Abstract
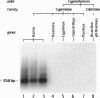
DNA is the first SINE isolated from zebrafish (Danio rerio) exhibiting all the hallmarks of these tRNA-derived elements. DANA is unique in its clearly defined substructure of distinct cassettes. In contrast to generic SINE elements, DANA appears to have been assembled by insertions of short sequences into a progenitor, tRNA-derived element. Once associated with each other, these subunits were amplified as a new transposable element with such a remarkable success that DANA-related sequences comprise approximately 10% of the modern zebrafish genome. At least some of the sequences comprised by the full-length element were capable of movement, forming a new group of mobile, composite transposons, one of which caused an insertional mutation in the zebrafish no tail gene. Being present only in the genus Danio, and estimated to be as old as the genus itself, DANA may have played a role in Danio speciation by massive amplification and genome-wide dispersion. There are extensive DNA polymorphisms between zebrafish populations and strains detected by PCR amplification using primers specific to DANA, suggesting that the DANA element will be useful as a molecular tool for genetic and phylogenetic analyses.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brosius J. Retroposons--seeds of evolution. Science. 1991 Feb 15;251(4995):753–753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- Coltman D. W., Wright J. M. Can SINEs: a family of tRNA-derived retroposons specific to the superfamily Canoidea. Nucleic Acids Res. 1994 Jul 25;22(14):2726–2730. doi: 10.1093/nar/22.14.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels G. R., Deininger P. L. Repeat sequence families derived from mammalian tRNA genes. 1985 Oct 31-Nov 6Nature. 317(6040):819–822. doi: 10.1038/317819a0. [DOI] [PubMed] [Google Scholar]
- Finnegan D. J. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989 Apr;5(4):103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- He L., Zhu Z., Faras A. J., Guise K. S., Hackett P. B., Kapuscinski A. R. Characterization of AluI repeats of zebrafish (Brachydanio rerio). Mol Mar Biol Biotechnol. 1992 Apr;1(2):125–135. [PubMed] [Google Scholar]
- Ivics Z., Izsvák Z., Hackett P. B. Enhanced incorporation of transgenic DNA into zebrafish chromosomes by a retroviral integration protein. Mol Mar Biol Biotechnol. 1993 Jun;2(3):162–173. [PubMed] [Google Scholar]
- Izsvák Z., Ivics Z., Hackett P. B. Characterization of a Tc1-like transposable element in zebrafish (Danio rerio). Mol Gen Genet. 1995 May 10;247(3):312–322. doi: 10.1007/BF00293199. [DOI] [PubMed] [Google Scholar]
- Johnson S. L., Midson C. N., Ballinger E. W., Postlethwait J. H. Identification of RAPD primers that reveal extensive polymorphisms between laboratory strains of zebrafish. Genomics. 1994 Jan 1;19(1):152–156. doi: 10.1006/geno.1994.1026. [DOI] [PubMed] [Google Scholar]
- Kaukinen J., Varvio S. L. Artiodactyl retroposons: association with microsatellites and use in SINEmorph detection by PCR. Nucleic Acids Res. 1992 Jun 25;20(12):2955–2958. doi: 10.1093/nar/20.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido Y., Aono M., Yamaki T., Matsumoto K., Murata S., Saneyoshi M., Okada N. Shaping and reshaping of salmonid genomes by amplification of tRNA-derived retroposons during evolution. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2326–2330. doi: 10.1073/pnas.88.6.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. G., Loomis C., Cowan N. J. Sequence of an expressed human beta-tubulin gene containing ten Alu family members. Nucleic Acids Res. 1984 Jul 25;12(14):5823–5836. doi: 10.1093/nar/12.14.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A., Biermann C. H., Ortí G. The phylogenetic position of the zebrafish (Danio rerio), a model system in developmental biology: an invitation to the comparative method. Proc Biol Sci. 1993 Jun 22;252(1335):231–236. doi: 10.1098/rspb.1993.0070. [DOI] [PubMed] [Google Scholar]
- Murata S., Takasaki N., Saitoh M., Okada N. Determination of the phylogenetic relationships among Pacific salmonids by using short interspersed elements (SINEs) as temporal landmarks of evolution. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):6995–6999. doi: 10.1073/pnas.90.15.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinder H., Bayer T. A., Gertzen E. M., Hoffmann W. Molecular analysis of the ependymin gene and functional test of its promoter region by transient expression in Brachydanio rerio. DNA Cell Biol. 1992 Jul-Aug;11(6):425–432. doi: 10.1089/dna.1992.11.425. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Saffer J. D., Lerman M. I. Unusual class of Alu sequences containing a potential Z-DNA segment. Mol Cell Biol. 1983 May;3(5):960–964. doi: 10.1128/mcb.3.5.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Merker S., van Eeden F. J., Halpern M. E., Kimmel C. B., Nüsslein-Volhard C. no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development. 1994 Apr;120(4):1009–1015. doi: 10.1242/dev.120.4.1009. [DOI] [PubMed] [Google Scholar]
- Sidhu M. S., Helen B. K., Athwal R. S. Fingerprinting human chromosomes by polymerase chain reaction-mediated DNA amplification. Genomics. 1992 Nov;14(3):728–732. doi: 10.1016/s0888-7543(05)80175-2. [DOI] [PubMed] [Google Scholar]
- Sinnett D., Deragon J. M., Simard L. R., Labuda D. Alumorphs--human DNA polymorphisms detected by polymerase chain reaction using Alu-specific primers. Genomics. 1990 Jul;7(3):331–334. doi: 10.1016/0888-7543(90)90166-r. [DOI] [PubMed] [Google Scholar]
- Soh J., Mariano T. M., Bradshaw G., Donnelly R. J., Pestka S. Generation of random internal deletion derivatives of YACs by homologous targeting to Alu sequences. DNA Cell Biol. 1994 Mar;13(3):301–309. doi: 10.1089/dna.1994.13.301. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Tsukada T., Notake M., Nakanishi S., Numa S. Structural analysis of repetitive DNA sequences in the bovine corticotropin-beta-lipotropin precursor gene region. Nucleic Acids Res. 1982 Mar 11;10(5):1459–1469. doi: 10.1093/nar/10.5.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- White S. E., Habera L. F., Wessler S. R. Retrotransposons in the flanking regions of normal plant genes: a role for copia-like elements in the evolution of gene structure and expression. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11792–11796. doi: 10.1073/pnas.91.25.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietkiewicz E., Labuda M., Sinnett D., Glorieux F. H., Labuda D. Linkage mapping by simultaneous screening of multiple polymorphic loci using Alu oligonucleotide-directed PCR. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8448–8451. doi: 10.1073/pnas.89.18.8448. [DOI] [PMC free article] [PubMed] [Google Scholar]