Abstract
The adenovirus (Ad) protein IX (pIX) is a minor component of the Ad capsid and is in part responsible for virion stability; virions lacking pIX are heat labile and lose their infectivity if the DNA content is greater than ∼35 kb. More recently, pIX has been identified as a transcriptional activator and, in transient-transfection assays, was shown to enhance expression from the E1A, E4, and major late Ad promoters by as much as 70-fold. In this study, we examined the role of pIX's ability to activate transcription during Ad replication. In transient-transfection assays, pIX had a minimal effect on expression from the E1A promoter, increasing expression by only 1.4-fold. We used helper-dependent Ad vectors, which had all Ad protein coding sequences deleted with the exception of E1A and which had capsids that either contained or lacked pIX, to show that pIX derived from decapsidation of the infecting virion does not influence expression of E1A. Similarly, expression of pIX from the Ad genome did not alter the expression levels of E1A. Viruses that had pIX deleted showed a threefold reduction in virus yield and expression of late genes compared to those of a similar virus which encoded pIX. This phenotype could not be rescued by growing the virus in cells which constitutively express pIX. Our results indicate that, although pIX can affect transcription from a variety of viral promoters, it does not appear to play a significant role in activation of Ad promoters during normal Ad replication.
The adenovirus (Ad) protein IX (pIX) is a minor component of the Ad virion that associates with the hexons that make up the facets of the icosahedron (1). There are 240 molecules of pIX per Ad virion, and within the virion, pIX is found as a trimer. pIX is expressed at an intermediate time during infection (after initiation of early gene expression but before expression of late genes); however, the levels of pIX increase dramatically after DNA replication has initiated (1). Although originally thought to be dispensable for virion formation (4), pIX was shown to be essential for the packaging of full-length viral DNA molecules (6). Deletion of pIX results in virions that are heat labile, with capsids that reportedly can no longer package full-length Ad DNA (36 kb) but, instead, can accommodate only 35 kb or less (∼97% of the wild-type genome). However, we have recently shown that large DNA molecules (up to at least 37.2 kb) can be accommodated in capsids deficient in pIX but that these virions are noninfectious (i.e., do not form plaques) for reasons which are still unclear (16).
In addition to its role as a structural protein, pIX has been implicated as a transcriptional activator and appears to be involved in virus-induced nuclear reorganization (10, 14). Whereas the N-terminal region of pIX is required for incorporation of the protein into virions, the C terminus, specifically a leucine repeat, is important for its transcriptional activity (14). In transient-transfection assays, pIX was shown to enhance expression from the Ad early region 1A (E1A), E4, and major late promoter, as well as the β-globin and herpes simplex virus type 1 thymidine kinase promoters (10). pIX does not have direct DNA binding activity but can homomultimerize (and potentially interact with other DNA binding or transcriptional proteins) by interaction through leucine coiled-coil structures (14). When expressed within a cell, pIX forms distinct clear amorphous structures within the nucleus. Recently, pIX has been shown to colocalize with promyelocytic leukemia protein in the nucleus (15).
The ability of pIX to activate the E1A promoter is intriguing. It has been suggested that pIX derived from virion decapsidation translocates and accumulates in the nucleus of the infected cell, beginning as early as 45 min postinfection (14). Thus, pIX may function to enhance expression from the E1A promoter in the newly infected cell, in a manner similar to that of VP16 during herpesvirus infection (reviewed in reference 5). The result would be that pIX could be a key factor responsible for kick-starting the virus life cycle.
In this study, we examine the role of pIX in the Ad life cycle. We show that, although pIX can affect expression from the E1A promoter in transient-transfection assays, pIX does not influence expression from this promoter during normal virus replication. Furthermore, deletion of pIX from an otherwise wild-type Ad results in only a small reduction in growth of the virus, suggesting that the ability of pIX to transcriptionally activate viral promoters is unimportant for viral growth.
MATERIALS AND METHODS
Cell and virus culture.
All cell culture media and reagents were obtained from Gibco Laboratories (Grand Island, N.Y.). 293 (7) and A549 (human lung carcinoma; ATCC CCL 185) cells were grown in monolayers in minimum essential medium supplemented with 100 U of penicillin per ml, 100 mg of streptomycin per ml, 2.5 mg of amphotericin B per ml, and 10% fetal bovine serum (complete medium). The 293-derived cell line that stably expresses the Cre recombinase, 293Cre4 (3), was propagated in complete medium supplemented with 0.4 mg of G418/ml. The 293-derived cell line that stably expresses the Ad type 5 (Ad5) pIX, 293pIXc4 (16), was grown in complete medium supplemented with 0.4 mg of hygromycin/ml. AdRP2050 helper virus has been described previously (16) and was grown and had its titers determined on 293 cells. The pIX-deletion helper virus, AdRP2105a, has been previously described (16). hdAdE1A has all Ad protein coding sequences deleted with the exception of E1A (up to 1,773 bp of the conventional Ad5 map [Fig. 1]) but contains Ad5 right and left inverted terminal repeats and packaging signal (8). hdAdE1A also contains the Escherichia coli lacZ gene under the regulation of the murine cytomegalovirus (MCMV) immediate-early promoter and simian virus 40 polyadenylation (pA) sequence. To maintain the size of this vector above the limit for efficient DNA packaging (∼28 kb) (13), hdAdE1A contains a ∼22-kb fragment of eukaryotic DNA derived from the human hypoxanthine-guanine phosphoribosyltransferase gene (11). Generation of helper-dependent Ad (hdAd) vector, whether amplified with AdRP2050 or with AdRP2105a, was performed as previously described (12). For hdAd vectors, the multiplicity of infection (MOI) is calculated as the number of lacZ-transducing particles (as determined on 293 cells) per cell. The level of helper virus in the preparations of hdAdE1ApIX+ and hdAdE1ApIX− was ∼0.01%, as determined by plaque assay on 293 and 293pIXc4 cells, respectively.
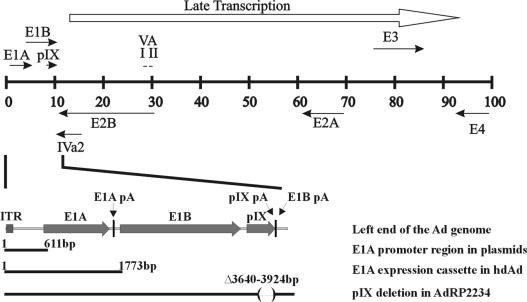
FIG. 1.
Simplified transcription map of the Ad5 genome. Also shown is an enlargement of the left end of the Ad genome and the regions utilized or deleted in the various constructs used in this study. ITR, inverted terminal repeat.
To examine the effect of pIX during Ad replication, we generated two viruses, AdRP2233 (E1+/E3−) and AdRP2234 (E1+/pIX−/E3−). AdRP2234 has a deletion of nucleotides 3640 to 3924 of the Ad5 genome, which removes most of the pIX coding sequence (this virus is predicted to express a truncated pIX protein of 15 amino acids). These viruses were constructed using a combination of traditional cloning and RecA+ bacterium-mediated recombination (2). The viruses were propagated and their titers were determined on 293 cells and, because of their size (∼35 kb), do not require pIX for virion formation. For analysis of virus growth, cells were infected at an MOI of 1 (calculated as the number of PFU per cell) with virus, and at various times postinfection, the cells and medium were harvested, sucrose was added to a final concentration of 4%, the cells were frozen until the end of the assay, and the titers for all of the samples from a single experiment were determined at the same time.
Transient-transfection studies.
All transfections were performed in 35-mm-diameter dishes, in replicates of five, with Superfect reagent (Qiagen), according to the manufacturer's instructions. Our pIX expression plasmid contains the Ad5 pIX coding sequence under the regulation of the human cytomegalovirus promoter and simian virus pA and is designated pCEP-pIX. For transfection of A549 cells, various quantities of pCEP-pIX were mixed with parental pCEP4 plasmid (Invitrogen) to give a total of 250 ng. This DNA was mixed with 1.5 μg of reporter plasmid which contained the luciferase gene under the regulation of the E1A promoter (Fig. 1). We also included 250 ng of an MCMV-lacZ-expressing plasmid which was used to correct the data for varying transfection efficiency. Twenty-four hours later, crude protein extracts were prepared from the cells by using reporter lysis buffer (Promega). The quantity of β-galactosidase and the amount of luciferase activity were determined using chemiluminescence assays (Tropix and Promega, respectively). Each transfection was performed twice with Superfect reagent (one representative data set is shown), and similar results were obtained using calcium phosphate precipitation.
Western blot analysis.
Protein extracts were prepared by infecting A549 cells at an MOI of 10 with hdAd-lacZ or hdAd-E1A or an MOI of 1 for AdRP2233 and AdRP2234. At various times postinfection, the cells were lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) protein loading buffer (62.5 mM Tris [pH 6.8], 25% glycerol, 2% SDS, 0.1% bromophenol blue, 5% 2-mercaptoethanol). After separation by 10% PAGE, the proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Immuno-Blot PVDF; Bio-Rad Laboratories, Hercules, Calif.), and the membrane was probed with an antibody specific for Ad5 E1A (MS-587-P1, 1/1,000; NeoMarkers, Fremont, Calif.), fiber (MS-1027-P0, 1/1,000; NeoMarkers), or tubulin (CP06, 1/5,000; Oncogene Research Products). Binding of the primary antibody was detected using a goat anti-mouse secondary antibody conjugated to horseradish peroxidase (Bio-Rad Laboratories) and visualized by chemiluminescence reaction (Amersham ECL Plus) and autoradiography.
Expression of pIX in 293pIXc4 and 293 cells.
To determine the level of pIX expression in 293pIXc4 cells, we used Western blot analysis to compare the intensity of the signal for pIX to that obtained for 293 cells that had been infected at various MOIs with an E1-deletion Ad which encoded pIX under endogenous regulation. Twenty-four hours after infection of the 293 cells, the cells were lysed with SDS-PAGE protein loading buffer and a protein sample was also prepared from uninfected 293pIXc4 cells. After separation by 10% PAGE, the proteins were transferred to a PVDF membrane, and the membrane was probed with an antibody which reacts with a variety of Ad capsid proteins, including pIX (ab6982, 1/10,000; Abcam) or tubulin. Detection of the primary antibody was as described above. To determine if 293 cells express pIX mRNA, we performed reverse transcription-PCR (RT-PCR) analysis. RNA was isolated from 293 or A549 cells with Trizol reagent (Gibco), as recommended by the manufacturer. RNA (100 ng) was converted to cDNA with Moloney murine leukemia virus reverse transcriptase (Roche) and random hexamers (Applied Biosciences), in a 20-μl reaction mixture under standard conditions. The cDNA (5 μl) was then subjected to PCR amplification (25 cycles) with a commercial kit (Invitrogen) and primers specific to pIX, which generated a 420-bp product. Negative controls consisted of PCRs which contained no template or RNA which had not been subjected to RT. In addition, control reactions were performed using synthetic oligonucleotides specific to β-actin, to verify the integrity of the cDNA. The resulting samples were subjected to electrophoresis on an 0.8% agarose gel and visualized by ethidium bromide staining.
RESULTS
Activity of the E1A promoter in transient-transfection assays with pIX.
In transient-transfection assays, pIX was shown to influence expression from several TATA-box-containing promoters and, most importantly, the E1A promoter (10, 14). The level of enhancement was 70-fold in COS-7 cells (10) but only 12-fold in A549 cells (14). We have confirmed that pIX can increase expression from the E1A promoter in A549 cells, but to a much lower degree than was previously observed (Fig. 2). Transfection of as little as 5 ng of the pIX expression plasmid resulted in an increase in expression from an E1A reporter plasmid (71.84 ± 3.64 versus 101.07 ± 4.03 RLU for cells transfected with 0 and 5 ng, respectively; P < 0.05) in A549 cells, although the increase was only 1.4-fold. A similar increase in luciferase expression was observed when 25 ng of pIX expression plasmid was included in the transfection; however, expression returned to control levels with 100 ng of plasmid. In previous studies, pIX was shown to affect various TATA-containing promoters, but only when expressed at very low levels (10); this is postulated to reflect the need for pIX transcriptional activity early during infection (i.e., before DNA replication), when the levels of pIX within the cells are very low. Therefore, in A549 cells, coexpression of pIX does affect expression from the E1A promoter, but only to a very small degree and only at low levels of pIX expression.
FIG. 2.
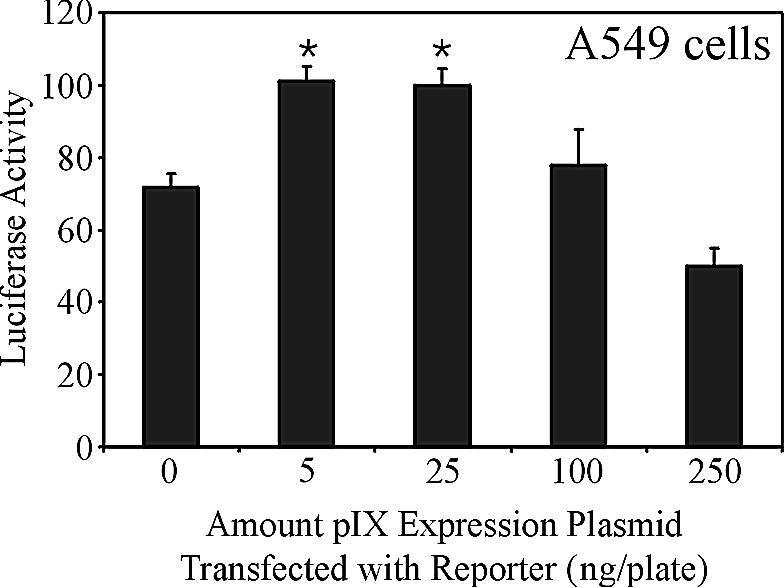
Transcriptional enhancement mediated by pIX. A549 cells were cotransfected with a pIX expression plasmid and a reporter plasmid containing the luciferase gene under the control of the E1A promoter. Twenty-four hours later, crude protein extracts were prepared and assayed for luciferase activity. The data have been normalized for transfection efficiency relative to a control MCMV-lacZ expression cassette included in the transfection. Shown is a data set representative of at least two independent transfections.
Effect of virion-derived pIX on expression of E1A.
pIX derived from decapsidation is reported to translocate to, and accumulate in, the nucleus soon after infection (10). It has been suggested that this population of pIX may function to enhance expression from Ad promoters located on the infecting DNA. At most, this would amount to 240 molecules of pIX from each infecting virion, but as we have shown, pIX appears to affect expression from the E1A promoter only when present at low levels within the cell. Recently, we developed a method for producing hdAd vectors with virions which lacked pIX (16), and we have described previously an hdAd encoding E1A under its endogenous promoter (Fig. 3A) (8). Therefore, we can generate an hdAd vector that contains E1A under normal regulation in virions which contain (12) or lack pIX. Using these reagents, we addressed the question of whether pIX from the incoming virion can affect expression of E1A. We generated hdAd vectors that encoded E1A in its normal position and under the regulation of its natural promoter, in addition to an MCMV-lacZ expression cassette, with capsids either containing or lacking pIX (designated hdAdE1ApIX+ and hdAdE1ApIX−, respectively). The lacZ expression cassette allows us to confirm equal levels of infection by the two viruses by direct cell counts in parallel plates after X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining. It should be noted that these vectors do not encode any other Ad protein except E1A.
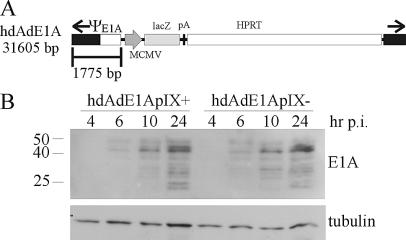
FIG. 3.
Expression of E1A from hdAd. (A) Structure of hdAdE1A, which encodes a lacZ expression cassette and expresses the E1A gene under the control of its endogenous promoter and encodes no other Ad proteins. (B) A549 cells were infected with E1A-expressing hdAd with virions that contained pIX (hdAdE1ApIX+) or lacked pIX (hdAdE1ApIX−). These hdAd vectors do not encode pIX; the pIX present in the viral capsid is provided by the helper virus during vector propagation with a helper virus that encodes or does not encode pIX. At various times postinfection (p.i.), protein samples were collected, separated by SDS-PAGE, and analyzed by immunoblotting with an E1A- or tubulin-specific antibody. Due to multiple splicing events and posttranscriptional modification, proteins derived from the E1A region range in size from ∼25 to 50 kDa. Numbers at left show molecular masses in kilodaltons. HPRT, hypoxanthine phosphoribosyltransferase.
A549 cells (in 35-mm-diameter dishes, five dishes for each treatment) were infected at an MOI of 1 with either hdAdE1ApIX− or hdAdE1ApIX+, and protein extracts were prepared from one dish of each set at 4, 6, 10, and 24 h postinfection. In addition, one dish from each treatment was stained with X-Gal at 24 h postinfection, and the number of blue cells was determined for 10 fields of view. The number of transduced cells was the same for each treatment: (1.92 ± 0.25) × 105 versus (2.01 ± 0.69) × 105 cells per dish for hdAdE1ApIX− and hdAdE1ApIX+, respectively. The protein samples were separated by SDS-PAGE, transferred to a PVDF membrane, and probed with anti-E1A antibody. As shown in Fig. 3B, we did not observe a difference in the kinetics or level of E1A expression between the two viruses. These data suggest that virion-derived pIX does not influence expression from the E1A promoter.
Effect of pIX on late gene expression and virus growth.
We next sought to determine if pIX influenced expression of Ad late genes and whether deletion of pIX would adversely affect growth of the virus. We constructed two viruses: the first virus is essentially wild type (AdRP2233, E1+/pIX+), with the exception that it has E3 deleted, while the second virus, AdRP2234, is identical to AdRP2233 but has the pIX coding sequence deleted. Neither AdRP2233 nor AdRP2234 requires pIX for virion formation, since they are below the DNA size that requires pIX (6) and form plaques equally well on 293 and 293pIXc4 cells (16). AdRP2234 was grown in either 293 or 293pIXc4 cells, generating capsids that lacked or contained pIX (AdRP2234 or AdRP2234pIX+, respectively). Growth of AdRP2234 in 293pIXc4 cells resulted in virions which contained pIX at levels identical to those for AdRP2233, as determined by Western blot analysis of proteins derived from purified Ad virions (data not shown).
To examine growth of these viruses, we infected A549 cells at an MOI of 1, as determined by titer determination on 293 cells, and harvested virus at various time points postinfection. The titers of the resulting virus were determined on 293 cells to examine the growth kinetics and overall yield of the virus (Fig. 4). We observed that the growth of AdRP2234 was reduced compared to that AdRP2233 and showed a similar yield of virus regardless of whether pIX was contained in the virions. At 36 h postinfection, we observed titers of (4.5 ± 0.98) × 108, (1.25 ± 0.49) × 108, and (0.95 ± 0.07) × 108 for AdRP2233, AdRP2234, and AdRP2234pIX+, respectively, illustrating a recovery of virus for AdRP2234 of approximately one-third that for AdRP2233. These data suggest that pIX encoded by the virus can affect the overall viral growth but that pIX from the incoming virion cannot.
FIG. 4.
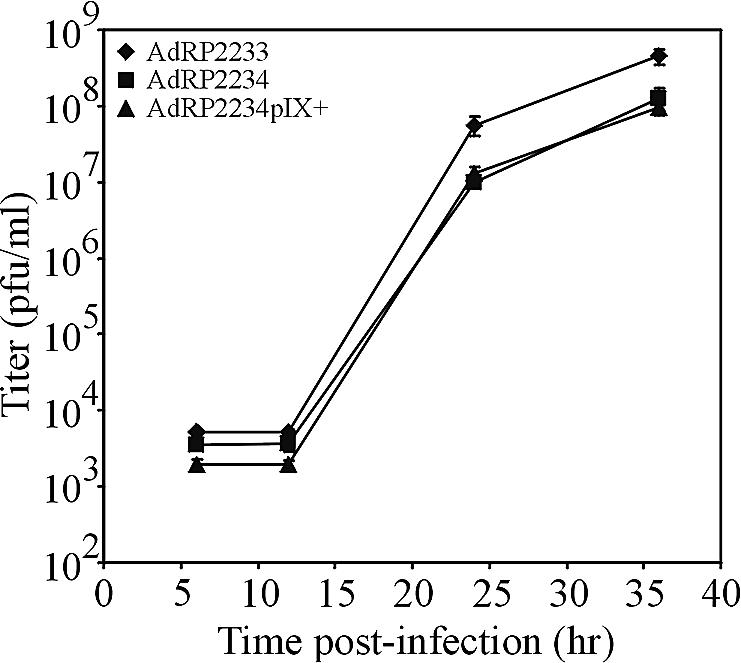
Growth of Ad vector lacking pIX. A549 cells were infected at an MOI of 1 with AdRP2233 (pIX+), AdRP2234 (pIX−), or AdRP2234 that had been grown on a pIX-complementing cell line for virions which contained pIX (AdRP2234pIX+). At various times postinfection, the medium and cells were collected and their titers were determined on 293 cells.
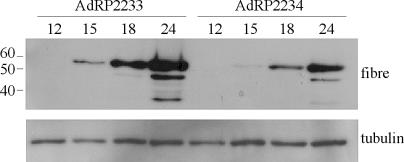
In an attempt to determine why deletion of pIX, from an otherwise wild-type virus, resulted in reduced recovery, we examined early (E1A) and late (fiber) gene expression. As expected, we did not observe a difference in the relative level of E1A expression between AdRP2233 and AdRP2234 (data not shown). However, we did observe a difference in expression of fiber, indicative of late protein expression, between the two viruses during the course of virus growth. As shown in Fig. 5, expression of fiber from AdRP2234 was reduced by 2.7-fold compared to that for AdRP2233 (based on densitometry analysis), consistent with our reduced recovery of virus (Fig. 4). Therefore, pIX may influence expression of late genes.
FIG. 5.
Expression of late protein from Ad vector with pIX deleted. A549 cells were infected at an MOI of 10 with AdRP2233 or AdRP2234. At various times postinfection (in hours, shown above the gels), the medium was removed and the cells were lysed with SDS-PAGE loading buffer. The protein samples were separated by SDS-10% PAGE, transferred to a PVDF membrane, and immunoblotted with antibody to Ad5 fiber or tubulin. Numbers at left are molecular masses in kilodaltons.
We next examined recovery of AdRP2233 and AdRP2234 in 293pIXc4 cells. If pIX is required for efficient late gene expression, we would expect to observe comparable levels of growth between the two viruses on this cell line, since it complements the pIX defect (16). These cells produce pIX at a level equivalent to that of 293 cells infected with an Ad at an MOI of approximately 3, when assayed 24 h postinfection (Fig. 6A). We infected 293pIXc4 cells in 35-mm-diameter dishes with 106 PFU of AdRP2233 or AdRP2234 (MOI of 1) and examined recovery of virus 24 h later. We observed reduced recovery of AdRP2234 relative to that of AdRP2233 (Fig. 6B), similar to that observed when virus was grown on the noncomplementing A549 cell line. The amount of recovered virus at 24 h was (11.5 ± 3.5) × 108 and (3.4 ± 1.3) × 108 PFU/ml for AdRP2233 and AdRP2234, respectively, or a 3.4-fold difference in titer. As expected, we observed that late gene expression was also reduced by about 2.8-fold for AdRP2234 relative to AdRP2233 (Fig. 6C). Therefore, constitutive expression of pIX does not improve the recovery of our pIX-deletion virus.
FIG. 6.
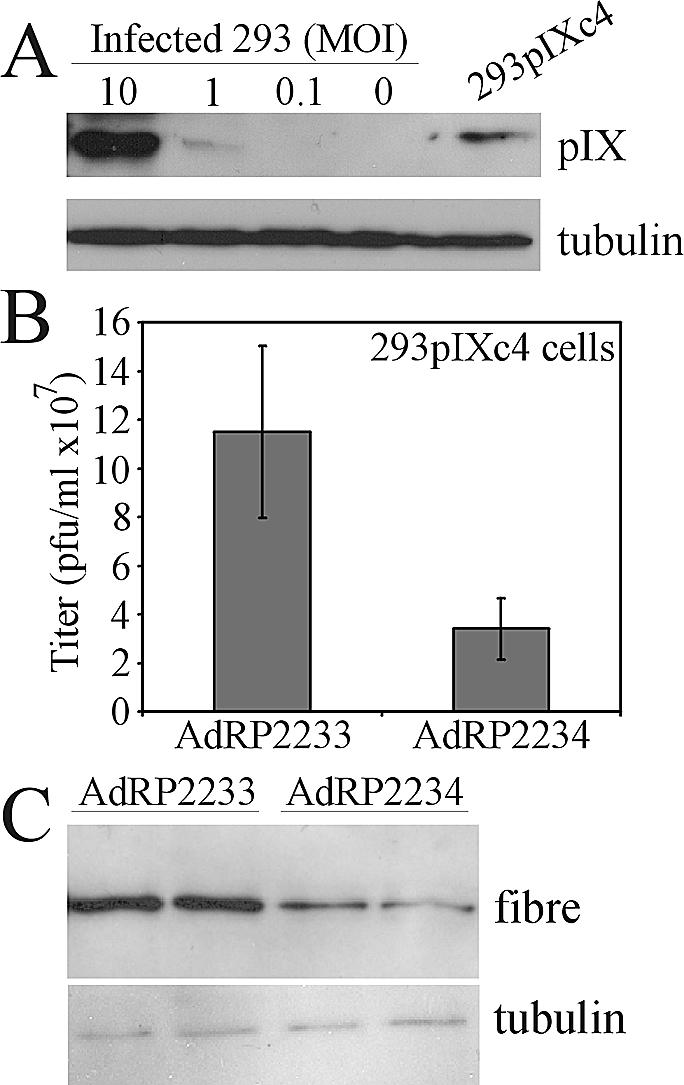
Growth of Ad vector with pIX deleted in 293pIXc4 cells. (A) The level of pIX expression in 293pIXc4 cells was verified by Western blot analysis with 293 cells infected with Ad at various MOIs as a standard. (B) 293pIXc4 cells were infected at an MOI of 1 with AdRP2233 or AdRP2234, the medium and cells were harvested 24 h later, and their titers were determined on 293 cells. (C) Protein samples were prepared in duplicate from cells infected with the indicated virus and analyzed by immunoblotting with fiber- or tubulin-specific antibody.
Interestingly, AdRP2234 appeared to grow slightly better on 293 cells than on 293pIXc4 cells, as determined by its relative titer to AdRP2233 (Fig. 7A), (3.5 ± 0.14) × 108 versus (2.0 ± 0.21) × 108 PFU/ml for AdRP2233 and AdRP2234, respectively, or only a 1.8-fold difference in virus recovery, and late gene expression was also improved (Fig. 7B). 293 cells do encode pIX (7, 9) and express the gene (as determined by RT-PCR [Fig. 7C]), but the quantity of pIX protein produced is very low and was not detected in our immunoblot assay (Fig. 6A). Thus, as we observed in our transient-transfection assays, pIX may influence Ad growth only when it is expressed at very low levels. However, in general pIX has only a minor effect on virus gene expression and growth.
FIG. 7.
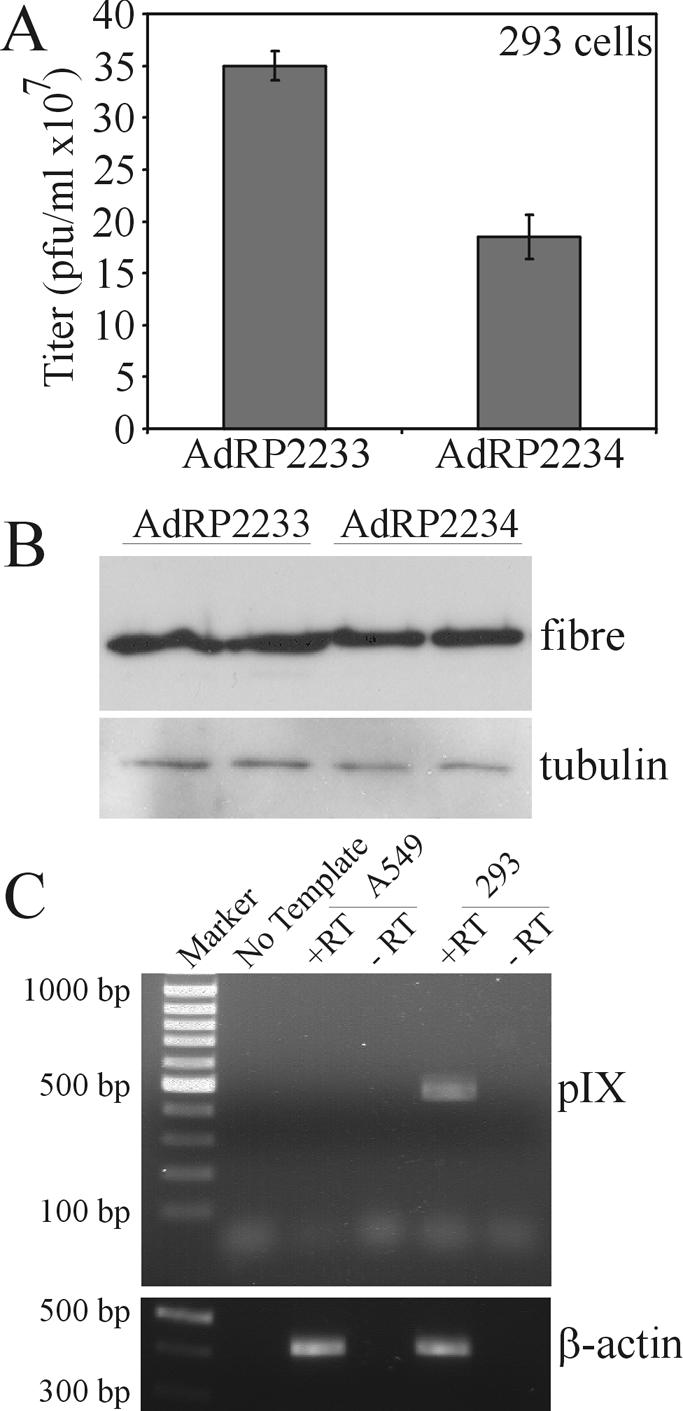
Growth of Ad vector with pIX deleted in 293 cells. (A) 293 cells were infected at an MOI of 1 with AdRP2233 or AdRP2234, the medium and cells were harvested 24 h later, and their titers were determined on 293 cells. (B) Protein samples were prepared in duplicate from cells infected with the indicated virus and analyzed by immunoblotting with fiber- or tubulin-specific antibody. (C) RNA was isolated from 293 or A549 cells and subjected to RT-PCR analysis with synthetic oligonucleotides specific for pIX. The resulting samples were separated on an 0.8% agarose gel. As a control, we performed RT-PCR analysis for β-actin.
DISCUSSION
Ad pIX has been implicated as a transcriptional activator and is reported to increase expression from the Ad E1A, E4, and major late promoters (10, 14). It has been suggested that pIX from the infecting virion migrates to the nucleus shortly after decapsidation (14) and that this pIX population may act to enhance expression from Ad early promoters. We have shown that hdAd vectors with capsids that contain or lack pIX and which encode E1A under its endogenous promoter show identical expression levels of E1A (Fig. 3). Also, replication of a virus which has the pIX coding sequence deleted is not affected by the presence or absence of pIX in the capsid (Fig. 4), nor is expression of E1A different for these two viruses. These data indicate that pIX derived from the capsid does not influence expression of Ad E1A nor, likely, that of E4. Although we have not examined the fate of virion-derived pIX after infection, if translocation of pIX occurs, it does not appear to affect expression of Ad early genes.
We have shown that pIX can act as a weak transcriptional activator when it is expressed transiently. In our experiments, pIX increased expression from a plasmid that encoded a reporter gene under the regulation of the E1A promoter, but only by 1.4-fold, which is considerably less than has been reported in other studies (10, 14). We note that the region of the Ad genome that we utilized in our studies contains sequences including all of the left inverted terminal repeat, packaging signal, E1A promoter, and the first 50 bp of the E1A coding sequence (nucleotides 1 to 611 of the conventional Ad map). In contrast, the promoter element used in the studies by Kedinger and coworkers corresponds to 100 to 560 bp of the Ad genome (10, 14). Since our test element contains additional DNA, it is possible that our E1A promoter element contains DNA sequences which bind a repressor protein, thereby reducing the enhancement effect mediated by pIX. However, since we did not observe any difference in the levels of E1A when we examined expression from “wild-type” pIX+ and pIX− viruses in A549 cells, we suggest that E1A is not normally influenced by expression of pIX. Temporally, we might not expect de novo-synthesized pIX to influence expression of E1A, since E1A is the first gene expressed off the infecting Ad genome and its expression precedes that of pIX by several hours.
We observed that recovery of viruses which did not encode pIX was reduced relative to that of a similar virus which encoded pIX, when grown on A549 cells, a cell line which does not complement the missing gene but does support the growth of E1+ Ad (Fig. 4). Reduced virus recovery correlated with reduced late gene expression (Fig. 5). This may indicate that, during normal Ad replication, pIX does have an effect on expression from the major late promoter. However, we could not “rescue” the poor growth of AdRP2234 by growing the virus on 293pIXc4 cells (Fig. 6). Interestingly, we observed that growth of our pIX-deletion virus, relative to that of pIX+ virus, was slightly better on normal 293 cells (compared to growth in 293pIXc4 cells), and the level of late gene expression from AdRP2234 was almost identical to that of AdRP2233 on this cell line (Fig. 7A and B). 293 cells do express pIX mRNA (Fig. 7C), although the level of protein expression may be very low. 293 cells may produce sufficient quantities to partially complement the pIX “transcriptional” defect in AdRP2234 but produce too little to complement the structural defect. In contrast, 293pIXc4 cells may produce too much pIX to allow it to perform its transcriptional activities efficiently, but these cells produce sufficient quantities to complement the structural requirements for pIX. pIX may be functionally or structurally different in cells that express low or high levels of the protein. The sequences responsible for pIX multimerization also appear crucial for pIX's transcriptional activating activity (14). Thus, at low levels of pIX, the protein may be predominantly a monomer with the activating domain available to enhance transcription. At a high protein concentration, pIX may preferentially multimerize, masking the activation domain. This scenario is consistent with our inability to show transcriptional enhancement mediated by capsid-derived pIX, since this pIX would likely be trimeric. The relationship between the level and timing of pIX gene expression and its effects on Ad promoters may be very complex. However, the fact remains that complete deletion of pIX from a virus results in only a two- to threefold decrease in virus recovery, suggesting that, if pIX does have a transcriptional role in normal Ad replication, it is not very important.
What is the normal role for pIX in the Ad life cycle? pIX is not an important transcriptional activator during Ad growth. We and others have shown that pIX does play a role in maintaining the structural integrity of the virion (4, 6, 16). Ghosh-Choudhury et al. (6) reported that pIX was required for the packaging of full-length Ad genomes, and capsids deficient in pIX could accommodate only approximately 35 kb of viral DNA, based on experiments which relied on the recovery of infectious virions. Recently, we reported that pIX-deficient capsids can package up to at least 37.2 kb of DNA, but such virions are rendered noninfectious for reasons which remain unclear (16). We are left with a mystery: what is the true function of pIX? Viruses which are small (∼35 kb) and lack pIX are fully infectious, as demonstrated in the present study; however, viruses which are large (∼37.2 kb) and lack pIX are completely noninfectious (16). In these large viruses, pIX is not necessary for capsid formation, and we have shown that pIX is not an important transcription factor for the virus. This suggests that pIX may be involved in some other crucial aspect of the virus life cycle. Although pIX is small, only 140 amino acids in Ad5, pIX appears to be a complex protein and may yet play multiple roles in the Ad life cycle.
Acknowledgments
We thank P. Joel Ross and Rashmi Kothary for critical evaluation of the manuscript.
This research was supported by funds provided by the Canadian Institutes of Health Research (CIHR), CIHR/Muscular Dystrophy Association of Canada/Amyotrophic Lateral Sclerosis Society of Canada Partnership Grant, the Jesse Davidson Foundation for Gene and Cell Therapy, Premier's Research Excellence Award, and a CIHR Institute Strategic Initiative. R.J.P. is a recipient of a CIHR New Investigator Award.
REFERENCES
- 1.Boulanger, P., P. Lemay, G. E. Blair, and W. C. Russell. 1979. Characterization of adenovirus protein IX. J. Gen. Virol. 44:783-800. [DOI] [PubMed] [Google Scholar]
- 2.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, L., M. Anton, and F. L. Graham. 1996. Production and characterization of human 293 cell lines expressing the site-specific recombinase Cre. Somat. Cell Mol. Genet. 22:477-488. [DOI] [PubMed] [Google Scholar]
- 4.Colby, W. W., and T. Shenk. 1981. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J. Virol. 39:977-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flint, J., and T. Shenk. 1997. Viral transactivating proteins. Annu. Rev. Genet. 31:177-212. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh-Choudhury, G., Y. Haj-Ahmad, and F. L. Graham. 1987. Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full-length genomes. EMBO J. 6:1733-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 8.Hubberstey, A. V., M. Pavliv, and R. J. Parks. 2002. Cancer therapy utilizing an adenoviral vector expressing only E1A. Cancer Gene Ther. 9:321-329. [DOI] [PubMed] [Google Scholar]
- 9.Louis, N., C. Evelegh, and F. L. Graham. 1997. Cloning and sequencing of the cellular-viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology 233:423-429. [DOI] [PubMed] [Google Scholar]
- 10.Lutz, P., M. Rosa-Calatrava, and C. Kedinger. 1997. The product of the adenovirus intermediate gene IX is a transcriptional activator. J. Virol. 71:5102-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parks, R. J., J. L. Bramson, Y. Wan, C. L. Addison, and F. L. Graham. 1999. Effects of stuffer DNA on transgene expression from helper-dependent adenovirus vectors. J. Virol. 73:8027-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parks, R. J., L. Chen, M. Anton, U. Sankar, M. A. Rudnicki, and F. L. Graham. 1996. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc. Natl. Acad. Sci. USA 93:13565-13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parks, R. J., and F. L. Graham. 1997. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J. Virol. 71:3293-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosa-Calatrava, M., L. Grave, F. Puvion-Dutilleul, B. Chatton, and C. Kedinger. 2001. Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J. Virol. 75:7131-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosa-Calatrava, M., F. Puvion-Dutilleul, P. Lutz, D. Dreyer, H. de The, B. Chatton, and C. Kedinger. 2003. Adenovirus protein IX sequesters host-cell promyelocytic leukaemia protein and contributes to efficient viral proliferation. EMBO Rep. 4:969-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sargent, K., P. Ng, C. Evelegh, F. L. Graham, and R. J. Parks. 2004. Development of a size-restricted pIX-deleted helper virus for amplification of helper-dependent adenovirus vectors. Gene Ther. 11:504-511. [DOI] [PubMed] [Google Scholar]