental Fig. S1A,F). Next, we crossed pGL2-BRI1 with pCOBL9-BRI1 (pGL2-BRI1;pCOBL9-BRI1 in bri1) to obtain BRI1 activity in all elongating epidermal cells (Fig. 1D). Interestingly, the mature short cortical cell length phenotype of pGL2-BRI1 was suppressed; cortical cell size parameters were similar to those of wild type (Fig. 1E–H). In addition, cortical cells of pGL2-BRI1;pCOBL9-BRI1 plants were slightly but significantly shorter when compared with the parental pCOBL9-BRI1 (Fig. 1G). In agreement, backcross of pGL2-BRI1 to wild-type plants expressing endogenous BRI1 [hereafter referred to as pGL2-BRI1(WT)] also suppressed root hypersensitivity to BL (Supplemental Fig. S1B). The inhibitory effect of BL on pGL2-BRI1 lines was not correlated with differential BRI1-GFP accumulation at the plasma membrane along the distinct root zones (Supplemental Fig. S2B). Thus, BR signaling has opposing effects on cell elongation, as demonstrated by the relative expression of BRI1 in the two elongating epidermal cell types, where BRI1 activity in hair cells restrains whole-root growth sensitivity to BL imposed by nonhair cells expressing BRI1.
High relative expression of BRI1 in nonhair cells elevates ethylene activity
Mechanisms underlying the inhibitory effect of BRs on root cell elongation are unknown. Comparison of the transcriptomic profile of pGL2-BRI1 versus wild-type plant root tips uncovered 150 differentially expressed genes, among which expression of 1-aminocyclopropane-1-carboxylate (ACC) synthase 9 (ACS9) in pGL2-BRI1 was enhanced as compared with wild type (Supplemental Table S1). ACS genes synthesize ACC, thereby catalyzing the rate-limiting step in ethylene biosynthesis. Because Arabidopsis root cell elongation is limited by ethylene, we wondered whether the pGL2-BRI1 response involves the established ethylene effect. While ACS9 has not been reported as a transcriptional BR target, its highly homologous gene, ACS5, is directly regulated by BES1/BZR2 and BZR1 (Oh et al. 2012). We therefore quantified the relative expression levels of these two genes in the two cell types of the root tips of BL-treated wild-type plants by using cell type-specific immunopurification of mRNAs in ribosome complexes (see the Materials and Methods; Mustroph et al. 2009b). Both hair and nonhair cells of wild-type plants exhibited similar responses to BL, manifested by elevations in transcript levels of both ACS5 and ACS9 and in agreement with similar BRI1 densities in these cells (Fig. 2A,B; Supplemental Fig. S3A,B).
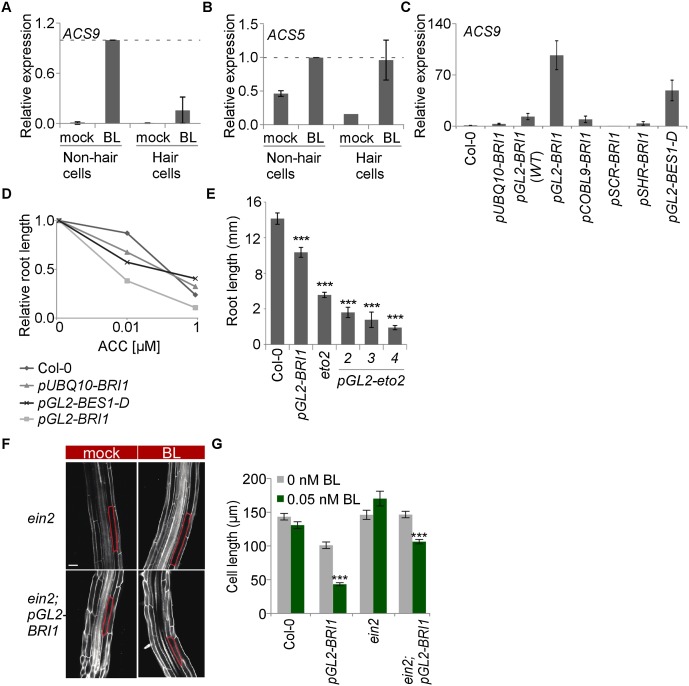
Figure 2.
Ethylene mediates the BRI1-triggered inhibitory effect on root cell elongation in nonhair cells. (A,B) Analysis of relative expression of ACS genes using immunopurified polysomal RNA from hair and nonhair cells of wild-type plants in the absence and presence of BL (mean ± SE; n = 2). (C) Analysis of relative expression of ACS9 from whole root tips of various transgenic lines. Note the high relative ACS expression levels in roots of nonhair cell targeted BRI1 and BES1-D (pGL2-BES1-D), while only a minimal response is detected in pUBQ10-BRI1 and in lines expressing pGL2-BRI1 in a background with endogenous BRI1 [pGL2-BRI1(WT)] (mean ± SE; n > 2). (D) Roots expressing BRI1 (pGL2-BRI1) and BES1-D (pGL2-BES1-D) in nonhair cells are hypersensitive to the ethylene precursor ACC as compared with wild type (Col-0) (mean ± SE; 27 < n < 42). (E) Expression of a dominant active version of ACS5 in nonhair cells (pGL2-eto2; independent transgenic lines are shown) is sufficient to inhibit whole-root growth, similar to the endogenous eto2 mutant. pGL2-BRI1 and wild-type plants served as controls (mean ± SE; 23 < n < 28). (F) The ethylene signaling component EIN2 is necessary for BRI1-driven inhibition of cell elongation. Mature cortical cell length is marked. Bar, 50 µm. (G) Average mature cortical cell length of roots untreated or treated with BL (mean ± SE; 29 < n < 49). (***) P < 0.001 with two tailed t-test.
Next, we compared the expression level of these ACS genes in various lines with BRI1 expression targeted to distinct tissues, including the endodermis and stele (pSCR-BRI1 and pSHR-BRI1, respectively) (Fig. 2C; Supplemental Fig. S3F; Hacham et al. 2011). pGL2-BRI1 roots had the highest basal ACS5 and ACS9 transcript levels, providing that BRI1 was not expressed in neighboring cells (Figs. 2C; Supplemental Fig. S3F,G). Thus, high relative expression of BRI1 in nonhair cells enhances the intensity of the BR response, as reflected by the induction of ACS gene expression. To determine whether this induction is mediated by BES1/BZR2 (hereafter referred to as BES1), we analyzed roots of pGL2-BES1-D plants, which express the dominant active version of BES1 in nonhair cells (in the wild-type background) (Supplemental Fig. S3J). Similar to pGL2-BRI1, pGL2-BES1-D roots exhibited high basal expression levels of the analyzed ACS genes and had short cortical cells (Fig. 2C; Supplemental Fig. 3F,K). In agreement with the rise in ACS gene expression, pGL2-BRI1 roots were hypersensitive to exogenously applied ACC, as manifested by intensified growth inhibition in response to low ACC concentrations, while roots of pGL2-BES1-D plants were less affected (Fig. 2D). Thus, BRI1 activity in nonhair cells elevates ACS genes at least partly via BES1.
Ethylene activity is both necessary and sufficient for the inhibitory BRI1 effect in nonhair cells
The enhanced sensitivity of pGL2-BRI1 roots to ACC raised the possibility that a local rise in ethylene production can inhibit cell elongation. To determine whether ethylene activity in nonhair cells is sufficient for inhibition of cell elongation in the inner cells, we established pGL2-eto2 lines, which express a dominant active form of ACS5 (Vogel et al. 1998). pGL2-eto2 roots were short as compared with those of wild type, demonstrating that ACC production in nonhair cells is sufficient to inhibit root length (Fig. 2E). To determine whether the ethylene signaling pathway is also necessary for BRI1-mediated inhibition of cell elongation in nonhair cells, we crossed pGL2-BRI1 lines with the ethylene-insensitive ETHYLENE-INSENSITIVE2 (ein2) mutant (Alonso et al. 1999). The short and wider cortical cell length of the parental pGL2-BRI1 and its root sensitivity to BL (Fig. 2F,G; Supplemental Fig. S4A) were largely suppressed in ein2;pGL2-BRI1, while the high basal expression level of ACS genes was maintained (Supplemental Fig. 3H,I). Suppression of root sensitivity to BL was also observed in progenies of a cross between pGL2-BRI1 and the auxin influx carrier mutant AUXIN1 (aux1), in agreement with the known interaction between ethylene and auxin, where AUX1 is required for full inhibition of cell expansion by ethylene (Supplemental Fig. S4B; Swarup et al. 2007). Thus, enhanced BRI1 activity in nonhair cells inhibits cortical cell elongation via activation of the ethylene signaling pathway.
High BRI1 expression in nonhair cells triggers high localized deposition of crystalline cellulose, which impacts root cell elongation
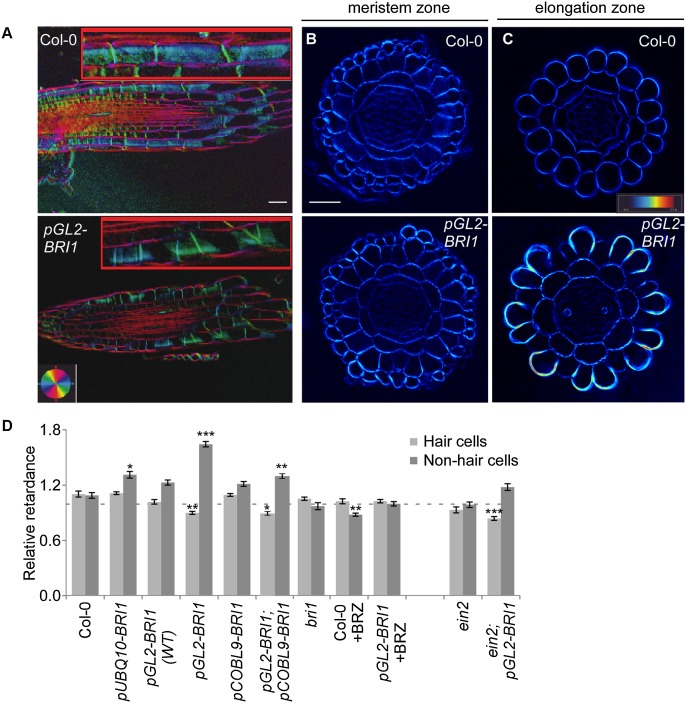
Unidirectional cell expansion is affected by the accumulation of crystalline cellulose and the angle of microfibril arrangement (Baskin 2005; Fujita et al. 2011). To determine whether the inhibitory effect of elevated BR activity in nonhair cells involves modulation of these structural parameters, we analyzed the cellulose microfibril orientations and the relative cellulose crystallinity levels in cross-sections of similar thickness of wild-type and pGL2-BRI1 plants using a computerized polarized light-based system (Materials and Methods; Abraham and Elbaum 2013). While no significant difference in the microfibril angle of all elongating epidermal walls was observed (Fig. 3A), we revealed elevated deposition of crystalline cellulose in nonhair cells entering the elongation zone in pGL2-BRI1 lines only (Figs. 3B–D; Supplemental Fig. S5A,B). In contrast, lines also expressing BRI1 in hair cells [pUBQ10-BRI1, pCOBL9-BRI1;pGL2-BRI1, and pGL2-BRI1(WT)] displayed reduced cellulose crystallinity in nonhair cells as compared with pGL2-BRI1. BRZ treatment abolished the accumulation of cellulose in pGL2-BRI1, in accordance with the demonstrated BR-dependent effect. Interestingly, the high crystallinity in pGL2-BRI1 nonhair cells was suppressed in ein2;pGL2-BRI1 and aux1;pGL2-BRI1 (Fig. 3D; Supplemental Fig. S5C), suggesting that high ethylene BR-activated signaling drives cell wall remodeling during rapid cell expansion.
Figure 3.
High relative BRI1 expression in nonhair cells triggers local accumulation of crystalline cellulose. (A–C) Polarized light microscopy images of longitudinal (A) and transverse (B,C) sections of the root elongation zone showing microfibril angle and quantity, respectively. (A) The inset highlights the signal at the cell wall surface. Note the similar microfibril angle in pGL2-BRI1 and wild-type (Col-0) roots, as inferred by the color key. Bar, 50 µm. (B) pGL2-BRI1 and wild type have similar levels of cellulose crystallinity in meristematic cells. Bar, 50 μM. (C) High accumulation of crystalline cellulose in the elongation zone of nonhair cells of pGL2-BRI1 as compared with wild type (note the light blue–yellow signal in the cell wall). (D) Quantification of retardance in the outer cell wall of hair and of nonhair cells. Values are expressed as the ratio of retardance between the outer epidermal cell wall and the inner cortical cell wall. Note that the high deposition of crystalline cellulose in nonhair cells was unique to pGL2-BRI1 roots in the bri1 background and required the ethylene signaling component EIN2 (mean ± SE; 40 < n < 600). (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 with two tailed t-test.
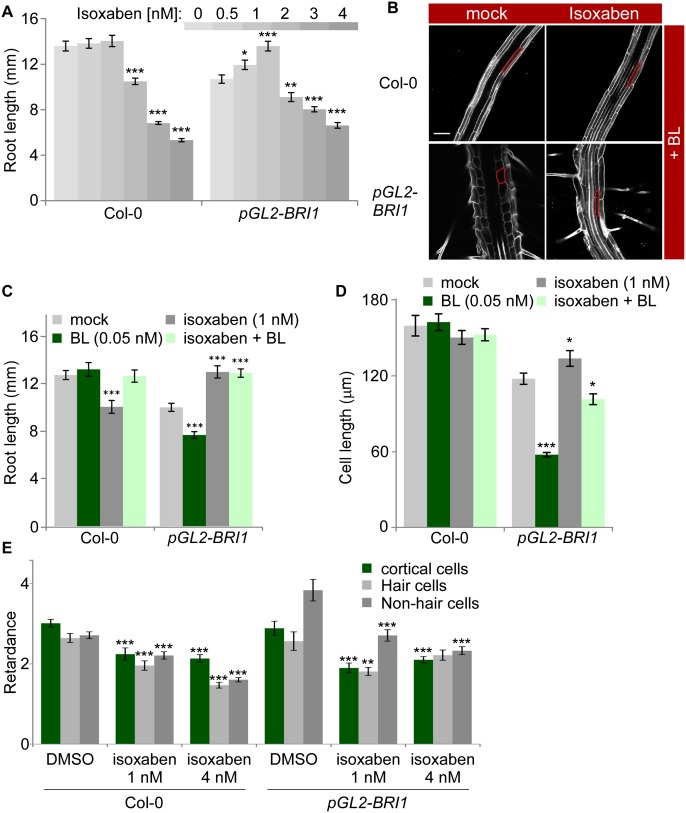
While mechanisms underlying high accumulation of crystalline cellulose are unknown, they could involve enhanced production of cellulose. We therefore reasoned that attenuation of cellulose production in pGL2-BRI1 roots would reduce the extent of inhibition of their unidirectional cell expansion. To test this hypothesis, we subjected pGL2-BRI1 roots to increased concentrations of isoxaben, a well-established inhibitor of cellulose synthesis (Desprez et al. 2002). As shown in Figure 4A, pGL2-BRI1 roots were more resistant to the inhibitory effect of the drug as compared with wild type and exhibited partial restoration of their typical short-length phenotype. Cellular analysis revealed that the longer root obtained upon treatment with 1 nM isoxaben was a result of improved cell elongation, while similar treatment caused wild-type cortical root cells to become wider (Fig. 4C,D; Supplemental Fig. S5D). Furthermore, pGL2-BRI1 roots grown on isoxaben were less hypersensitive to BL, as implicated in reduced inhibition of cortical cell length (Fig. 4B–D) and in agreement with lower crystalline cellulose levels in their nonhair cells (Fig. 4E). Thus, differential BRI1 activity impacts growth via local structural modulation of the cell wall.
Figure 4.
Moderate inhibition of cellulose production facilitates unidirectional growth in pGL2-BRI1 lines. (A) The effect of increasing concentrations of isoxaben on the root length of wild-type (Col-0) and pGL2-BRI1 roots (mean ± SE; 32 < n < 36). Note the enhanced effect of low isoxaben concentrations on pGL2-BRI1 root growth. (B) Confocal microscopy image of wild-type (Col-0) and pGL2-BRI1 roots untreated and treated with 1 nM isoxaben in the presence of BL, with the cortical cell highlighted in red. Bar, 50 µm. (C,D) Quantification of root length and mature cortical cell length of roots untreated and treated with 1 nM isoxaben in the absence and presence of BL (mean ± SE; 22 < n < 32 and 16 < n < 88, respectively). (E) Quantification of retardance in the inner cortical and outer cell wall of hair and of nonhair cells. Absolute values are shown. Note the small reduction in crystalline cellulose levels in nonhair cells of pGL2-BRI1 as compared with wild type in response to a mild isoxaben treatment (mean ± SE; 22 < n < 60). (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 with two tailed t-test.
Discussion
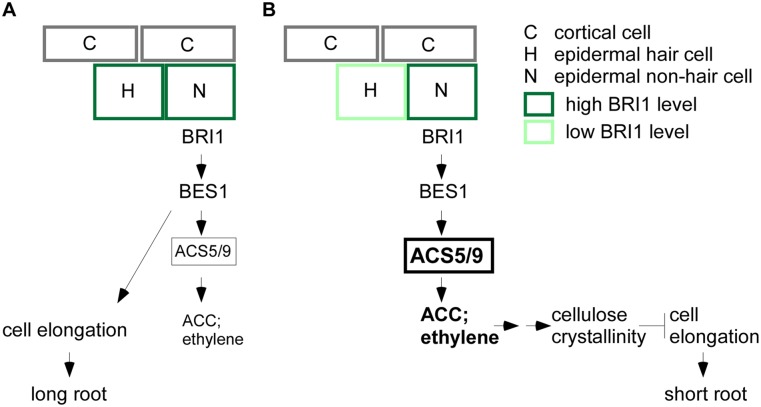
A key open issue in developmental biology questions how the individual cells of an organ reach their final size in a coordinated manner. Our study shows that the relative expression level of BRI1 in neighboring epidermal cells determines the outcome of its downstream signaling. Low relative BRI1 activity in hair cells leads to an enhanced response to BR signaling in nonhair cells, consequently triggering ACS genes at least in part via BES1/BZR2 (Fig. 5). As a result, the ethylene precursor ACC accumulates and enhances ethylene signaling, which in turn inhibits unidirectional cell expansion and stimulates local deposition of crystalline cellulose. Enhanced cellulose production fails to support cell elongation of nonhair cells and their interconnected neighboring cells.
Figure 5.
Schematic representation of BRI1-regulated root cell elongation. A model illustrating that the relative expression level of BRI1 in neighboring epidermal cells determines the intensity of its downstream signaling and subsequent whole-root growth via positive (A) and negative (B) effects (see the text).
We speculate that mechanisms coordinating BR signaling between hair and nonhair cells involve interwoven genetic and mechanical factors. An increasing list of signaling components has been shown to shift between adjacent cells through plasmodesmata, thereby inducing nonautonomous functions (Sevilem et al. 2012). Growth coordination is also set by mechanical feedback loops imposed by differential growth rates among adjacent cells (Hamant et al. 2008; Heisler et al. 2010; Uyttewaal et al. 2012). Stochastic differences in BR signaling strength between epidermal cells would potentially initiate this feedback, as in a self-organizing system. Impaired cell wall homeostasis has also been shown to affect growth via cell wall integrity sensors (Hematy and Hofte 2008; Wolf et al. 2012a). The proposed involvement of signaling from the cell wall in controlling growth was supported by a recent study in which cell wall perturbation was sufficient to activate BR responses via a yet unknown mechanism (Wolf et al. 2012b). In this scenario, accumulation of crystalline cellulose would act as a compensatory signal.
The crystalline to amorphous cellulose ratio is important for plant growth and morphogenesis, since it inherently impinges on the mechanical properties of the cell wall (Schindelman et al. 2001; Xu et al. 2008; Fujita et al. 2011; Abraham and Elbaum 2013; Liu et al. 2013). X-ray diffraction studies demonstrated that a high degree of crystalline cellulose correlates with attenuated unidirectional cell expansion in the growing regions of inflorescence stems in a process that does not involve changes in cellulose microfibril orientation (Fujita et al. 2011). In agreement with these findings, our study showed a normal angle of cellulose microfibrils in nonhair cells and a high crystalline cellulose associated with their reduced unidirectional growth. Our study went beyond mere establishment of a correlation by demonstrating the dependency of reduced unidirectional growth and sensitivity to BRs on enhanced cellulose production.
Perturbing organ growth in a cell type-specific manner, as compared with loss-of-function studies per se, challenges the robustness of the system, revealing novel aspects previously unpredicted by models (e.g., Kierzkowski et al. 2013). Our work also supports the importance of using and developing tools for tissue-specific structural and biochemical modifications of the cell wall (e.g., Peaucelle et al. 2011) in combination with spatiotemporal perturbation of hormonal signaling pathways in attempts to obtain novel insights into final size determination.
Materials and methods
Plant material, growth conditions, and chemical treatments
All Arabidopsis thaliana lines were on the Columbia (Col-0) background. ein2 and eto2 seeds were obtained from the Arabidopsis Biological Resource Center (CS8844) and Nottingham Arabidopsis Stock Centre (N8059), respectively. aux1-21 was a gift from M. Bennett (Marchant and Bennett 1998). Transgenic lines were as in Hacham et al. (2011), except for pBRI1-BRI1-GFP (Geldner et al. 2007) and pGL2-eto2 as well as pCOBL9-BRI1 (established in this study). Plant agar media were as described in Hacham et al. (2011), supplemented with 0.2% sucrose. Plates with sterilized seeds were stratified in the dark for 2 d at 4°C and then transferred to 22°C in continuous light (∼70 μmol m−2 s−1) for 7 d. For chemical and hormone treatments, 3-d-old seedlings were transferred to the relevant supplemented medium and analyzed after an additional 4 d. BRZ, BL, and isoxaben were dissolved in 100% dimethyl sulfoxide (DMSO). BRZ was added to a final concentration of 2 μM.
Vector constructs and transgenic lines
Plants were transformed by the standard floral dip method using Agrobacterium containing the pMLBART or pART27 binary vector. The promoter fragment upstream of the COBL9 coding sequences was amplified from genomic DNA and cloned to the polylinker of pBJ36. The coding sequence of the eto2 gene was amplified from cDNA, prepared from eto2 mutant RNA, and cloned into the 3′ end of pGL2 in pBJ36 (Supplemental Table S2). To establish transgenic lines for polysomal RNA isolation from hair and nonhair cells, the Flag-RPL18 fragment was amplified from pGATA:HF-RPL18 (Mustroph et al. 2009b) and subcloned by KpnI/KpnI into the 3′ end of pGL2 and pCOBL9 in pBJ36. Primer sequences used for amplification and the corresponding restriction sites for pBJ36 insertion are listed in Supplemental Table S2. Transgenic lines were selected for BASTA or kanamycin resistance. The homozygous bri1 background was verified using CAPS marker digested with PmeI.
Root growth analysis
For root elongation measurements, 7-d-old seedlings were scanned, and root length was measured using ImageJ software and the NeuronJ plugin. Meristematic cell number, represented by the number of cortical cells, was determined from confocal microscopy images. The number of independent experiments and the two-tailed t-test calculations (Microsoft Excel) are listed in Supplemental Table S3.
Confocal microscopy
Fluorescence signals were detected using an LSM 510 META confocal laser-scanning microscope (Zeiss) with a 25× water immersion objective lens (NA 0.8). Roots were imaged in water supplemented with 10 μg/mL propidium iodide (PI). PI and GFP were viewed at excitation wavelengths of 488 nm and 561 nm, respectively. Fluorescence emission was collected at 575 nm for PI and between 500 and 530 nm bandpass for GFP.
Quantification of fluorescence signal
To determine the fluorescence profile of BRI1-GFP, Z stack images were acquired from three overlapping zones of the root using the same confocal settings for all transgenic lines. The analysis was performed with Fiji software (http://fiji.sc/fiji). Images were stitched using the Stitching plugin. The resulting image was projected in the Z-axis using average projection. A segmented line was then used to mark the region of interest along the epidermis. For Supplemental Figure S3F, the GFP signal was quantified using ImageJ. The polygon tool was used to mark regions with fluorescence signal, and background fluorescence was subtracted from the measurement. The number of independent experiments and the two-tailed t-test calculations (Microsoft Excel) are listed in Supplemental Table S3.
RNA extraction and expression analysis
Total RNA extraction and quantitative real-time PCR assays were performed as described in Hacham et al. (2011). Immunopurification of Flag-tagged polysomes from root cells was performed as described in Mustroph et al. (2009a). Immunopurified RNA was then linearly amplified using the MessageAmp II aRNA kit (Ambion) and similarly subjected to quantitative real-time PCR. The number of independent experiments is listed in Supplemental Table S3.
Microarray experiment
RNA was extracted from root tips of 7-d-old seedlings and hybridized to an Affymetrix Arabidopsis ATH1 array. Samples were run in a fluidic station (FS-450) and scanned using a GeneChip Scanner (300 7G). Data were quantitated by the Affymetrix expression console.
Anatomical cross-sections for polscope
Seedlings were fixed and stained in 1.25% glutaraldehyde, 0.05 M sodium cacodylate, 0.05% methylene blue, 0.05% borax, and 0.05% azure overnight at 4°C. Fixed seedlings were next dehydrated with ethanol and soaked for a few days in Historesin infiltration medium (Leica) and blocked with Historesin hardener, according to the manufacturer’s instructions. Ultramicrotome (LKB 8800) and homemade glass knives were used to section samples at ∼3-μm width. Sections were restained with diluted methylene blue solution, heat-dried, and covered with Immu-Mount (Thermo Scientific) and a coverslip.
Polarized light analysis
The LC-PolScope image processing system (CRi, Inc.), an automated method that detects small variations in light retardance, was used for the analysis of crystalline cellulose. This method is based on the property of the crystalline parts of cellulose microfibrils to split the light beam and retard part of the light (for example, see Iyer et al. 1968). The light retardation is strongest for microfibrils that lie perpendicularly to the direction of light propagation. For microfibrils with similar orientation, the higher the crystallinity level, the larger the light retardance (Abraham and Elbaum 2013). Images were captured using a Nikon Eclipse 80i microscope equipped with a camera and a liquid crystal from an Abrio imaging system. Abrio version2.2.0.1 software was used to analyze images and extract retardance values. Retardance values for the outer cell wall of epidermal cells were normalized to the values obtained for the inner cortical cell wall to minimize subtle differences in sample thickness. In Figure 4E, retardance values were presented without normalization. Cross-sections of the elongation zone (identified by the end of root cap cells), from at least three independent roots were analyzed. The number of independent experiments and the two-tailed t-test calculations (Microsoft Excel) are listed in Supplemental Table S3.
Acknowledgments
We appreciate the technical assistance of A. Sisso and E. Okopnik. We also thank M. Duvshani-Eshet and N. Dahan (Life Sciences and Engineering Infrastructure Unit and the Russell Barrie Nanotechnology Institute [RBNI] at Technion), M. Bennett (University of Nottingham), and J. Bailey-Serres (University of California at Riverside) for sharing published material. This research was supported by grants from FP7-PEOPLE-IRG-2008, Binational Agriculture Research and Development (BARD; IS-4246–09), and Israel Science Foundation (ISF; 1498/09).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.239335.114.
References
- Abraham Y, Elbaum R 2013. Quantification of microfibril angle in secondary cell walls at subcellular resolution by means of polarized light microscopy. New Phytol 197: 1012–1019 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR 1999. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Baskin TI 2005. Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Bradford KJ, Trewavas AJ 1994. Sensitivity thresholds and variable time scales in plant hormone action. Plant Physiol 105: 1029–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Song S, Dhugga KS, Rafalski JA, Benfey PN 2007. Combining expression and comparative evolutionary analysis. The COBRA gene family. Plant Physiol 143: 172–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD 2011. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ 2005. Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refregier G, Desnos T, Aletti E, Py N, Pelletier S, Hofte H 2002. Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol 128: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K 1994. Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474 [Google Scholar]
- Fridman Y, Savaldi-Goldstein S 2013. Brassinosteroids in growth control: how, when and where. Plant Sci 209: 24–31 [DOI] [PubMed] [Google Scholar]
- Fujita M, Himmelspach R, Hocart CH, Williamson RE, Mansfield SD, Wasteneys GO 2011. Cortical microtubules optimize cell-wall crystallinity to drive unidirectional growth in Arabidopsis. Plant J 66: 915–928 [DOI] [PubMed] [Google Scholar]
- Geldner N, Hyman DL, Wang X, Schumacher K, Chory J 2007. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garcia MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-Garcia S, Russinova E, Cano-Delgado AI 2011. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859 [DOI] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S 2011. Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jonsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, et al. 2008. Developmental patterning by mechanical signals in Arabidopsis. Science 322: 1650–1655 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jonsson H, Traas J, Meyerowitz EM 2010. Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol 8: e1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hematy K, Hofte H 2008. Novel receptor kinases involved in growth regulation. Curr Opin Plant Biol 11: 321–328 [DOI] [PubMed] [Google Scholar]
- Iyer KRK, Neelakantan P, Radhakrishnan T 1968. Birefringence of native cellulosic fibers. I. Untreated cotton and ramie. J Polym Sci, A-2, Polym Phys 6: 1747–1758 [Google Scholar]
- Kierzkowski D, Lenhard M, Smith R, Kuhlemeier C 2013. Interaction between meristem tissue layers controls phyllotaxis. Dev Cell 26: 616–628 [DOI] [PubMed] [Google Scholar]
- Le J, Vandenbussche F, Van der Straeten D, Verbelen JP 2001. In the early response of Arabidopsis roots to ethylene, cell elongation is up- and down-regulated and uncoupled from differentiation. Plant Physiol 125: 519–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF, Shang-Guan KK, Zhang BC, Liu XL, Yan MX, Zhang LJ, Shi YY, Zhang M, Qian Q, Li JY, et al. 2013. Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet 9: e1003704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Bennett MJ 1998. The Arabidopsis AUX1 gene: a model system to study mRNA processing in plants. Plant Mol Biol 36: 463–471 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW 1996. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Mussig C, Shin GH, Altmann T 2003. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol 133: 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Juntawong P, Bailey-Serres J 2009a. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol 553: 109–126 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J 2009b. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY 2012. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Hofte H 2011. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol 21: 1720–1726 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN 2012. Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles L, Stepanova A, Alonso J 2013. Molecular mechanisms of ethylene–auxin interactions. Mol Plant 6: 1734–1737 [DOI] [PubMed] [Google Scholar]
- Ruzicka K, Ljung K, Vanneste S, Podhorska R, Beeckman T, Friml J, Benkova E 2007. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Chory J 2008. Growth coordination and the shoot epidermis. Curr Opin Plant Biol 11: 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Peto C, Chory J 2007. The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202 [DOI] [PubMed] [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN 2001. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilem I, Miyashima S, Helariutta Y 2012. Cell-to-cell communication via plasmodesmata in vascular plants. Cell Adhes Migr 7: 27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM 2007. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. 2010. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Perry P, Hagenbeek D, Van Der Straeten D, Beemster GT, Sandberg G, Bhalerao R, Ljung K, Bennett MJ 2007. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19: 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomas S, Beemster GT, Bennett MJ 2012. Hormonal regulation of root growth: integrating local activities into global behaviour. Trends Plant Sci 17: 326–331 [DOI] [PubMed] [Google Scholar]
- Uyttewaal M, Burian A, Alim K, Landrein B, Borowska-Wykret D, Dedieu A, Peaucelle A, Ludynia M, Traas J, Boudaoud A, et al. 2012. Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell 149: 439–451 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Woeste KE, Theologis A, Kieber JJ 1998. Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc Natl Acad Sci 95: 4766–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste KE, Vogel JP, Kieber JJ 1999. Factors regulating ethylene biosynthesis in etiolated Arabidopsis thaliana seedlings. Physiol Plant 105: 478–484 [Google Scholar]
- Wolf S, Hematy K, v H 2012a. Growth control and cell wall signaling in plants. Annu Rev Plant Biol 63: 381–407 [DOI] [PubMed] [Google Scholar]
- Wolf S, Mravec J, Greiner S, Mouille G, v H 2012b. Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol 22: 1732–1737 [DOI] [PubMed] [Google Scholar]
- Xu SL, Rahman A, Baskin TI, Kieber JJ 2008. Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20: 3065–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. 2011. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]