Abstract
Aim:
Activating transcription factor 6 (ATF6) is a key signal transducer of endoplasmic reticulum stress (ER stress). This study was conducted to clarify the potential role of ATF6 in the insulin signaling pathway under chronic ER stress.
Methods:
ER stress of HEK293 cells was induced with tunicamycin (2 μg/mL). The cells were transfected with ATF6α or ATF6β. The phosphorylation level of insulin receptor (IR) was analyzed using Western blot. The changes in ER stress and ERK signaling pathways were explored using Western blot and quantitative real-time PCR.
Results:
Tunicamycin-induced chronic ER stress attenuated IR tyrosine phosphorylation in a time-dependent manner, whereas over-expression of ATF6 protected IR from desensitization. ATF6 modulation of IR suppression was associated with insulin-stimulated extracellular signal-regulated kinase (ERK) phosphorylation. The treatment of the cells with a specific ERK inhibitor U0126 (10 μmol/L) mimicked the effect of ATF6 over-expression and restored the insulin-stimulated IR phosphorylation. The treatment of the cells with the ERK activator epidermal growth factor (EGF, 200 ng/mL) decreased the protection effect of ATF6 on IR.
Conclusion:
Our results demonstrate that ATF6 may serve as a potential therapeutic target for the treatment of insulin resistance and type 2 diabetes.
Keywords: activating transcription factor 6, ER stress, insulin receptor, MEK/ERK pathway, type 2 diabetes
Introduction
Type 2 diabetes is a severe threat to worldwide public health. Its main features include relative insulin deficiency, insulin resistance and hyperglycemia1. Over the past few years, tremendous progress has been made in elucidating the molecular mechanisms involved in the origin and development of insulin resistance, although many gaps still remain to be filled.
The endoplasmic reticulum (ER) is an organelle where protein folding, post-translational modification and oligomerization take place. This reticulum system senses alterations of intracellular homeostasis provoked by stimuli, subsequently inducing an unfolded protein response (UPR)2. The UPR decreases protein translation, increases chaperone expression and accelerates the degradation of misfolded proteins via three main signal transducers to alleviate cellular stress and restore ER homeostasis. These transducers include inositol-requiring enzyme-1α (IRE-1α), PKR-like ER kinase (PERK), and activating transcription factor 6 (ATF6). When ER stress fails to be attenuated, apoptosis will be activated and cells are subjected to chronic stimulation2.
Recently, there has been a special interest in ER stress because of its central role in the pathogenesis of type 2 diabetes, and valuable results have been discovered3, 4, 5, 6. For example, it was found that molecular indicators of ER stress, such as PERK, eukaryotic initiation factor 2α (eIF2α) and glucose-regulated protein 78 (GRP78), increased in liver and adipose tissues of dietary and genetic murine obesity models. In addition, administration of tunicamycin, an ER stress inducer, results in suppression of insulin receptor signaling (insulin receptor substrate 1 (IRS-1) tyrosine and AKT serine phosphorylation) in liver cells3. ER stress was also discovered to repress the insulin signaling pathway through activation of the IRE-1α and c-Jun N-terminal kinase (JNK) pathways3, 7, 8. Over-expression of X-box binding protein 1 (XBP-1), another important ER stress signal transducer, down-regulates JNK activity, enhances insulin receptor signaling pathway activity and restores glucose homeostasis3. The attenuation of ER stress by chemical chaperone [eg, 4-phenyl butyric acid (PBA) and taurine-conjugated derivative (TUDCA)] treatment or oxygen-regulated protein 150 kDa (Orp150) over-expression has been confirmed effective in increasing insulin sensitivity and ameliorating diabetes in vivo4, 5, 6. Although targeting ER stress has been hinted at as a promising therapeutic strategy for anti-diabetic agent discovery, further investigations should remain in consideration of the complexity of the ER stress network.
Activating transcription factor 6 (ATF6) is a 90-kDa ER-anchoring protein with cytoplasmic and transmembrane domains. The cytoplasmic domain contains a transcriptional activation domain (TAD) and a b-ZIP DNA binding motif9, 10. Two isoforms of ATF6 have been determined to date, ATF6α and ATF6β. Although the N-terminals of these two isoforms share conserved b-ZIP domains, ATF6α and ATF6β differ greatly in their transcriptional abilities. This is because of their dissimilar transcriptional activation domains11, which leads to different functions on target genes12, 13. When unfolded proteins accumulate in the ER by chemical stimulation or in physiopathological conditions14, 15, ATF6 is released from the ER lumen by sequential cleavage of Site 1 (S1P) and Site 2 proteases (S2P)9. This produces a 50-kDa fragment that moves to the nucleus to activate chaperone genes, such as GRP78, GRP94 and protein disulfide isomerase (PDI), thus ameliorating ER stress16.
Regarding the potent involvement of ER stress in the pathogenesis of type 2 diabetes17, 18, 19 and the determined function of ATF6 in the management of cell homeostasis15, 16, we attempted to clarify the potential role of ATF6 in the insulin signaling pathway. We discovered that both ATF6α and ATF6β attenuate tunicamycin stimulated IR desensitization through an ERK-related pathway. It is expected that members of the ATF6 family could be potential therapeutic targets for anti-obesity and type 2 diabetes drug discovery.
Materials and methods
Materials
Restriction enzymes were purchased from New England Biolabs, NJ, USA. Cell culture plastic ware was from Corning Inc, MA, USA. DMEM culture medium and fetal bovine serum were from Invitrogen, CA, USA. Tunicamycin, insulin, epidermal growth factor, U0126, MTT [3-(4,5-dimethyl-2-yl)-2,5-diphenyl tetrazolium bromide] and SB203580 were obtained from Sigma, USA. RNAiso, RT reagent Kit and SYRB Premix Ex Taq were purchased from TaKaRa, Japan. Anti-ATF6α (full and cleavage) antibody was from Imgenex Corp, San Diego, USA. Anti-IR, anti-phospho-IGF-I receptor β (Tyr1135/1136)/insulin receptor β (Tyr1150/1151), anti-AKT, anti-phospho-AKT (Ser473), anti-p44/42 Erk1/2, anti-phospho-p44/42 Erk1/2 (Thr202/Tyr204), anti-SAPK/JNK, anti-phospho-SAPK/JNK (Thr183/Tyr185), anti-p38 MAPK, anti-phospho-p38 MAPK (Thr180/Tyr182), anti-GRP78/Bip, anti-GAPDH, anti-eIF2α-pSer51 and anti-IRE1α-pSer74 antibodies were purchased from Cell Signaling Technology, MA, USA. Plasmids pcDNA3.1-hATF6α and β were kindly provided by Prof Kazutoshi MORI (Department of Biophysics, Graduate School of Science, Kyoto University, Kyoto, Japan).
Plasmid constructs
ATF6α and ATF6β genes were amplified by Pfu polymerase (Tiangen) from the plasmids pcDNA3.1-hATF6α and pcDNA3.1-hATF6β. The genes were then ligated into Xho I/Hind III-digested and Xho I/EcoR I-digested pEYFP-C1 vectors, respectively.
Stable transfection
HEK293 cells were plated into 3.5 cm plates and transfected with 3 μg pEYFP-C1/pEYFP-C1-hATF6α/pEYFP-C1-hATF6β by using 8 μL Lipofectamine™ 2000 (Invitrogen Inc, California, USA). Medium was changed after 5 h. On the next day, cells were harvested and plated into 10-cm plates in the ratio of 1:400 and incubated in 10% DMEM/FBS. Medium was refreshed every 2 d. After 2 weeks, resistant foci of clones were selected, verified and maintained for further study.
Western blot analysis
Supernatants of the whole cell extracts were fractionated by SDS-PAGE and transferred to Hybond-c nitrocellulose membrane (Amersham Bioscience, Sweden). The membranes were blocked for 1 h at room temperature and then incubated overnight at 4 °C in TBST buffer (5% milk) containing antibody. On the next day, the membranes were incubated for an hour at room temperature in TBST buffer (5% milk) containing anti-rabbit IgG or anti-mouse IgG (Jackson-ImmunoResearch, West Grove, PA, USA). Blots were visualized by incubation with SuperSignal West Dura chemiluminescence kit (Pierce Biotechnology, Rockford, USA) and exposed to light-sensitive film. The experiments were performed in triplicate, and bands were quantified using Image-Pro Plus software and statistically analyzed. Each band was calculated as “intensity×area” using the Image-Pro Plus software (MediaCybernetics, Micromecanique, France). SD was calculated from the three replications of the experiment.
RNA preparation and quantitative real-time PCR
Total RNA was extracted from transiently transfected HEK293 cells or stable cell lines (6-well) using RNAiso (TaKaRa, Japan) reagent according to the kit instruction. Complementary DNA was synthesized using an RT reagent kit (TaKaRa, Japan). Real-time PCR was performed using SYBR Premix Ex Taq (TaKaRa, Japan) on DNA Engene OpticonTM 2 System (MJ Research, Waltham, MA, USA). The PCR cycle was 95 °C for 10 s, 58 °C for 45 s, and 72 °C for 30 s. The primers are listed in Table 1.
Table 1. Primer sequences of selected genes for quantitative real-time PCR.
| Gene | Primer sequence |
|---|---|
| ATF6α | sense 5′-TCAACTCAGCACGTTCCTGAGG-3′ antisense 5′-GCACATTCACCACATGGTTAGG-3′ |
| ATF6β | sense 5′-ATCCAGCGAGGCTCTTGGGGTAGG-3′ antisense 5′-TGGCATTGGGACAGTGGTTAGCAC-3′ |
| β-actin | sense 5′-GATGAGATTGGCATGGCTTT-3′ antisense 5′-GAGAAGTGGGGTGGCTT-3′ |
| GRP94 | sense 5′-ATGTTTCCCGCGAGACTCTTC-3′ antisense 5′-GAGTGGTCTTCAATCACACCAA-3′ |
| PDI | sense 5′-GACGGAGTACCCTACGCTCAA-3′ antisense 5′-AGAAGCCAATGACCACTAGGT-3′ |
| ERdj4 | sense 5′-GGTGTGCCAAAATCGGCATC-3′ antisense 5′-GCACTGTGTCCAAGTGTATCATA-3′ |
MTT assay
Stable cell lines were plated in 96-well culture plates (10 000 cells/well). After overnight culture, the cells were treated with tunicamycin (0, 2, 5, and 10 μg/mL) for 12 h. MTT stock solution was added at a final concentration of 0.5 mg/mL. Cells were then cultured for another 4 h. The resulting formazan crystals were dissolved in dimethylsulfoxide (DMSO, 200 μL) for 10 min. Absorbance was then recorded at 490 nm on a microplate spectrophotometer (Bio-Rad, USA). The effect of tunicamycin on cell viability was assessed as percentage of cell viability compared to DMSO-treated control cells, which were arbitrarily assigned as 100% viable.
Statistical analysis
All data are shown as mean±standard deviation (SD). Data were analyzed using either a one-way ANOVA with an appropriate post hoc test comparison of multiple groups or an unpaired student's t test for comparison of two groups as described in the figure legends (Graphpad Prism software).
Results
Excessive ER stress desensitized insulin receptor signaling
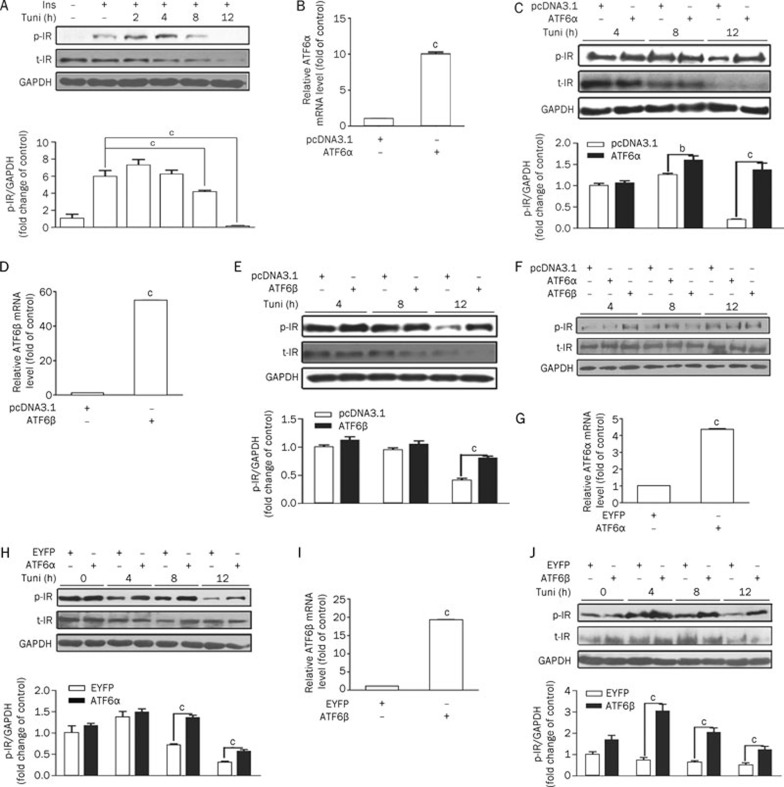
As reported, the ER stress inducer tunicamycin significantly inhibited insulin signal transduction by suppression of insulin-induced IRS-1 and AKT phosphorylation in Fao liver cells3. Here we examined the potential effects of tunicamycin on the insulin receptor in the HEK293 cell line. This cell line is often used as a model cell line in preliminary investigations related to ER stress, MAPK and insulin signaling pathways20, 21. Compared with the DMSO-treated group, short-term (2–4 h) stimulation with tunicamycin increased insulin-induced IR (Tyr1150/1151) phosphorylation. Prolonged tunicamycin treatment (8–12 h) attenuated such insulin-induced IR activities (Figure 1A). In addition, we also observed that the total amount of IR decreased after long-term tunicamycin treatment. These results indicate that short-term stimulation with tunicamycin could activate IR signaling, whereas excessive ER stress will desensitize this signaling in HEK293 cells.
Figure 1.
ATF6 protects the insulin pathway from ER-stress induced desensitization. (A) Excessive ER stress-desensitized insulin receptor signaling. When confluence reached 80%, HEK293 cells were refreshed with serum-free DMEM supplemented with tunicamycin (2 μg/mL) for the time indicated before 5-min insulin (167 nmol/L) stimulation. (B and D) The transient over-expression efficiency of ATF6α (B)/β (D). HEK293 cells were transiently transfected with pcDNA3.1 or pcDNA3.1-hATF6α/β. After 5 h, cells were refreshed with 10% DMEM/FBS and then harvested for real-time RT-PCR on the next day. (C and E) ATF6α (C)/β (E) transient over-expression protected the insulin pathway from ER stress-induced desensitization. HEK293 cells were transiently transfected with pcDNA3.1 or pcDNA3.1-hATF6α/β for 5 h and then refreshed with serum-free DMEM supplemented with tunicamycin (2 μg/mL) for the indicated time before 5-min insulin (167 nmol/L) stimulation. The cells were harvested to analyze the phosphorylated and total IR. (F) ATF6α/β's effect on the insulin pathway without tunicamycin treatment. HEK293 cells were transiently transfected with pcDNA3.1 or pcDNA3.1-hATF6α/β. After 5 h, cells were refreshed with serum-free DMEM for 4, 8, and 12 h before 5-min insulin (167 nmol/L) stimulation. (G and I) The stable over-expression efficiency of ATF6α (G)/β (I). ATF6 over-expression stable cell lines (HEK293-EYFP-C1-ATF6α and HEK293-EYFP-C1-ATF6β) and control cells (HEK293-EYFP-C1) were plated into 6-well culture plates (100 000 cells/well) and then harvested for real-time PCR on the third day. (H and J) ATF6α (H)/β (J) stable over-expression protects the insulin pathway against ER stress-induced desensitization. HEK293 (EYFP-C1), HEK293 (EYFP-C1-ATF6α) and HEK293 (EYFP-C1-ATF6β) were separately treated with tunicamycin (2 μg/mL) and then stimulated for 5 min with insulin (167 nmol/L). The phosphorylated and total IR were analyzed. bP<0.05; cP<0.01.
ATF6 protected the insulin pathway from ER stress-induced desensitization
ATF6 optimized long-term ER stress and protected cells from damage through regulating a number of genes responsible for protein folding and degradation22, 23, 24. Bearing that in mind, we thus investigated whether members of the ATF6 family could protect IR activity from excessive ER stress. For this assay, after transiently transfected with pcDNA3.1-ATF6α/ATF6β, HEK293 cells were treated with tunicamycin for 4–12 h. The over-expression efficiency is shown in panels B and D of Figure 1. As shown in panels C and E of Figure 1, IR phosphorylation was highly responsive to insulin stimulation from 8 h in ATF6α-transfected cells and from 12 h in ATF6β-transfected cells. To further confirm that ATF6α/ATF6β protection of IR phosphorylation in response to insulin induction only appeared under long-term ER stress stimulation, IR phosphorylation in ATF6α/ATF6β overexpressing cells without tunicamycin treatment were examined at 4, 8, and 12 h. As shown in Figure 1F, ATF6α/ATF6β overexpression failed to alter IR phosphorylation without tunicamycin treatment. Moreover, tunicamycin markedly decreased the total IR, while over expression of ATF6α (or ATF6β) failed to prohibit IR degradation (Figure 1C and 1E). To further verify this result, an ATF6α (or ATF6β) stable cell line with EYFP-C1 as a control was constructed and phosphorylation of IR was examined. The over-expression efficiency is shown in Figure 1G and 1I. As indicated in Figure 1H and 1J, obvious increase of IR phosphorylation induced by insulin was shown from 8-h tunicamycin stimulation in the ATF6α stable cell line and from 4-h tunicamycin supplementation in the ATF6β stable cell line. Compared with the transiently transfected cells, the protection of IR phosphorylation from degradation appeared at an earlier time in the ATF6 stable cell lines. This might be caused by a more efficient response to tunicamycin stimulation in stable cell lines with persistent ATF6 expression.
Therefore, all of our results suggest that the ATF6 family could protect IR activity from long-term ER stress stimulation, while such an effect may not be associated with the regulation of total IR protein level.
ATF6 protection against insulin resistance did not merely link with its function in ER stress
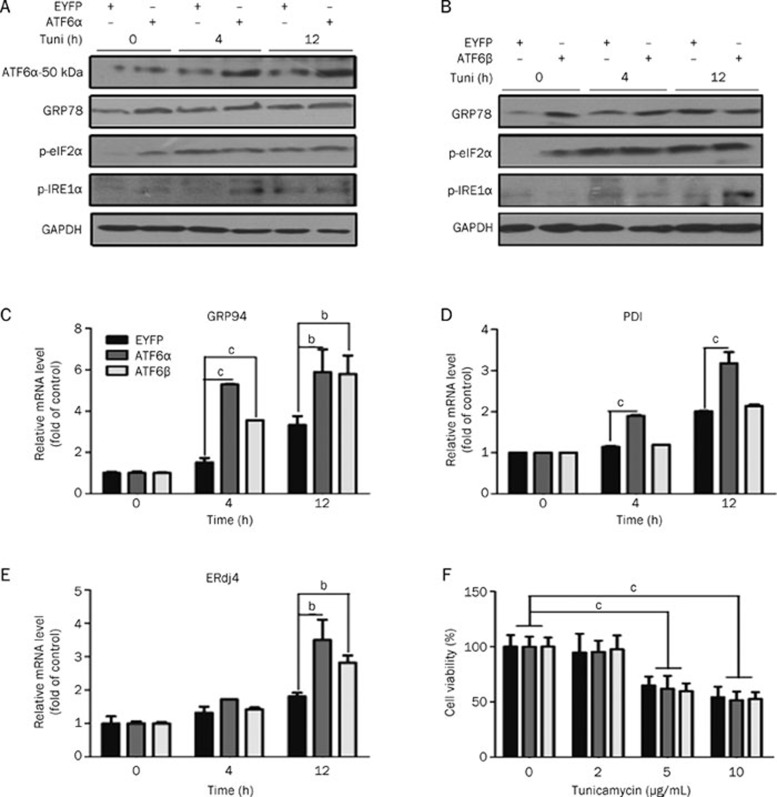
ATF6 is one of the three main transducers in UPR that up-regulate a series of genes to relieve the overload of unfolded protein in the ER, thus ameliorating long-term ER stress damage24, 25, 26, 27. Here, expression of three ATF6 target genes (GRP78, GRP94 and PDI) was examined in tunicamycin stimulated HEK293 stable cell lines. The results in Figure 2A–2D and Figure S1 indicate that the stable over-expressing ATF6α (or ATF6β) cells respond to tunicamycin stimulation potently as reflected by the increased expression levels of these genes as early as 4 h after stimulation. Additionally, we also examined whether the PERK-eIF2α and IRE1α-XBP-1 pathways could be modulated by ATF6 over-expression. As shown in Figure 2A and 2B, the PERK-phosphorylated eIF2α phosphorylation did not differ in the stable cells with ATF6α or ATF6β over-expression. Phosphor-IRE1α and mRNA levels of ERdj4 (one of the IRE1α-XBP-1 downstream genes) were up-regulated (Figure 2A, 2B, and 2E) at 12 h. These results are consistent with previously reported results28. Therefore, we propose that ATF6 function in the insulin pathway might be associated with resistance to ER stress. Considering that the cell death induced by ER stress could act as an assessment to evaluate sensitivity to ER stress29, we applied an MTT assay in stable ATF6α (or ATF6β) over-expressing cells. The results indicate that ATF6 over-expressing cells failed to counteract tunicamycin-induced cell death in HEK293 cells within 12 h (Figure 2F). Therefore, all of the above-mentioned results suggest that the regulation of the ATF6 family on IR activity might not solely depend on its modulation of UPR.
Figure 2.
ATF6 regulation on IR was not solely dependent on its protective role in ER stress. (A and B) HEK293 (EYFP-C1), HEK293 (EYFP-C1-ATF6α), and HEK293 (EYFP-C1-ATF6β) were cultured with tunicamycin for the indicated time before harvest. The ATF6α-50 kDa (cleavage form of ATF6α), GRP78, phosphorylated eIF2α and IRE1α were then analyzed. (C–E) HEK293 (EYFP-C1), HEK293 (EYFP-C1-ATF6α), and HEK293 (EYFP-C1-ATF6β) were cultured in 6-well plates and stimulated with tunicamycin (2 μg/mL) for the indicated time before harvest. Total RNA extraction and mRNA quantification of GRP94 (C), PDI (D), and ERdj4 (E) were performed as described in “Materials and methods”. (F) MTT assays were applied to HEK293 (EYFP-C1), HEK293 (EYFP-C1-ATF6α), and HEK293 (EYFP-C1-ATF6β) as described in “Materials and methods”. bP<0.05, cP<0.01.
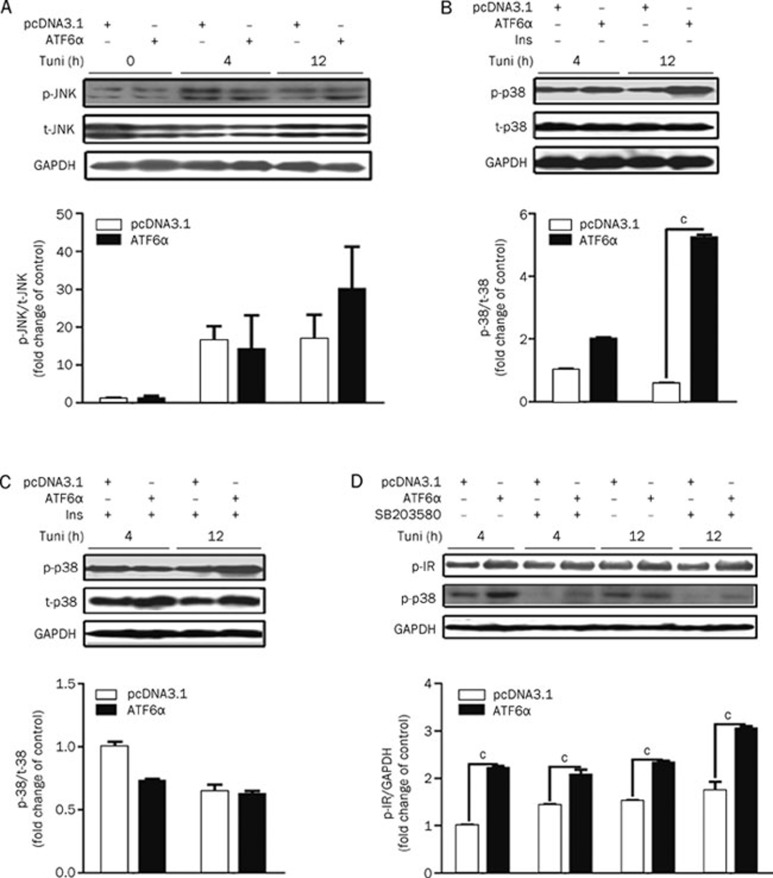
ATF6 failed to attenuate tunicamycin-induced JNK pathway activation
As reported, the JNK pathway plays a central role in connecting ER stress with type 2 diabetes. Upon activation by ER stress through IRE1α30, JNK can directly phosphorylate IRS-1 at Ser307 leading to the reduction of insulin-induced IRS-1 tyrosine phosphorylation and a decrease in activity of the insulin pathway31, 32. Given that ATF6 might protect the insulin pathway by attenuating JNK pathway activity, the related assays were performed. The results in Figure 3A indicate that stimulation with tunicamycin for 4–8 h strongly elevated JNK phosphorylation, while ATF6α could not reverse such elevation. These results suggest that ATF6α might not participate in the regulation of JNK signaling.
Figure 3.
ATF6 protection was not involved in the JNK and p38 signaling pathways. (A) HEK293 cells were transiently transfected with pcDNA3.1/pcDNA3.1-hATF6α. After tunicamycin (2 μg/mL) treat-ment for the indicated time, the total/phosphorylated JNK was analyzed. (B and C) HEK293 cells were transiently transfected with pcDNA3.1/pcDNA3.1-hATF6α. After tunicamycin (2 μg/mL) treatment with (C) or without (B) insulin (167 nmol/L) stimulation for 5 min, the phosphorylated and total p38 were analyzed. (D) After transfection, HEK293 cells were treated with tunicamycin (2 μg/mL) together with SB203580 (5 μmol/L) for the indicated time and insulin (167 nmol/L) for 5 min. Then, the phosphorylated IR was analyzed. cP<0.01.
ATF6 protection on IR phosphorylation from excessive tuni-camycin stimulation is not connected with the p38 pathway
Besides the JNK pathway, the MKK3/6-p38 and MEK1/2-ERK pathways are also associated with several key proteins within the insulin receptor signaling pathway (eg, IR, IRS, and AKT)33, 34, 35, 36, 37. We thereby hypothesized that the ERK or p38 pathways might be involved in ATF6's protection on insulin signaling from excessive ER stress stimulation. To evaluate this hypothesis, basal and insulin-stimulated p38 phosphorylation were tested in control or ATF6α transfected HEK293 cells with 4–12 h incubation with tunicamycin. An obvious increase in p38 phosphorylation was observed in ATF6α-transfected cells treated with tunicamycin. There were no apparent differences in p38 activity found between control and ATF6α transfected cells with insulin stimulation (Figures 3B and 3C). In addition, SB203580 (specific p38 inhibitor) did not alter the phosphorylation of IR with long-term tunicamycin treatment -(Figure 3D). These results indicate that the p38 signaling pathway might not contribute to the protective effects of ATF6. Similar results were also obtained by over-expressing ATF6β (data not shown).
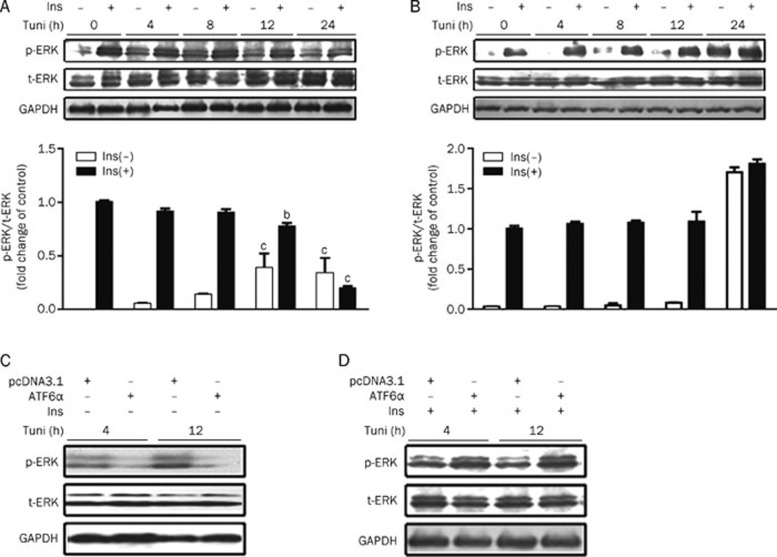
ATF6 regulation against insulin receptor signaling is associated with the ERK pathway
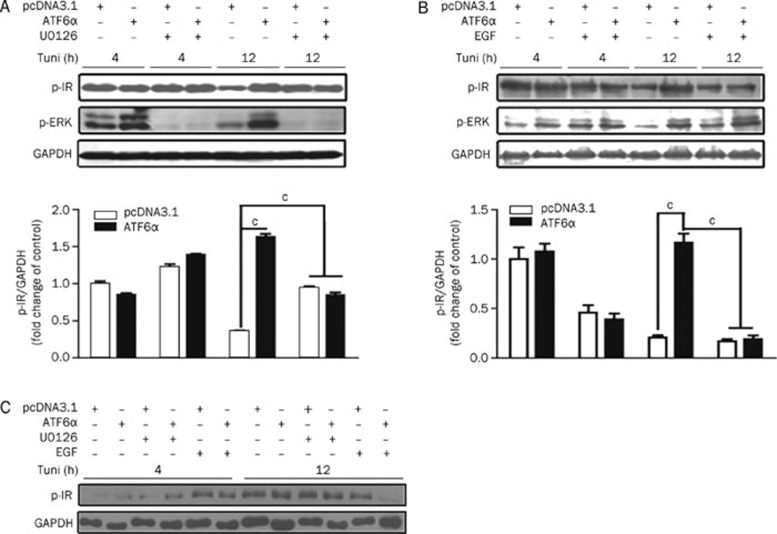
To further explore the potential signaling pathway for ATF6 regulation of the insulin receptor, basal and insulin-stimulated ERK phosphorylation were tested in control or ATF6α transfected HEK293 cells incubated with tunicamycin for 0–24 h. The results indicate that insulin-stimulated ERK phosphorylation was significantly attenuated in cells exposed to tunicamycin at 12 h (Figure 4A), while its sensitivity in ATF6α-overexpressing cells maintained stability except with 24 h tunicamycin incubation (Figure 4B). Furthermore, the basal level of ERK phosphorylation was largely inhibited in ATF6α-overexpressing cells as compared with control cells (Figure 4C), whereas ATF6α was able to enhance the level of insulin-induced ERK phosphorylation (Figure 4D) with tunicamycin treatment. Therefore, our findings demonstrate that ATF6 modulates ERK activity induced by tunicamycin stimulation. To further confirm the role of ERK in ATF6 effects on IR, the IR activity was then evaluated in control and ATF6α transient transfected cells with or without U0126 (specific ERK inhibitor) treatment. As shown in Figure 5A, IR phosphorylation was greatly dampened by long-term tunicamycin incubation in control but not in ATF6α transient transfected cells. As expected, U0126 treatment almost completely inhibited ERK activity and restored the insulin-stimulated IR phosphorylation in control cells. On the contrary, epidermal growth factor (EGF, ERK activator) treatment together with tunicamycin stimulation largely decreased the protective effects of ATF6α on IR (Figure 5B). However, the results shown in Figure 5C are without tunicamycin treatment and suggest that the above ERK-associated hypothesis was tenable under the instance of tunicamycin stimulation. Similar results were also obtained by over-expressing ATF6β (data not shown). Accordingly, our results imply that ATF6 protects IR activity through attenuation of basal ERK activity (at 4–12 h) that is subject to excessive ER stress stimulation.
Figure 4.
ATF6 could modulate ERK activity induced by tunicamycin simulation. (A and B) HEK293 cells were transiently transfected with pcDNA3.1 (A)/pcDNA3.1-hATF6α (B). After tunicamycin (2 μg/mL) treatment with or without insulin (167 nmol/L) stimulation for 5 min, phosphorylated and total ERK were analyzed. (C and D) After transfection, tunicamycin (2 μg/mL) treated HEK293 cells were stimulated with (D) or without (C) insulin (167 nmol/L). Then, phosphorylated and total ERK were analyzed. bP<0.05, cP<0.01 compared with control groups (0 h) with or without insulin stimulation, respectively.
Figure 5.
ATF6 regulation of IR signaling pathway activity is associated with ERK pathway. (A and B) After transfection, HEK293 cells were treated with tunicamycin (2 μg/mL) together with U0126 (10 μmol/L) (A) or EGF (200 ng/mL) (B) for the indicated time and insulin for 5 min (167 nmol/L). Then, the phosphorylated IR was analyzed. (C) HEK293 cells were transiently transfected with pcDNA3.1/pcDNA3.1-hATF6α. After 5 h, cells were refreshed and incubated with U0126 (10 μmol/L) or EGF (200 ng/mL) in serum-free DMEM for 4, 8, and 12 h without tunicamycin stimulation before 5-min insulin (167 nmol/L) stimulation. cP<0.01.
Discussion
In recent years, accumulating evidence has demonstrated that the ER may serve as an “interpreter” by sensing a number of exogenous or endogenous stimuli such as inflammatory cytokines38, 39, abnormal energy fluxes15 and oxidative stimuli40. The ER is a complicated and systemic network, and excessive ER stress may result in insulin resistance3, 41, 42 and islet β-cell dysfunction43. Currently, increasing chaperones has been reported to help improve insulin resistance6. Here we focused on ATF6 for its potent service as a critical signal transducer in the ER stress system. ATF6 is responsible for up-regulating main chaperone systems27 and plays a pivotal role in XBP-1 generation44, which is essential to normal insulin signaling pathway functioning and glucose homeostasis3. In addition, the genetic report has even identified ATF6 as a susceptibility gene of type 2 diabetes in Pima Indians45.
As previously published43, 46, short-term (from 0 to 4 h) ER stress stimulation could increase AKT activity, while long-term (from 8 to 12 h) stimulation could suppress AKT activity. This reflects the dual character of ER stress: the adaptive/moderate level stress (featured by increased chaperone proteins and decreased misfolded proteins) and the destructive/excessive level stress (featured by activating JNK, apoptosis signal-regulating kinase (ASK))2, 3, 30, 47. We found that tyrosine-phosphorylated IR was first elevated and then decreased when stimulated with tunicamycin, while total IR decreased after tunicamycin incubation (Figure 1A) based on the results of p-IR/GAPDH ratio quantification instead of the p-IR/t-IR ratio because t-IR was too weak to quantify in 12 h. IR is an important upstream regulator of AKT and its change may partly account for the variation of AKT activity. We discovered that the ATF6 family could protect IR activity from tunicamycin stimulation. To further investigate whether ATF6 protection on IR could affect its downstream molecules, phosphorylation of AKT was examined. As shown in Figure S2, ATF6 also protects AKT from phosphorylation degradation induced by tunicamycin.
The contribution of ER stress transducer and related genes were evaluated in HEK293 cell lines to investigate the mechanism of ATF6 protection on the insulin pathway. Although the phosphorylation of IRE1α and cleavage of ATF6 could be modulated in ATF6 over-expressing cells, we failed to find any differences between ATF6 over-expression and control cells in the MTT assay. These results suggest that ATF6 over-expression does not improve cell sensitivity to ER stress and regulation of the ATF6 family on IR activity might not be dependent on its protective roles in ER stress. Recently, more new target genes of the ATF6 family have been discovered, including ER-stress related genes (eg, GRP78, GRP94, PDI, XBP-121) and others [eg, Sterol regulatory element binding protein 2 (SREBP2)48]. It is reported that 250 genes could be up-regulated by tunicamycin treatment and at least 45 were ATF6α-dependent, although most of these genes have not yet been fully investigated23.
The MAPK signaling pathways MKK4/7-SAPK/JNK, MKK3/6-p38, and MEK1/2-ERK have been reported to play key roles in insulin resistance induced by excessive ER stress33, 34, 35, 36, 37, 38. Therefore, the phosphorylation of JNK, p38 and ERK were examined in ATF6 over-expressing cells. Our results indicate that the MEK1/2-ERK pathway is associated with the ATF6 protection mechanism. Before our investigation, several studies reported that MEK exerted negative effects on the insulin signaling pathway via down-regulation of the expression of IR/IRS, decreasing tyrosine phosphorylation of IR/IRS, increasing serine phosphorylation of IRS and inhibiting the subsequent activations of PI3K and AKT33, 34, 35. Several diabetes-inducing agents have also been demonstrated as causing insulin resistance in a MEK-dependent manner35, 20. ER stress was also reported to induce activation of ERK46, and ATF6 was shown to counteract the effects of ER stress23, 24. Based on this prior evidence and our results (Figures 4 and 5), it could be concluded that excessive ER stress could increase the basal, but decrease the insulin-induced, activity of ERK. We thus propose the potential effects of ATF6 to be as follows. Long-time ER stress stimulation could induce a constant “basal” activation of the ERK pathway, which might have a negative feedback causing the desensitization of IR activity. ATF6 could counteract this negative process through down-regulation of the basal ERK activity and up-regulation of insulin-induced ERK activity until 12 h, leading to the restoration of insulin signaling pathway activity. Although our results also showed that ATF6 could enhance p38 activity, it seems that p38 is not involved in ATF6 modulation of IR phosphorylation (Figure 3). This is consistent with the finding that p38 stimulates insulin-induced glucose uptake through GLUT4 activation instead of IR/IRS/AKT-dependent transporter translocation36, 37.
As reported, ATF6 family members exhibit different influences on GRP78 transcription with a hypothetical mechanism that ATF6α, as a strong but labile transcription factor, and ATF6β, as a weak but stable transcription factor, work collaboratively to address different ER stress signals11, 12. However, we found that ATF6α and ATF6β demonstrated almost the same effects on IR activity. Thus, we hypothesized that no matter what characteristics they possess, these two isotypes might have similar influences on ER quality and cellular physiology in the process of adaptation to long-time stress since chronic stimulation would fully induce their activities.
Contrary to the opinion that the ATF6 family contributes to alleviating cellular stress9, 16, 24, the studies by Seo et al suggest that ATF6 plays an important role in the development of β-cell dysfunction49. Although ATF6 was shown to suppress insulin gene expression in INS-1 cells, this process might be a feedback to prevent hyperinsulinmia in vivo. In addition, this report also indicated that ATF6 expression was increased in the pancreatic islets of diabetic rats compared with non-diabetic counterparts. However, this study did not confirm that the increase in ATF6 expression might cause or aggregate diabetes. Based on our current work, we suggest that ATF6 family members could be up-regulated in response to cellular stress to counteract the diabetic stress.
Recently, more studies have suggested that the signaling pathway involved in ER stress could be used as a potential target for anti-type 2 diabetes drug design4, 5, 6, 38, 39. Here we discovered that ATF6 family members protect the insulin receptor pathway from long-term tunicamycin treatment (4–12 h) that mimics the effects of chronic ER stress, implying that ATF6 could be a potential target for increasing insulin sensitivity. However, further investigations need to be carried out to clarify the underlying mechanism by which ATF6 regulates ERK activity and other diabetes-related pathways.
Author contribution
Xu SHEN, Hua-liang JIANG, Jing CHEN, and Vatcharin RUKACHAISIRIKUL designed the research; Hong SHEN, Yu ZHANG, Li-li CHEN, and Xu WANG performed experiments in investigating the effect of tunicamycin on IR; Xuan TANG and Hong SHEN studied the effect of ATF6 on IR under long-term ER stress by transient transfection and stable transfection with ATF6, and they also performed related assays to reveal the potential molecular mechanisms; Xu SHEN, Hua-liang JIANG, and Jing CHEN supervised the project; and Xu SHEN, Jing CHEN, Hong SHEN, and Xuan TANG contributed to the manuscript writing.
Acknowledgments
We gratefully acknowledge Prof Kazutoshi MORI for the plasmids of pcDNA3.1-hATF6α and pcDNA3.1-hATF6β. This work was supported by the State Key Program of Basic Research of China (No 2010CB912501 and 2007CB914304), the National Natural Science Foundation of China (No 90713046 and 30890044), Shanghai Basic Research Project from the Shanghai Science and Technology Commission (No 11ZR1444500) and Open Research Fund of the Key Laboratory of Biotherapy, West China Hospital, West China Medicine School, Sichuan University (No SKLB200901).
Footnotes
Supplementary figures are available at Acta Pharmacologica Sinica website of NPG.
Supplementary Information
GRP78 protein level responded to tunicamycin stimulation more potentially in ATF6 over-expression stable cells than in control cells.
ATF6 protected AKT phosphorylation from desensitization induced by tunicamycin incubation.
References
- Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–33. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–61. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–40. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa K, Miyazaki M, Matsuhisa M, Takano K, Nakatani Y, Hatazaki M, et al. The endoplasmic reticulum chaperone improves insulin resistance in type 2 diabetes. Diabetes. 2005;54:657–63. doi: 10.2337/diabetes.54.3.657. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Kaneto H, Kawamori D, Yoshiuchi K, Hatazaki M, Matsuoka TA, et al. Involvement of endoplasmic reticulum stress in insulin resistance and diabetes. J Biol Chem. 2005;280:847–51. doi: 10.1074/jbc.M411860200. [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang X, Zuberi A, Hwang D, Quon MJ, Lefevre M, et al. Inhibition of insulin sensitivity by free fatty acids requires activation of multiple serine kinases in 3T3-L1 adipocytes. Mol Endocrinol. 2004;18:2024–34. doi: 10.1210/me.2003-0383. [DOI] [PubMed] [Google Scholar]
- Herschkovitz A, Liu YF, Ilan E, Ronen D, Boura-Halfon S, Zick Y. Common inhibitory serine sites phosphorylated by IRS-1 kinases, triggered by insulin and inducers of insulin resistance. J Biol Chem. 2007;282:18018–27. doi: 10.1074/jbc.M610949200. [DOI] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Davé UP, Prywes R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–64. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. ER stressregulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Thuerauf DJ, Morrison LE, Hoover H, Glembotski CC. Coordination of ATF6–mediated transcription and ATF6 degradation by a domain that is shared with the viral transcription factor, VP16. J Biol Chem. 2002;277:20734–9. doi: 10.1074/jbc.M201749200. [DOI] [PubMed] [Google Scholar]
- Thuerauf DJ, Morrison L, Glembotski C. Opposing roles for ATF6alpha and ATF6beta in endoplasmic reticulum stress response gene induction. J Biol Chem. 2004;279:21078–84. doi: 10.1074/jbc.M400713200. [DOI] [PubMed] [Google Scholar]
- Thuerauf DJ, Marcinko M, Belmont PJ, Glembotski CC. Effects of the isoform-specific characteristics of ATF6 alpha and ATF6 beta on endoplasmic reticulum stress response gene expression and cell viability. J Biol Chem. 2007;282:22865–78. doi: 10.1074/jbc.M701213200. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kauppinen A, Suuronen T, Kaamiranta K, Ojala J. ER stress in Alzheimer's disease: a novel neuronal trigger for inflammation and Alzheimer's pathology. J Neuroinflammation. 2009;6:41. doi: 10.1186/1742-2094-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Sanyal AJ. Endoplasmic reticulum stress and the unfolded protein response. Clin Liver Dis. 2009;13:581–90. doi: 10.1016/j.cld.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Dobson CM. Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol. 2004;15:3–16. doi: 10.1016/j.semcdb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Kammoun HL, Hainault I, Ferré P, Foufelle F. Nutritional related liver disease: targeting the endoplasmic reticulum stress. Curr Opin Clin Nutr Metab Care. 2009;12:575–82. doi: 10.1097/MCO.0b013e32833189db. [DOI] [PubMed] [Google Scholar]
- Fonseca SG, Burcin M, Gromada J, Urano F. Endoplasmin reticulum stress in beta-cells and development of diabetes. Curr Opin Pharmacol. 2009;9:763–70. doi: 10.1016/j.coph.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Kaufman RJ. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev. 2008;29:317–33. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa Y, Yoshizumi M, Fujita Y, Ali N, Kanematsu Y, Ishizawa K, et al. ERK1/2 activation by angiotensin II inhibits insulin-induced glucose uptake in vascular smooth muscle cells. Exp Cell Res. 2005;308:291–9. doi: 10.1016/j.yexcr.2005.04.028. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–67. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JA, Dorner AJ, Edwards CA, Hendershot LM, Kaufman RJ. Immunoglobulin binding protein (BiP) function is required to protect cells from endoplasmic reticulum stress but is not required for the secretion of selective proteins. J Biol Chem. 1997;272:4327–34. doi: 10.1074/jbc.272.7.4327. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, et al. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–64. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–76. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–9. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–99. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641–51. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C, Kaplowitz N. ER stress: Can the liver cope. J Hepatol. 2006;45:321–33. doi: 10.1016/j.jhep.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Nakayaa H, Zhao J, Ei-Fakhrany A, Isosaki M, Satoh H, Kyotani Y, et al. Neuroprotective effects of praipexole against tunicaycin-induced cell death in PC12 cells. Clin Exp Pharacol Physiol. 2009;36:1183–5. doi: 10.1111/j.1440-1681.2009.05224.x. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–6. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Hirosumi J, Tuncman G, Chang L, Görgün CZ, Uysal KT, Maeda K, et al. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–6. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- Aguirre V, Uchida T, Yenush L, Davis R, White MF. The c-Jun NH2-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser(307) J Biol Chem. 2000;275:9047–54. doi: 10.1074/jbc.275.12.9047. [DOI] [PubMed] [Google Scholar]
- Fujishiro M, Gotoh Y, Katagiri H, Sakoda H, Ogihara T, Anai M, et al. Three mitogen-activated protein kinases inhibit insulin signaling by different mechanisms in 3T3-L1 adipocytes. Mol Endocrinol. 2003;17:487–97. doi: 10.1210/me.2002-0131. [DOI] [PubMed] [Google Scholar]
- De Fea K, Roth RA. Modulation of insulin receptor substrate-1 tyrosine phosphorylation and function by mitogen-activated protein kinase. J Biol Chem. 1997;272:31400–6. doi: 10.1074/jbc.272.50.31400. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Berg AH, Lewis RY, Lisanti MP, Scherer PE. Tumor necrosis factor alpha-mediated insulin resistance, but not dedifferentiation, is abrogated by MEK1/2 inhibitors in 3T3-L1 adipocytes. Mol Endocrinol. 2000;14:1557–69. doi: 10.1210/mend.14.10.0542. [DOI] [PubMed] [Google Scholar]
- Sweeney G, Somwar R, Ramlal T, Volchuk A, Ueyama A, Klip A. An inhibitor of p38 mitogen-activated protein kinase prevents insulin-stimulated glucose transport but not glucose transporter translocation in 3T3-L1 adipocytes and L6 myotubes. J Biol Chem. 1999;274:10071–8. doi: 10.1074/jbc.274.15.10071. [DOI] [PubMed] [Google Scholar]
- Somwar R, Kim DY, Sweeney G, Huang C, Niu W, Lador C, et al. GLUT4 translocation precedes the stimulation of glucose uptake by insulin in muscle cells: potential activation of GLUT4 via p38 mitogen-activated protein kinase. Biochem J. 2001;359:639–49. doi: 10.1042/0264-6021:3590639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS. Inflammation and endoplasmic reticulum stress in obesity and diabetes. Int J Obes (Lond) 2008;32:S52–4. doi: 10.1038/ijo.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–17. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RF, Terada LS. Focal oxidant and Ras signaling on the ER surface activates autophagy. Autophagy. 2010;6:828–9. doi: 10.1128/MCB.01445-09. [DOI] [PubMed] [Google Scholar]
- Hotamisligil G. Role of endoplasmic reticulum stress and c-Jun NH2-terminal kinase pathways in inflammation and origin of obesity and diabetes. Diabetes. 2005;54:S73–8. doi: 10.2337/diabetes.54.suppl_2.s73. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Ohsugi M, Liu Z, Fatrai S, Bernal-Mizrachi E, Permutt MA. Endoplasmic reticulum stress-induced apoptosis is partly mediated by reduced insulin signaling through phosphatidylinositol 3-kinase/Akt and increased glycogen synthase kinase-3beta in mouse insulinoma cells. Diabetes. 2005;54:968–75. doi: 10.2337/diabetes.54.4.968. [DOI] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–66. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thameem F, Farook VS, Bogardus C, Prochazka M. Association of amino acid variants in the activating transcription factor 6 gene (ATF6) on 1q21-q23 with type 2 diabetes in Pima Indians. Diabetes. 2006;55:839–42. doi: 10.2337/diabetes.55.03.06.db05-1002. [DOI] [PubMed] [Google Scholar]
- Hu P, Han Z, Couvillon AD, Exton JH. Critical role of endogenous Akt/IAPs and MEK1/ERK pathways in counteracting endoplasmic reticulum stress-induced cell death. J Biol Chem. 2004;279:49420–9. doi: 10.1074/jbc.M407700200. [DOI] [PubMed] [Google Scholar]
- Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, et al. ASK1 is essential for endoplasmic reticulum stress–induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–55. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Lu M, Mori K, Luo S, Lee AS, Zhu Y, et al. ATF6 modulates SREBP2-mediated lipogenesis. EMBO J. 2004;23:950–8. doi: 10.1038/sj.emboj.7600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HY, Kim YD, Lee KM, Min AK, Kim MK, Kim HS, et al. Endoplasmic reticulum stress-induced activation of activating transcription factor 6 decreases insulin gene expression via up-regulation of orphan nuclear receptor small heterodimer partner. Endocrinology. 2008;149:3832–41. doi: 10.1210/en.2008-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GRP78 protein level responded to tunicamycin stimulation more potentially in ATF6 over-expression stable cells than in control cells.
ATF6 protected AKT phosphorylation from desensitization induced by tunicamycin incubation.