Abstract
The need for scalable strategies to probe the biological consequences of candidate cancer genes has never been more pressing. The zebrafish, with its capacity for high-throughput transgenesis, in vivo imaging and chemical/genetic screening, has ideal features for undertaking this task. Unique biological insights from zebrafish have already led to the identification of novel oncogenic drivers and small molecules being used to treat the human cancer. This review summarizes the recent main findings and describes pertinent areas where the zebrafish can greatly contribute to our understanding of cancer biology and treatment.
Current Opinion in Genetics & Development 2014, 24:38–45
This review comes from a themed issue on Cancer genomics
Edited by David J Adams and Ultan McDermott
For a complete overview see the Issue and the Editorial
Available online 27th December 2013
0959-437X/$ – see front matter, © 2013 The Authors. Published by Elsevier Ltd. All rights reserved.
Introduction
The wealth of genetic and transcriptomic data in cancer biology, accumulated through international cancer efforts such as The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC), present unprecedented opportunities for identifying therapeutically meaningful targets. A major challenge in genomic approaches has been the lack of appropriate model systems in which to test these on a large scale. Owing to its small size, heavy brood, and rapid maturation time, the zebrafish has emerged as an important new cancer model that complements what can traditionally be achieved in mice and cell culture systems. Advances in transgenic and mutagenesis strategies have already led to a wide variety of zebrafish cancer models with distinct capabilities for high-throughput screening and in vivo imaging [1•,2,3•,4–16]. Despite significant progress in the past 10 years, however, the unique role of zebrafish in cancer research has still yet to be defined. Here, we review recent major achievements in the zebrafish cancer field in light of the available models and advances in genomic techniques. We conclude by discussing future areas of research where zebrafish efforts will be the most effective.
Blood tumors
Numerous leukemic lines have been generated since the first zebrafish model of leukemia was reported in 2003, in a landmark paper showing that expression of mouse c-Myc in transgenic zebrafish unleashed rapid leukemia development [1•]. Consisting of a variety of T or B-cell lymphoblastic (ALL) and myeloid (AML) malignancies, zebrafish leukemia is typically modeled through the expression of a frequently mutated proto-oncogene (such as c-Myc [1•], TEL-AML [4] and NOTCH1 [6]) under the rag2 promoter in developing lymphocytes. A major advantage of this system is the tagging of a fluorescent marker to the gene of interest, enabling powerful real-time tracking of lymphocyte migration and proliferation.
An illustrative example of this tool is an elegant work by Feng et al., in studying a Bcl-2;Myc zebrafish model of lymphoblastic lymphoma (T-LBL) [17]. In this study, Feng et al. monitored the local metastatic behavior of Discosoma red (ds-RED) tagged zebrafish lymphocytes in transparent casper fish, which had vasculature defined by enhanced green fluorescence protein (EGFP). Through live imaging of these cells, the authors were able to determine that lymphoblast autophagy was responsible for preventing their intravasion into the marrow, a hallmark transition of T-LBL to acute T-ALL. Cross-testing in zebrafish and human T-LBL cell lines revealed that this autophagy was caused by high levels of S1P1, which when suppressed resulted in widespread dissemination of the disease (Table 1).
Table 1.
Tools for modeling cancer in zebrafish.
| Purpose | Tool | Examples |
|---|---|---|
| Mutagenesis for forward genetic screens | N-ethyl-N-nitrosourea (ENU) [56] | [3•,5,57] |
| Retroviral-based insertional mutagenesis [58] | [59] | |
| Transgenesis | Tol2 transposon [60,61] | [16] |
| Transgenesis for inducible gene expression | GAL4/UAS [62] | [63] |
| Heat-shock Cre/loxP [64,65] | [11,20,66,67] | |
| Tet-on [68] | [69] | |
| LexPR system [70] | [14] | |
| Site-specific mutagenesis | Talens [71] | [72] |
| Zinc finger nucleases [73,74] | [75] | |
| CRISPr [50] | [52] | |
| Selective expression of mutant alleles in somatic tissue | Plasmid injection (miniCoopR shuttle vector system) [28••] | [28] |
In another study, live imaging of zebrafish embryos enabled Ridges et al. to identify a selective inhibitor of lymphocyte proliferation that is remarkably effective against human T-ALL xenografts [18••]. Ridges et al. screened over 26 000 chemicals for activity that could diminish fluorescent-tagged lymphocyte development in zebrafish larvae. One compound, lenaldekar, induced long-term remission in a zebrafish T-ALL model with encouraging responses in efficacy and toxicity when targeted against human xenografts in mice. While the drug's mechanism remains to be determined, this study provides a key example of the application of zebrafish for pre-clinical drug discovery.
In contrast to T-ALL, efforts to study acute myeloid leukemia (AML), the most lethal and commonly diagnosed leukemia, have not been as successful. To our knowledge, there is one zebrafish AML model and it is based on expression of the MOZ/TIF2 (MYST3/NCOA2) fusion gene under spi1 control in the kidney, where hematopoiesis occurs in zebrafish [19]. Attempts to model AML from proto-oncogenes KRASG12D [20], NUP98-HOXA9 [21] and AML1-ETO [22] have instead led to new models of myeloproliferative neoplasms (MPN) that for unknown reasons do not advance to AML. While the early MPN phenotypes provide valuable read-outs for chemical-genetic screening [23•], their inability to progress to AML may indicate biological differences in this system that warrant further investigation.
In spite of these and other exciting discoveries, there remain areas of active challenge in modeling leukemia in zebrafish. These include to what extent the models truly recapitulate basic aspects of the human disease, to what extent they can be used as models for interrogating genomic changes, and how they can be most effectively used to identify new drug targets across a wider range of disease types. In the coming years, large scale testing of candidate drivers (culled from the TCGA type efforts) in zebrafish leukemic lines will be necessary for these models to further demonstrate their worth.
Solid tumors
Improved transgenic strategies have enhanced the complexity and diversity of solid tumor models in zebrafish, many of which were established through N-ethyl-N-nitrosourea (ENU) mutagenesis screens of mutations in specific genes of interest, such as the important tumor suppressor genes tp53, apc and pten [3•,5,24]. Here we focus on two rapidly growing areas of solid tumor model research: melanoma and embryonal rhabdomyosarcoma.
Melanoma
The first experimental confirmation that oncogenic BRAFV600E (BRAF), mutated in 40–50% of human melanomas [25–27], can promote nevi (moles) and melanoma formation was demonstrated in zebrafish [7]. Since then, similar findings have been shown with NRASQ61K [8] although this model remains less exploited thus far. The simplicity of visualizing melanoma development in these models has led to their widespread adoption and several important, proof-of-principle experiments.
Using the BRAF model, Ceol et al. [28••] tested the oncogenicity of 30 candidate melanoma cancer genes found in a region recurrently amplified in human metastatic melanoma [29]. Genes were overexpressed in melanocytes through the injection of a miniCoopR shuttle vector system into BRAF and p53 mutant embryos. By monitoring for accelerated tumor onset, Ceol et al. were able to identify that SETDB1, a histone transferase, is an oncogene that causes more aggressive melanoma development in zebrafish. This work was the first to demonstrate the feasibility of high-throughput screening of candidate cancer genes in zebrafish and establishes a basis for guiding future approaches aiming to filter down large cancer datasets.
In another application of this line, BRAF expression was associated with a distinct gene signature that resembled expression profiles of embryonic neural crest stem/progenitor cells, thereby motivating White et al. [30••] to screen for suppressors of this embryonic phenotype. A class of compounds, called inhibitors of dihydroorotate dehydrogenase (DHODH), was found to selectively abrogate neural crest development in zebrafish as well as melanoma growth in mouse xenografts and human cell lines. Currently being followed in Phase I/II clinical trials, the DHODH inhibitor leflunomide is a pivotal demonstration of how an embryonic phenotype can be translated to findings about the human disease and lead molecules from zebrafish research into clinical investigation.
Detailed live imaging of melanocytes in a temperature sensitive mitfa (mitfavc7) mutant has provided novel insights into the direct consequences of mitfa activity on tumorigenesis. Reduced mitfa activity caused a dramatic increase in melanocyte cell division [31] and was found to directly affect tumor morphology and formation in the BRAF model [32•]. As these findings could be reversed with the restoration of mitfa's activity, this work substantiates the notion that mitfa is a modifier of BRAF-driven melanoma and provides a functional link between low MITF expression in patients with their poor melanoma prognosis.
Embryonal rhabdomyosarcoma
Recent studies using a KRASG12D-driven model of embryonal rhabdomyosarcoma (ERMS) [11] have highlighted the importance of the cell of origin as a determinant of ERMS. For example, Ignatius et al. [33] used dynamic cellular imaging of a mosaic transgenic rag2-KRASG12D model to track the movement and evolution of ERMS cell subpopulations in embryonic and adult zebrafish. Their findings revealed new roles for differentiated ERMS cells in tumor growth and suggest that mechanisms governing their homeostatic maintenance in regulating growth could be relevant considerations in developing potential therapeutic treatment.
In a similar approach, using promoters representing various stages of muscle development (cdh15, rag2, mylz2), Storer et al. [34] drove expression of KRASG12D and observed that tumors that originated from the more progenitor like cells were more invasive and undifferentiated. These tumors were found to closely recapitulate subgroups of human ERMS based on differentiation status and harbor unique signaling pathways in each subgroup. Confirmation of these pathways as therapeutic targets awaits further study but demonstrates how cross-species oncogenomics can be used to guide therapeutic targeting strategies.
Important insights have also been described in other zebrafish models that cannot be described here [35–39] (reviewed in [40••,41••,42,43]). It is apparent though that some tumor types are better modeled in zebrafish than others. Major areas that have not been as well developed include reliable, penetrant models of pancreatic adenocarcinoma and intestinal carcinoma. While some attempts in this direction have been made [44], these and other diverse solid tumors will require further development.
Comparative oncogenomic approaches
One of the biggest challenges in experimental cancer research is to demonstrate that the model in question recapitulates the human disease. While zebrafish tumors generally resemble their intended human cancers on a histological level [1•,7,8,24], there remain differences in tumor spectrum, incidence and onset [3•,5,24] that are still not well understood. An emerging mode of comparison is through new genomic technologies, which, with careful exploitation, may also point to genetic events that are important for malignant human tumor evolution.
Several studies have begun to compare genomic aberrations in zebrafish cancer to those in human. Rudner et al. [45] employed high-density array comparative genomic hybridization (aCGH) to zebrafish and human T-ALL and found a small number of repeatedly altered genes in zebrafish that also recur in human. Greater overlap was shown in samples from advanced stages of the disease, indicating a heightened conservation for genes under selective pressure. In another study, Zhang et al. [46] sequenced a large cohort of zebrafish malignant peripheral nerve sheath tumors (MPNSTs) and distinguished amplified genes that were shared with the human disease. While the identification of these commonly mutated genes is a promising first step, their experimental validation will be critical toward demonstrating their biological significance.
Our group recently investigated the full spectrum of coding mutations in a zebrafish cancer through exome sequencing of melanomas derived from BRAF and NRAS-driven transgenic lines [76]. In probing for secondary genetic events important for melanoma development, we found that the mutation burden in zebrafish melanomas was sparse compared to human cancer, and equally heterogeneous to the point that cross-species comparisons were difficult. Despite the mutation load, we were able to quantify the multi-hit model of these engineered cancers and highlight a potential new cooperating event with BRAF and p53 mutation through the protein kinase A-cyclic AMP pathway. The work provides the first insights into the mutagenic processes of an engineered zebrafish cancer and will be instructive in guiding future studies of this type in zebrafish.
In particular, it is clear from our experience that there are technical challenges in adapting sequencing tools to zebrafish that require substantial optimization and development. The tremendous diversity both within and between zebrafish strains [47,48], nearly a magnitude greater than that of human, combined with the duplicated genome and other species-specific differences can complicate alignment and overwhelm somatic mutation algorithms with false calls. For this reason, extensive confirmation of these mutations is paramount to avoid errors and to ensure that the data are suitable for meaningful analyses.
While these issues are being addressed, genomic pursuits in zebrafish can focus on modalities that are more robust to nuances in alignment, such as genomic copy number changes and transcriptome profiles based on RNA-seq. The latter strategy provides the additional advantage of capturing a wider range of aberrations — important given the heterogeneity — that together converge on a single expression phenotype. This and optimization of available tools will provide researchers far greater scope for evaluating the relevance of zebrafish cancer and in prescribing new targets and strategies for investigating the human disease.
Future prospects and challenges
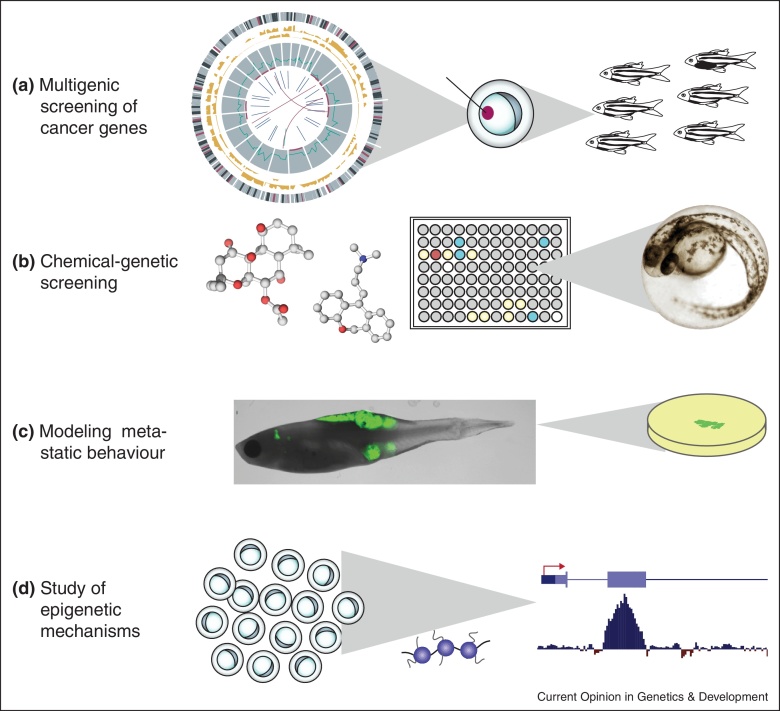
The zebrafish field has seen major growth over the past 10 years, as rapid application of transgenic and chemical screening techniques have placed the fish in a unique category of cancer models. But while creating and analyzing models of human cancer is useful, it ultimately is not significantly advantageous to that done in mouse models. For the fish to offer truly novel and important insights into human cancer will require major innovations in technology and scale. Several areas are particularly amenable to study in the zebrafish, as outlined below (Figure 1).
Figure 1.
Important areas of zebrafish application in cancer research. (a) Multigenic screening involves the parallel testing of the oncogenic potential of candidate cancer genes by injecting plasmids harboring gene of interests into embryos and monitoring for accelerated tumor onset in adult fish [28••]. (b) Chemical libraries can be screened for activity in live zebrafish embryos using early embryonic phenotypic markers in 96 well plates. (c) Tumor metastasis can be followed through the injection of GFP-labelled cell cultures in transparent zebrafish, called casper [17]. (d) One method of studying cancer epigenetics in fish is to perform chromatin immunoprecipitation (ChIP) upon FACs sorted embryos followed by sequencing or expression profiling. Embryos can be rapidly collected in tens of thousands of batches using the i-spawn [55].
Multigenic changes in cancer
It is increasingly recognized that most human cancers are wildly heterogeneous at genetic, and likely, epigenetic, levels. To fully capture this complexity will require in vivo models that can express not just one to four altered genes, but potentially dozens. The increasing sophistication in making knockouts using TALENS [49,49] and the Cas9/CRISPr [50] genome editing system has made it possible to target nearly any candidate cancer gene in the in vivo setting. Although CRISPr was initially thought to be primarily useful for generating germline mutations [50,51], more recent work has highlighted its capacity for inducing somatic, biallelic disruptions in the F0 injected fish [52]. This is a tremendous advantage in zebrafish, since thousands of embryos per day can be generated, each of which can conceptually be injected with a CRISPr and phenotypes directly assessed without going to the next generation. In a typical fish facility containing 2000–10 000 adult pairs of fish, the capacity to test hundreds of candidate genes serially or in parallel dwarfs what can be achieved in mouse models. It seems likely that large-scale genetic screens using this methodology in zebrafish will be forthcoming in the near future, complementing what has been done using ENU screens.
Chemical screening
Traditionally it has been difficult to perform large-scale chemical screens in vivo. However, numerous studies have now shown that the zebrafish is highly amenable to large-scale screens, testing thousands of compounds using detailed, in vivo phenotypic readouts. Although the majority of these screens have relied upon ‘proxy’ embryonic phenotypes (i.e. an embryonic gene program thought related to an adult cancer phenotype), better models of young fish with bona fide cancer will make screening directly in young fish possible.
Modeling metastasis
Responsible for nearly all deaths from solid tumors, the capacity to accurately model metastasis in vivo is essential to improving cancer survival. Our group (RW) has developed a transparent adult zebrafish, casper, that offers very high sensitivity for imaging each of the steps of metastasis [53]. Combining the optical superiority of this model with all of the other key technologies (transgenesis, transplantation, chemical screens, CRISPr's), and with the pool of available mutants generated from the Zebrafish Mutation Project [54], the zebrafish offers a completely unique model in which to deeply probe the biology of metastasis.
Epigenetics changes in cancer
A few studies (e.g. the discovery of SETDB1 in melanoma [28••] as mentioned above) have just begun to explore how the zebrafish can be used to understand epigenetic contributions to cancer. This clearly emerging field will greatly benefit from the genetic and chemical screening tools available in the fish. Improvements in performing core biochemical techniques (i.e. ChIP-seq, methyl-seq, RNA-seq) along with zebrafish cell lines and antibodies will potentially allow for probing of how epigenetic changes contribute to cancer phenotypes. Rapid and large-scale transgenesis, particularly with inducible systems, will be a key method to determine the temporal dynamics of such changes, which will differ from purely genetic changes seen in many tumor types.
Conclusion
As we enter the post-genomics era, the stage is set for zebrafish researchers to capitalize on the strengths of this model system and make significant contributions to cancer research. Already, zebrafish have shown great potential through proof-of-principle experiments involving high-throughput screening [18••,23•,28••,30••] and detailed live imaging [17,31,33,34] of embryonic and adult phenotypes. New genomic technologies have provided greater resolution for performing analyses of zebrafish cancer but require careful application and interpretation. In order to fully maximize the potential of zebrafish in cancer research, strategic areas, such as systematic and scalable methods of functional gene interrogation, using the multitude of existing models, should become a priority. Such focused efforts will inevitably lead zebrafish toward an impact on cancer research that is far more vital and productive.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We would like to thank Chris Dooley for critically reading the manuscript; Niccolò Bolli for useful discussions and Felix Krüger for providing the chemical structures in Figure 1. JY and DLS are supported by the Wellcome Trust.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
References
- 1•.Langenau D.M., Traver D., Ferrando A.A., Kutok J.L., Aster J.C., Kanki J.P., Lin S., Prochownik E., Trede N.S., Zon L.I. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]; This study was the first demonstration of cancer in an induced transgenic zebrafish line.
- 2.Yang H.W., Kutok J.L., Lee N.H., Piao H.Y., Fletcher C.D.M., Kanki J.P., Look A.T. Targeted expression of human MYCN selectively causes pancreatic neuroendocrine tumors in transgenic zebrafish. Cancer Res. 2004;64:7256–7262. doi: 10.1158/0008-5472.CAN-04-0931. [DOI] [PubMed] [Google Scholar]
- 3•.Berghmans S., Look T.A. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci U S A. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification and characterization of the tp53 zebrafish mutant, which has been vital in demonstrating cooperativity in tumour formation of new zebrafish models.
- 4.Sabaawy H.E., Azuma M., Embree L.J., Tsai H.-J., Starost M.F., Hickstein D.D. TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103:15166–15171. doi: 10.1073/pnas.0603349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faucherre A., Taylor G.S., Overvoorde J., Dixon J.E., Hertog J.D. Zebrafish pten genes have overlapping and non-redundant functions in tumorigenesis and embryonic development. Oncogene. 2007;27:1079–1086. doi: 10.1038/sj.onc.1210730. [DOI] [PubMed] [Google Scholar]
- 6.Chen J., Jette C., Kanki J.P., Aster J.C., Look A.T., Griffin J.D. NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia. 2007;21:462–471. doi: 10.1038/sj.leu.2404546. [DOI] [PubMed] [Google Scholar]
- 7.Patton E.E., Widlund H.R., Kutok J.L., Kopani K.R., Amatruda J.F., Murphey R.D., Berghmans S., Mayhall E.A., Traver D., Fletcher C.D.M. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 8.Dovey M., White R.M., Zon L.I. Oncogenic NRAS cooperates with p53 loss to generate melanoma in zebrafish. Zebrafish. 2009;6:397–404. doi: 10.1089/zeb.2009.0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neumann J.C., Dovey J.S., Chandler G.L., Carbajal L., Amatruda J.F. Identification of a heritable model of testicular germ cell tumor in the zebrafish. Zebrafish. 2009;6:319–327. doi: 10.1089/zeb.2009.0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frazer J.K., Meeker N.D., Rudner L., Bradley D.F., Smith A.C.H., Demarest B., Joshi D., Locke E.E., Hutchinson S.A., Tripp S. Heritable T-cell malignancy models established in a zebrafish phenotypic screen. Leukemia. 2009;23:1825–1835. doi: 10.1038/leu.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langenau D.M., Keefe M.D., Storer N.Y., Guyon J.R., Kutok J.L., Le X., Goessling W., Neuberg D.S., Kunkel L.M., Zon L.I. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21:1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santoriello C., Gennaro E., Anelli V., Distel M., Kelly A., Köster R.W., Hurlstone A., Mione M. Kita driven expression of oncogenic HRAS leads to early onset and highly penetrant melanoma in zebrafish. PLoS ONE. 2010;5:e15170. doi: 10.1371/journal.pone.0015170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S., Leach S.D. Screening pancreatic oncogenes in zebrafish using the Gal4/UAS system. In: William Detrich H., Westerfield M., Zon L.I., editors. Methods in Cell Biology. Academic Press; New York: 2011. pp. 367–381. (Chapter 15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen A.T., Emelyanov A., Koh C.H.V., Spitsbergen J.M., Parinov S., Gong Z. An inducible krasV12 transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Dis Model Mech. 2011;5:63–72. doi: 10.1242/dmm.008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin J., Padmanabhan A., de Groh E.D., Lee J.-S., Haidar S., Dahlberg S., Guo F., He S., Wolman M.A., Granato M. Zebrafish neurofibromatosis type 1 genes have redundant functions in tumorigenesis and embryonic development. Dis Model Mech. 2012;5:881–894. doi: 10.1242/dmm.009779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leacock S., Basse A., Chandler G., Kirk A., Rakheja D., Amatruda J. A zebrafish transgenic model of Ewing's sarcoma reveals conserved mediators of EWS-FLI1 tumorigenesis. Dis Model Mech. 2012;5:95–106. doi: 10.1242/dmm.007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng H., Stachura D.L., White R.M., Gutierrez A., Zhang L., Sanda T., Jette C.A., Testa J.R., Neuberg D.S., Langenau D.M. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell. 2010;18:353–366. doi: 10.1016/j.ccr.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Ridges S., Heaton W.L., Joshi D., Choi H., Eiring A., Batchelor L., Choudhry P., Manos E.J., Sofla H., Sanati A. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood. 2012;119:5621–5631. doi: 10.1182/blood-2011-12-398818. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows the application of combined biological imaging and chemical screening of zebrafish embryos for drug discovery in the identification of a chemical compound that selectively inhibits human T-ALL xenograft growth in mice.
- 19.Zhuravleva J., Paggetti J., Martin L., Hammann A., Solary E., Bastie J.-N., Delva L. MOZ/TIF2-induced acute myeloid leukaemia in transgenic fish. Br J Haematol. 2008;143:378–382. doi: 10.1111/j.1365-2141.2008.07362.x. [DOI] [PubMed] [Google Scholar]
- 20.Le X., Langenau D., Keefe M., Kutok J., Neuberg D., Zon L. Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc Natl Acad Sci U S A. 2007;104:9410–9415. doi: 10.1073/pnas.0611302104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forrester A.M., Grabher C., McBride E.R., Boyd E.R., Vigerstad M.H., Edgar A., Kai F.-B., Da’as S.I., Payne E., Look A.T. NUP98-HOXA9-transgenic zebrafish develop a myeloproliferative neoplasm and provide new insight into mechanisms of myeloid leukaemogenesis. Br J Haematol. 2011;155:167–181. doi: 10.1111/j.1365-2141.2011.08810.x. [DOI] [PubMed] [Google Scholar]
- 22.Yeh J.R.J., Munson K.M., Chao Y.L., Peterson Q.P., MacRae C.A., Peterson R.T. AML1-ETO reprograms hematopoietic cell fate by downregulating scl expression. Development. 2007;135:401–410. doi: 10.1242/dev.008904. [DOI] [PubMed] [Google Scholar]
- 23•.Yeh J.-R.J., Munson K.M., Elagib K.E., Goldfarb A.N., Sweetser D.A., Peterson R.T. Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat Chem Biol. 2009;5:236–243. doi: 10.1038/nchembio.147. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides an example of a chemical-genetic screening approach leading to the identification of COX-2 as a small molecule modifier of the AML-ETO oncogene driven embryonic hematopoietic phenotype.
- 24.Haramis A.-P.G., Hurlstone A., van der Velden Y., Begthel H., van den Born M., Offerhaus G.J.A., Clevers H.C. Adenomatous polyposis coli-deficient zebrafish are susceptible to digestive tract neoplasia. EMBO Rep. 2006;7:444–449. doi: 10.1038/sj.embor.7400638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 26.Pollock P.M., Harper U.L., Hansen K.S., Yudt L.M., Stark M., Robbins C.M., Moses T.Y., Hostetter G., Wagner U., Kakareka J. High frequency of BRAF mutations in nevi. Nat Genet. 2002;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 27.Brose M.S., Volpe P., Feldman M., Kumar M., Rishi I., Gerrero R., Einhorn E., Herlyn M., Minna J., Nicholson A. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 28••.Ceol C.J., Houvras Y., Jane-Valbuena J., Bilodeau S., Orlando D.A., Battisti V., Fritsch L., Lin W.M., Hollmann T.J., Ferré F. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides proof-of-principle demonstration of high-throughput candidate cancer gene screening in zebrafish in identifying SETDB1 as a driver of enhanced melanoma formation.
- 29.Garraway L.A., Widlund H.R., Rubin M.A., Getz G., Berger A.J., Ramaswamy S., Beroukhim R., Milner D.A., Granter S.R., Du J. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 30••.White R.M., Cech J., Ratanasirintrawoot S., Lin C.Y., Rahl P.B., Burke C.J., Langdon E., Tomlinson M.L., Mosher J., Kaufman C. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates the translation of a zebrafish embryonic phenotype to human disease leading to the first chemical compound (leflunomide) from a zebrafish study to enter Phase I/II clinical trials.
- 31.Taylor K.L., Lister J.A., Zeng Z., Ishizaki H., Anderson C., Kelsh R.N., Jackson I.J., Patton E.E. Differentiated melanocyte cell division occurs in vivo and is promoted by mutations in Mitf. Development. 2011;138:3579–3589. doi: 10.1242/dev.064014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Lister J.A., Capper A., Zeng Z., Mathers M.E., Richardson J., Paranthaman K., Jackson I.J., Patton E.E. A conditional Zebrafish MITF mutation reveals MITF levels are critical for melanoma promotion versus regression in vivo. J Investig Dermatol. 2013 doi: 10.1038/jid.2013.293. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses a temperature-sensitive mitfa mutant (mitfavc7) to show how differential levels of mitfa activity can modulate BRAF-driven melanoma formation, morphology and regression.
- 33.Ignatius M.S., Chen E., Elpek N.M., Fuller A.Z., Tenente I.M., Clagg R., Liu S., Blackburn J.S., Linardic C.M., Rosenberg A.E. In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell. 2012;21:680–693. doi: 10.1016/j.ccr.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storer N.Y., White R.M., Uong A., Price E., Nielsen G.P., Langenau D.M., Zon L.I. Zebrafish rhabdomyosarcoma reflects the developmental stage of oncogene expression during myogenesis. Development. 2013;140:3040–3050. doi: 10.1242/dev.087858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hettmer S., Liu J., Miller C.M., Lindsay M.C., Sparks C.A., Guertin D.A., Bronson R.T., Langenau D.M., Wagers A.J. Sarcomas induced in discrete subsets of prospectively isolated skeletal muscle cells. Proc Natl Acad Sci U S A. 2011;108:20002–20007. doi: 10.1073/pnas.1111733108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shive H.R., West R.R., Embree L.J., Azuma M., Sood R., Liu P., Hickstein D.D. brca2 in zebrafish ovarian development, spermatogenesis, and tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:19350–19355. doi: 10.1073/pnas.1011630107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albacker C.E., Storer N.Y., Langdon E.M., DiBiase A., Zhou Y., Langenau D.M., Zon L.I. The histone methyltransferase SUV39H1 suppresses embryonal rhabdomyosarcoma formation in Zebrafish. PLoS ONE. 2013;8:e64969. doi: 10.1371/journal.pone.0064969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le X., Pugach E.K., Hettmer S., Storer N.Y., Liu J., Wills A.A., DiBiase A., Chen E.Y., Ignatius M.S., Poss K.D. A novel chemical screening strategy in zebrafish identifies common pathways in embryogenesis and rhabdomyosarcoma development. Development. 2013;140:2354–2364. doi: 10.1242/dev.088427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He S., Lamers G.E., Beenakker J.-W.M., Cui C., Ghotra V.P., Danen E.H., Meijer A.H., Spaink H.P., Snaar-Jagalska B.E. Neutrophil-mediated experimental metastasis is enhanced by VEGFR inhibition in a zebrafish xenograft model. J Pathol. 2012;227:431–445. doi: 10.1002/path.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.White R., Rose K., Zon L. Zebrafish cancer: the state of the art and the path forward. Nature. 2013;13:624–636. doi: 10.1038/nrc3589. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is a comprehensive review of the development of the zebrafish cancer field with respect to cancer models, techniques and their application to cancer research.
- 41••.Liu S., Leach S.D. Zebrafish models for cancer. Annu Rev Pathol Mech Dis. 2011;6:71–93. doi: 10.1146/annurev-pathol-011110-130330. [DOI] [PubMed] [Google Scholar]; This review provides an excellent, detailed overview of current zebrafish models and the transgenic and mutagenesis strategies used to build them.
- 42.Chen E.Y., Langenau D.M. Zebrafish models of rhabdomyosarcoma. In: William Detrich H., Westerfield M., Zon L.I., editors. Methods in cell biology. Academic Press; New York: 2011. pp. 383–402. (Chapter 16) [Google Scholar]
- 43.Mione M.C., Trede N.S. The zebrafish as a model for cancer. Dis Model Mech. 2010;3:517–523. doi: 10.1242/dmm.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park S.W., Davison J.M., Rhee J., Hruban R.H., Maitra A., Leach S.D. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology. 2008;134:2080–2090. doi: 10.1053/j.gastro.2008.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudner L.A., Brown K.H., Dobrinski K.P., Bradley D.F., Garcia M.I., Smith A.C.H., Downie J.M., Meeker N.D., Look A.T., Downing J.R. Shared acquired genomic changes in zebrafish and human T-ALL. Oncogene. 2011;30:4289–4296. doi: 10.1038/onc.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang G., Hoersch S., Amsterdam A., Whittaker C.A., Beert E., Catchen J.M., Farrington S., Postlethwait J.H., Legius E., Hopkins N. Comparative oncogenomic analysis of copy number alterations in human and zebrafish tumors enables cancer driver discovery. PLoS Genet. 2013;9:e1003734. doi: 10.1371/journal.pgen.1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M., Collins J.E., Humphray S., McLaren K., Matthews L. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown K.H., Dobrinski K.P., Lee A.S., Gokcumen O., Mills R.E., Shi X., Chong W.W.S., Chen J.Y.H., Yoo P., David S. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci U S A. 2012;109:529–534. doi: 10.1073/pnas.1112163109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bedell V.M., Wang Y., Campbell J.M., Poshusta T.L., Starker C.G., Krug R.G., II, Tan W., Penheiter S.G., Ma A.C., Leung A.Y.H. In vivo genome editing using a high-efficiency TALEN system. Nature. 2013;490:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J-RJ, Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Kaini P., Sander J.D., Joung J.K., Peterson R.T., Yeh J.-R.J. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS ONE. 2013;8:e68708. doi: 10.1371/journal.pone.0068708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jao L.-E., Wente S.R., Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White R.M., Sessa A., Burke C., Bowman T., LeBlanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C.E. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kettleborough R.N.W., Busch-Nentwich E.M., Harvey S.A., Dooley C.M., de Bruijn E., van Eeden F., Sealy I., White R.J., Herd C., Nijman I.J. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2014;496:494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adatto I., Lawrence C., Thompson M., Zon L.I. A new system for the rapid collection of large numbers of developmentally staged zebrafish embryos. PLoS ONE. 2011;6:e21715. doi: 10.1371/journal.pone.0021715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beckwith L.G., Moore J.L., Tsao-Wu G.S., Harshbarger J.C., Cheng K.C. Ethylnitrosourea induces neoplasia in zebrafish (Danio rerio) Lab Invest. 2000;80:379–385. doi: 10.1038/labinvest.3780042. [DOI] [PubMed] [Google Scholar]
- 57.Goessling W., North T.E., Lord A.M., Ceol C., Lee S., Weidinger G., Bourque C., Strijbosch R., Haramis A.-P., Puder M. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev Biol. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- 58.Amsterdam A., Burgess S., Golling G., Chen W., Sun Z., Townsend K., Farrington S., Haldi M., Hopkins N. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amsterdam A., Sadler K.C., Lai K., Farrington S., Bronson R.T., Lees J.A., Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:e139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawakami K., Shima A., Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci U S A. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suster M.L., Kikuta H., Urasaki A., Asakawa K., Kawakami K. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol Biol. 2009;561:41–63. doi: 10.1007/978-1-60327-019-9_3. [DOI] [PubMed] [Google Scholar]
- 62.Kawakami K., Takeda H., Kawakami N., Kobayashi M., Matsuda N., Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 63.Anelli V., Santoriello C., Distel M., Köster R., Ciccarelli F., Mione M. Global repression of cancer gene expression in a zebrafish model of melanoma is linked to epigenetic regulation. Zebrafish. 2009;6:417–424. doi: 10.1089/zeb.2009.0612. [DOI] [PubMed] [Google Scholar]
- 64.Birling M.-C., Gofflot F., Warot X. Site-specific recombinases for manipulation of the mouse genome. Methods Mol Biol. 2009;561:245–263. doi: 10.1007/978-1-60327-019-9_16. [DOI] [PubMed] [Google Scholar]
- 65.Thummel R., Burket C.T., Brewer J.L., Sarras M.P.J., Li L., Perry M., McDermott J.P., Sauer B., Hyde D.R., Godwin A.R. Cre-mediated site-specific recombination in zebrafish embryos. Dev Dyn. 2005;233:1366–1377. doi: 10.1002/dvdy.20475. [DOI] [PubMed] [Google Scholar]
- 66.Langenau D.M., Feng H., Berghmans S., Kanki J.P., Kutok J.L., Look A.T. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2005;102:6068–6073. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng H., Langenau D.M., Madge J.A., Quinkertz A., Gutierrez A., Neuberg D.S., Kanki J.P., Thomas Look A. Heat-shock induction of T-cell lymphoma/leukaemia in conditional Cre/lox-regulated transgenic zebrafish. B J Haematol. 2007;138:169–175. doi: 10.1111/j.1365-2141.2007.06625.x. [DOI] [PubMed] [Google Scholar]
- 68.Huang C.-J., Jou T.-S., Ho Y.-L., Lee W.-H., Jeng Y.-T., Hsieh F.-J., Tsai H.-J. Conditional expression of a myocardium-specific transgene in zebrafish transgenic lines. Dev Dyn. 2005;233:1294–1303. doi: 10.1002/dvdy.20485. [DOI] [PubMed] [Google Scholar]
- 69.Li Z., Huang X., Zhan H., Zeng Z., Li C., Spitsbergen J.M., Meierjohann S., Schartl M., Gong Z. Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet-on xmrk transgenic zebrafish. J Hepatol. 2012;56:419–425. doi: 10.1016/j.jhep.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 70.Emelyanov A., Parinov S. Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev Biol. 2008;320:113–121. doi: 10.1016/j.ydbio.2008.04.042. [DOI] [PubMed] [Google Scholar]
- 71.Huang P., Xiao A., Zhou M., Zhu Z., Lin S., Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 72.Moore F.E., Reyon D., Sander J.D., Martinez S.A., Blackburn J.S., Khayter C., Ramirez C.L., Joung J.K., Langenau D.M. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs) PLoS ONE. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meng X., Noyes M.B., Zhu L.J., Lawson N.D., Wolfe S.A. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doyon Y., McCammon J.M., Miller J.C., Faraji F., Ngo C., Katibah G.E., Amora R., Hocking T.D., Zhang L., Rebar E.J. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sood R., Carrington B., Bishop K., Jones M., Rissone A., Candotti F., Chandrasekharappa S.C., Liu P. Efficient methods for targeted mutagenesis in zebrafish using zinc-finger nucleases: data from targeting of nine genes using CompoZr or CoDA ZFNs. PLoS ONE. 2013;8:e57239. doi: 10.1371/journal.pone.0057239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yen J., White R.M., Wedge D.C., Van Loo P., de Ridder J., Capper A., Richardson J., Jones D., Raine K., Watson I.R. The genetic heterogeneity and mutational burden of engineered melanomas in zebrafish models. Genome Biology. 2013;14:R113. doi: 10.1186/gb-2013-14-10-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]