Abstract
The membrane glycoproteins Gn and Gc of Hantaan virus (HTNV) (family Bunyaviridae) are modified by N-linked glycosylation. The glycoproteins contain six potential sites for the attachment of N-linked oligosaccharides, five sites on Gn and one on Gc. The properties of the N-linked oligosaccharide chains were analyzed by treatment with endoglycosidase H, peptide:N-glycosidase F, tunicamycin, and deoxynojirimycin and were confirmed to be completely of the high-mannose type. Ten glycoprotein gene mutants were constructed by site-directed mutagenesis, including six single N glycosylation site mutants and four double-site mutants. We determined that four sites (N134, -235, -347, and -399) on Gn and the only site (N928) on Gc in their ectodomains are utilized, whereas the fifth site on Gn (N609), which faces the cytoplasm, is not glycosylated. The importance of individual N-oligosaccharide chains varied with respect to folding and intracellular transport. The oligosaccharide chain on residue N134 was found to be crucial for protein folding, whereas single mutations at the other glycosylation sites were better tolerated. Mutation at glycosylation sites N235 and N399 together resulted in Gn misfolding. The endoplasmic reticulum chaperones calnexin and calreticulin were found to be involved in HTNV glycoprotein folding. Our data demonstrate that N-linked glycosylation of HTNV glycoproteins plays important and differential roles in protein folding and intracellular trafficking.
Hantaan virus (HTNV) is the prototype of the genus Hantavirus in the family Bunyaviridae and is a causative agent of hemorrhagic fever with renal syndrome. Like other viruses in this family, HTNV has a tripartite, single-stranded negative-sense RNA genome. The small, medium, and large genomic RNA segments encode the nucleocapsid (N) protein, the precursor to the virion envelope glycoproteins (GPC), and the virion-associated RNA polymerase (L [large] protein), respectively (33, 34). GPC is cotranslationally cleaved to generate two proteins termed Gn and Gc (33), presumably by cellular signal peptidase at the pentapeptide motif WAASA (21), and mature Gn and Gc have apparent molecular weights of 68,000 and 55,000, respectively (35). (Gn and Gc are designated according to their location relative to the N and C termini of GPC and were previously referred to as G1 and G2.) Both Gn and Gc are type I integral transmembrane proteins with their own signal sequences (33). Our recent work indicated that the Gc signal sequence is an integral part of the Gn cytoplasmic tail and is required for Golgi targeting of both proteins (38). Similar to most other viruses in the family, HTNV glycoproteins Gn and Gc form a heterodimer and accumulate in the Golgi complex, where viruses mature and assemble (reviewed in reference 39). It is accepted that the heterodimerization of Gn and Gc is essential for their correct folding and transport to the Golgi complex (32, 38, 40).
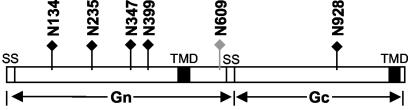
HNTV Gn and Gc proteins are both modified by N-linked glycosylation. They possess six potential N-linked glycosylation sites (Fig. 1), five on Gn (at N residues 134, 235, 347, 399, and 609) and one on Gc (N928) (37). The N glycosylation sites are highly conserved among all hantaviruses, suggesting that N glycosylation is crucial for the conformation and functions of the proteins. HTNV glycoproteins are sensitive to endoglycosidase H (endo H) treatment, indicating that the N-glycan chains are predominantly of the high-mannose type (1, 36). However, there is disagreement as to whether the glycans are completely of the high-mannose type (1) or whether some of them have acquired endo H resistance (32, 36).
FIG. 1.
Schematic of HTNV GPC and potential N-linked glycosylation sites. The GPC is represented by the bar, with signal sequences (SS) and transmembrane domains (TMD) shown as open boxes and filled boxes, respectively. Lollipops indicate the locations of the potential N glycosylation sites on Gn and Gc (five in Gn and one in Gc), and the filled ones point to sites that in this study were shown to contain oligosaccharides.
N-linked glycosylation is important not only for correct protein folding but also for glycoprotein function (16, 18, 27, 44). Enveloped viruses may contain one or more types of integral membrane proteins, and the majority of them are modified by the addition of N-linked carbohydrate chains (7). N-linked glycosylation is associated with the diverse functions of viral glycoproteins, such as receptor binding, mediation of membrane fusion and penetration into cells, directing virus morphogenesis at the budding site, and working as antigens to elicit a protective immune response (3, 7).
In this study, we reexamined the properties of the N-linked oligosaccharides on HTNV glycoproteins and confirmed that the N-glycans are completely endo H sensitive. To determine the utilization of the potential N glycosylation sites and study their role in protein folding and intracellular trafficking of the glycoproteins, we constructed 10 mutants bearing either single or double glycosylation site mutations. Our data show that the individual sites of N glycosylation have different effects on protein folding and intracellular trafficking of the glycoproteins. Two endoplasmic reticulum (ER) chaperones, calnexin (CNX) and calreticulin (CRT), were found associated with the glycoproteins and presumably play a role in HTNV glycoprotein folding.
MATERIALS AND METHODS
Cells and viruses.
HeLaT4+cells and Vero E6 cells (ATCC C1008) were grown in Dulbecco's modified Eagles's medium containing 10% fetal bovine serum (FBS). BSR-T7 cells, which stably express T7 RNA polymerase (4), were kindly provided by K. K. Conzelmann (Max-von-Pettenkofer Institut, Munich, Germany) and were grown in Glasgow modified Eagle's medium containing 10% FBS. vTF7-3, a recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase (9), was a gift from B. Moss, National Institutes of Health, Bethesda, Md. vT-HTN M, a recombinant vaccinia virus that expresses HTNV glycoproteins (Gn and Gc), was constructed in this laboratory (unpublished data).
Antibodies.
Monoclonal antibodies (MAbs) recognizing HTNV glycoproteins were kindly provided by C. S. Schmaljohn (Virology Division, U.S. Army Medical Research Institute for Infectious Diseases, Frederick, Md.). MAbs 8B6, 3D5, 16D2, and 6D4 are specific for Gn, and MAbs 11E10, 8E10, 16E6, and HCO2 are specific for Gc (2). A rabbit polyclonal antibody against GM130, a cis-Golgi matrix protein (26), was provided by M. Lowe (School of Biological Science, University of Manchester, Manchester, United Kingdom). Anti-FLAG M2 MAb, rabbit polyclonal anti-CRT antibody, and goat anti-rabbit immunoglobulin G (IgG) conjugated with fluorescein isothiocyanate (FITC) were purchased from Sigma. Goat anti-mouse IgG conjugated with Cy5 was purchased from Amersham Pharmacia Biotech (Buckingham, United Kingdom), and rabbit anti-CNX polyclonal antibody was purchased from StressGen Biotechnologies Corp. (Victoria, Canada).
Plasmid construction and mutagenesis.
HTNV M-segment cDNA was cloned into pGEM-1 under the control of the T7 promoter and used as the template for site-directed PCR mutagenesis using Turbo-Pfu DNA polymerase (Stratagene). To construct the N glycosylation site mutants, one (for single mutation) or two (for double mutations) of the asparagine (N) residues at the potential sites on Gn and Gc were replaced with glutamine (Q). Briefly, PCR was performed with the primer pairs being annealed to the template DNA (pGEM-HTN M) in an outward orientation, and the amplified product was treated with DpnI and T4 polynucleotide kinase (New England Biolabs) and then recircularized with T4 DNA ligase. The resulting six single N glycosylation site mutants were designated N134Q, N235Q, N347Q, N399Q, N609Q, and N928Q according to the position of the substitution. The coding regions of mutants N134Q, N235Q, N347Q, N399Q, and N928Q were also cloned into pTM1 (25) for expression in BSR-T7 cells. Four double glycosylation site mutants (N235/399Q, N235/928Q, N347/928Q, and N399/928Q) were constructed in pGEM-1 either by site-directed mutagenesis (for N235/299Q) or by DNA cloning using the above single-site mutants.
Since most of the MAbs against HTNV glycoproteins do not react, or react poorly, with denatured and misfolded proteins (37, 42), we constructed an M-segment cDNA expressing GPC tagged with the FLAG (DYLDDDDL) epitope, which was inserted into the N terminus of Gn between residues 24 (aspartic acid, D) and 25 (methionine, M) just downstream of the signal peptide. All mutants were verified by sequence analysis. The primers used and the details of PCR are available from the authors on request.
Infection and transfection of cells.
For immunoprecipitation experiments, subconfluent monolayers of Vero E6 cells were grown in 35-mm-diameter petri dishes, and for immunofluorescence experiments, BSR-T7 and HeLaT4+ cells were grown on 13-mm-diameter cover slips. Cells were infected with vTF7-3 at 5 PFU/cell for 60 min and then transfected with plasmid DNA as described previously (20). Briefly, for cells grown on 35-mm-diameter dishes, 2 μg of DNA and 15 μl of liposomes were diluted in 500 μl of OptiMEM (BRL-Life Technologies, Paisley, United Kingdom), and for cells grown on cover slips, 0.5 μg of plasmid DNA and 6 μl of cationic liposome were diluted in 250 μl of OptiMEM. The DNA-liposome mixtures were incubated for 10 min at room temperature before being added to cells that had been washed previously with OptiMEM. At 3 h posttransfection, 0.5 ml of Dulbecco's modified Eagle's medium containing 10% FBS was added and incubation was continued at 37°C.
DNJ and tunicamycin treatments.
1-Deoxynojirimycin (DNJ) (Calbiochem) was dissolved in distilled H2O at a concentration of 100 mM, and tunicamycin (Boehringer Mannheim) was dissolved in dimethyl sulfoxide at a concentration of 1 mg/ml. At 3 h posttransfection (or 4 h postinfection with vT-HTN M), the inhibitors were added to the medium to give a final concentration of 1 or 2 mM for DNJ or 2 μg/ml for tunicamycin. The inhibitors were also present during radiolabeling for immunoprecipitation.
Metabolic radiolabeling and immunoprecipitation.
Cells were incubated for 1 h in starvation medium lacking methionine prior to radiolabeling with [35S]methionine (Amersham Pharmacia Biotech) at 7 h posttransfection with plasmid constructs or 8 h postinfection with vT-HTN M. For the pulse-chase assay cells grown in 35-mm-diameter dishes were labeled for 20 min with 80 μCi of [35S]methionine (200 μCi/ml), and for immunoprecipitation cells were labeled for a further 15 h at 37°C with 50 μCi of [35S]methionine (100 μCi/ml). To radiolabel the sugar moiety of HTNV glycoproteins, vT-HTN M-infected cells were labeled for 4 h with [3H]mannose (200 μCi/ml; Amersham Pharmacia Biotech) at 8 h postinfection. Cells were then lysed on ice with 300 μl of nondenaturing radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.4], 1% Triton X-100, 300 mM NaCl, 5 mM EDTA) containing a cocktail of protease inhibitors (Roche) and 20 mM N-ethylmaleimide. Cell lysates were clarified by centrifugation for 10 min at 4°C at 16,000 × g, and HTNV glycoproteins were immunoprecipitated with anti-Gn and anti-Gc MAbs or anti-FLAG conjugated to protein A-agarose (Sigma). The beads were washed five times with buffer containing 0.1% Triton X-100 and once with ice-cold phosphate-buffered saline (PBS), and the bound proteins were either analyzed by sodium dodecyl sulfate (SDS)-8 to 10% polyacrylamide gel electrophoresis (PAGE) or subjected to digestion with endo H and peptide:N-glycosidase F (PNGase F).
endo H and PNGase F digestion.
Immunoprecipitates were denatured in 30 μl of denaturing buffer (0.5% SDS and 1% β-mercaptoethanol) at 100°C for 10 min and cooled to room temperature. The denatured samples were then subjected to digestion with 150 mU of endo H (New England Biolabs) in a 40-μl reaction mixture containing 50 mM sodium citrate (pH 5.5), 0.5% SDS, and 1% β-mercaptoethanol or with 4 mU of PNGase F (New England Biolabs) in a 40-μl reaction mixture containing 50 mM sodium phosphate (pH 7.5), 0.5% SDS, 1% NP-40, and 1% β-mercaptoethanol for 20 h at 37°C. The treated samples were analyzed by SDS-10% PAGE under reducing conditions.
Immunofluorescence staining.
The immunofluorescence assay was performed as previously described (38). Briefly, at 5 h posttransfection of HeLaT4+ cells or at 48 h posttransfection of BSR-T7 cells, cycloheximide was added to the culture medium to a final concentration of 50 μg/ml and cells were incubated for a further 4 h. The cells were then fixed for 10 min with 4% paraformaldehyde and permeabilized by incubation in PBS containing 0.1% Triton X-100 for 30 min. The cells were reacted for 30 min with anti-Gn and -Gc MAbs (at a 1:500 dilution) and anti-GM130 (at a 1:600 dilution). After thorough washing with PBS, the cells were stained for 30 min with Cy5-conjugated anti-mouse IgG and fluorescein isothiocyanate-conjugated anti-rabbit IgG. The localization of viral antigens and Golgi proteins was examined using a Zeiss LSM confocal microscope.
RESULTS
Analysis of N glycosylation of HTNV glycoproteins.
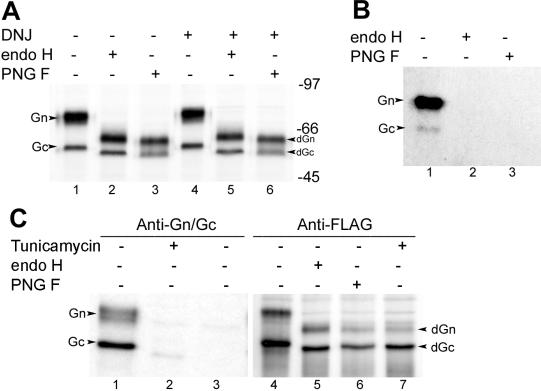
We first reexamined the endo H and PNGase F sensitivity of HTNV Gn and Gc expressed from a recombinant vaccinia virus containing full-length M-segment cDNA. Consistent with previously published reports (1, 36), treatment of the glycoproteins with endo H and PNGase F resulted in a significant electrophoretic mobility shift of both proteins, corresponding to apparent molecular weight losses of 12,000 for Gn and 3,000 for Gc (Fig. 2A, lanes 1 to 3). However, as mentioned previously by Schmaljohn et al. (36) and Ruusula et al. (32), we noticed that Gn after digestion by endo H was clearly slightly larger (approximately 1,000) than Gn after digestion by PNGase F (Fig. 2A, lanes 2 and 3). To investigate the significance of the mobility shift between the Gn proteins deglycosylated by endo H and PNGase F, we reanalyzed the oligosaccharide chains with the help of two inhibitors, DNJ and tunicamycin. DNJ inhibits ER α-glucosidases I and II and thus blocks the sequential removal of the terminal three glucose residues from the core oligosaccharide on the nascent glycoproteins in the lumen of the ER (8, 10, 41), thus preventing the further trimming of N-linked sugars (13, 14). We observed that DNJ treatment generated higher-molecular-weight forms of Gn and Gc proteins (Fig. 2A, lane 4), confirming that the trimming of N-glycans of both proteins had been inhibited. However, the digestion patterns generated by endo H and PNGase F in the presence of DNJ were identical to those without DNJ treatment in that the endo H-digested Gn was still slightly larger than that digested by PNGase F (Fig. 2A, lanes 5 and 6). These results indicate that the slight difference in molecular mass of Gn was not due to the acquisition of endo H resistance but to the presence of N-acetylglycosamine residues left after endo H treatment.
FIG. 2.
Biochemical analyses of N-glycan chains on HTNV glycoproteins Gn and Gc. (A) DNJ blocks N-glycan trimming of Gn and Gc. Vero E6 cells were infected with vT-HTN M and radiolabeled with 50 μCi of [35S]methionine for 15 h in the absence (lanes 1 to 3) or presence (lanes 4 to 6) of 2 mM DNJ, and cell lysates were immunoprecipitated with anti-Gn and -Gc MAbs. The resulting precipitates were subjected to digestion with endo H and PNGase F for 20 h as indicated and analyzed by SDS-10% PAGE under reducing conditions. The deglycosylated forms of Gn and Gc are marked dGn and dGc, respectively. (B) endo H and PNGase F digestion of HTNV glycoproteins labeled with [3H]mannose. Vero E6 cells infected with vT-HTN M were labeled with 200 μCi of [3H]mannose for 4 h, and cell lysates were reacted with anti-Gn and -Gc MAbs, followed by digestion with endo H and PNGase F. The gel was exposed for 60 days. (C) Effect of tunicamycin on glycosylation of HTNV glycoproteins. Vero E6 cells were infected with vTF7-3 and then mock transfected (lane 3) or transfected with HTNV M-segment cDNA tagged with FLAG. Cells were labeled with 50 μCi of [35S]methionine for 15 h in the absence or presence of tunicamycin (2 μg/ml) and immunoprecipitated with anti-Gn and -Gc MAbs (lanes 1 to 3) or anti-FLAG and anti-Gc antibodies (lanes 4 to 7). The immunoprecipitates were subjected to digestion with endo H or PNGase F as indicated.
To further confirm the results presented above, we labeled the sugar moiety of HTNV glycoproteins with [3H]mannose followed by digestion with endo H and PNGase F. As shown in Fig. 2B, no radioactive signal remained after treatment with either glycosidase, indicating that the N-glycan chains on both Gn and Gc are of the high-mannose type.
We also compared the electrophoretic mobilities of Gn and Gc proteins synthesized unglycosylated in vivo in the presence of tunicamycin to those in which N-glycan was removed in vitro by digestion with endo H and PNGase F (Fig. 2C). Consistent with previously published data (36), the HTNV MAbs did not react with Gn and reacted only poorly with Gc synthesized in the presence of tunicamycin (Fig. 2C, lane 2), suggesting that the attachment of N-linked glycan chains is essential for the correct folding of HTNV glycoproteins. To visualize the glycoproteins, especially Gn, that were unglycosylated in the presence of tunicamycin, we constructed a FLAG-tagged GPC in which the FLAG epitope (DYLDDDDL) was added at the N terminus of Gn. The glycoproteins produced from this construct were correctly glycosylated and cleaved (Fig. 2C, lane 4) and were transported to the Golgi complex (results not shown). By comparing the electrophoretic mobilities of the proteins, we found that Gn and Gc, either deglycosylated with endo H and PNGase F (Fig. 2C, lane 5 and 6) or synthesized in the presence of tunicamycin (lane 7), were nearly identical in size.
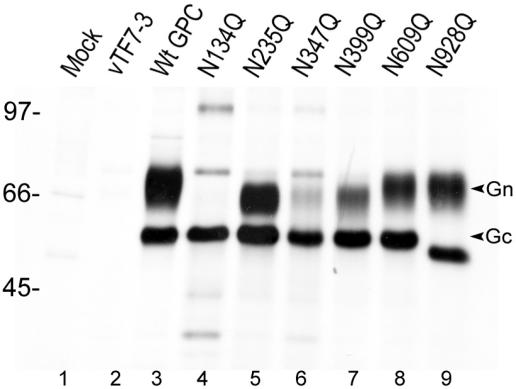
Determination of the N-linked glycosylation sites used.
There are six potential N-linked glycosylation sites on HTNV glycoproteins: five sites on Gn (at N residues 134, 235, 347, 399, and 609) and one on Gc (N928) (Fig. 1). The site at N609 was presumed not to be used since it is on the cytoplasmic side of the membrane. To determine the utilization of each individual site for the attachment of oligosaccharide chains, we generated six mutations in the HTNV M cDNA, individually replacing each asparagine (N) residue by glutamine (Q). The mutant glycoproteins were labeled with [35S]methionine, immunoprecipitated with anti-Gn and anti-Gc MAbs, and analyzed by SDS-PAGE under reducing conditions (Fig. 3). None of the mutations affected cleavage of the GPC, as both Gn and Gc forms were observed, except for mutant N134Q, where only mature Gc could be identified. Mutations at three glycosylation sites on Gn (at residues N235, N347, and N399) and the one on Gc (at residue N928) resulted in increased electrophoretic mobility corresponding to molecular weight losses of approximately 3,000 compared to wild-type Gn and Gc (Fig. 3, lanes 3, 5 to 7, and 9). No obvious band corresponding to Gn protein was observed for mutant N134Q, but multiple bands were seen (Fig. 3, lane 4), reminiscent of aggregation and degradation of a misfolded protein. The data suggest that the addition of a glycan chain at the first N glycosylation site is vital for the correct folding and stability of Gn protein. The same multiple bands were also seen to a lesser extent for the N347Q mutant, where a decreased amount of Gn was detected (lane 6). By contrast, the mobility of Gn made by the mutant N609Q (the fifth potential glycosylation site) was, as expected, indistinguishable from that of wild-type Gn (Fig. 3, lane 8). In conclusion, all four potential N glycosylation sites on the ectodomain of Gn (at residues 134, 235, 347, and 399) and the one potential site on Gc (N928) are glycosylated, whereas the site at residue N609 of Gn that faces the cytoplasm is not used.
FIG. 3.
Expression of single N glycosylation site mutants of HTNV glycoproteins. Vero E6 cells were infected with vTF7-3 followed by transfection with wild type (Wt) or mutated HTNV M cDNA. Cells were labeled with 50 μCi of [35S]methionine for 15 h and then immunoprecipitated with anti-Gn and -Gc MAbs. Immunoprecipitates were analyzed by SDS-10% PAGE under reducing conditions.
Effect of N glycosylation on HTNV glycoprotein trafficking.
One of the characteristic features of the mature HTNV glycoproteins is their ability to target to the Golgi complex (29), and Golgi targeting has been shown to be dependent on the heterodimerization of Gn and Gc (32, 38, 40). This feature was exploited to assess the role of N-linked glycosylation in protein folding and intracellular trafficking of the glycoproteins. In this study the single glycosylation site mutations were expressed by direct transfection of BSR-T7 cells with the mutant cDNAs (Fig. 4), and the glycoprotein cDNAs with double glycosylation site mutations were expressed in Vero E6 cells by using the vaccinia virus-T7 expression system (Fig. 5). Our results demonstrated that the individual sites had different effects on protein folding and Golgi targeting. Mutation at the first glycosylation site resulted in a typical ER staining pattern of the expressed glycoproteins (Fig. 4B). The glycoproteins expressed from the N347Q mutant partially colocalized with the Golgi marker (Fig. 4D), indicating the Golgi targeting ability was also compromised to some extent. However, the glycoproteins expressed from the other three mutants (N235Q, N399Q, and N928Q) were able to target to the Golgi complex and colocalized with the Golgi marker, GM130 (Fig. 4C, E, and F), suggesting that the single mutations at these sites were well tolerated with no obvious effect on protein folding and intracellular transport. Localization of these single N glycosylation mutants was also studied in Vero E6 cells, using the vaccinia virus-T7 expression system, and the results were consistent with our observations for BSR-T7 cells (data not shown).
FIG. 4.
Intracellular localization of HTNV glycoproteins with a single N glycosylation site mutation. BSR-T7 cells were transfected with glycosylation site mutant cDNA and doubly stained with a mixture of anti-Gn and anti-Gc MAbs and an antiserum against the Golgi marker GM130. Merged confocal microscopic images are also shown, with HTNV glycoproteins stained red, the Golgi complex stained green, and colocalization shown as yellow.
FIG. 5.
Intracellular localization of HTNV glycoproteins with a double N glycosylation site mutation. HeLaT4+ cells were transfected with cDNAs containing double glycosylation site mutations and were doubly stained with a mixture of anti-Gn and -Gc MAbs and an antiserum against the Golgi marker GM130. Merged confocal microscopic images are also shown, with HTNV glycoproteins stained red, the Golgi complex stained green, and colocalization shown as yellow.
Since single mutations at two sites on Gn (N235 and N399) and the one on Gc (N928) showed no obvious effect on intracellular transport, we next made four double mutations to see the additive effect of these sites. Glycoproteins expressed from mutant N235/399Q, in which the second (N235) and fourth (N399) sites on Gn were changed, showed a typical ER staining pattern (Fig. 5A), and no colocalization with GM130 was seen. Glycoproteins expressed from the double mutants (N235/928Q and N347/928Q), each with a mutation in both Gn and Gc, were shown to partially colocalize with the Golgi marker (Fig. 5B and C). In contrast, glycoproteins synthesized by mutant N399/928Q appeared to target to the Golgi complex efficiently (Fig. 5D).
Effect of N glycosylation on HTNV glycoprotein folding.
The effect of N glycosylation on protein folding of Gn and Gc was assessed indirectly by comparing the immunoreactivity of wild-type glycoproteins and the N glycosylation mutants with a panel of MAbs against HTNV Gn (8B6, 3D5, 16D2, and 6D4) and Gc (11E10, 8E10, 16E6, and HCO2) (2). The glycoproteins were expressed in Vero E6 cells, using the vaccinia virus-T7 system, and radiolabeled with [35S]methionine, and cell lysates were divided into four aliquots. Each aliquot was reacted with a pair of anti-Gn and Gc MAbs (Fig. 6). Anti-Gn MAbs 3D5 and 16D2 and anti-Gc MAb 8E10 were less efficient at precipitating the appropriate target wild-type glycoprotein than the other MAbs (Fig. 6A). Consistent with results shown in Fig. 4, mutation of the first glycosylation site (N134) on Gn led to the loss of reactivity with all four anti-Gn MAbs and reduced reactivity with two anti-Gc MAbs (11E10 and 8E10) (Fig. 6B). Mutation of the second (N235) and fourth (N399) glycosylation sites on Gn had no major effect on the reactivity patterns with the MAbs (Fig. 6C and E). The mutation at site three on Gn (N347) resulted in poor reactivity with the four anti-Gn MAbs and anti-Gc MAb 11E10 (Fig. 6D), whereas lack of N-glycans on Gc (N928Q) appeared to have little effect on the reactivity of the expressed glycoproteins with the panel of MAbs (Fig. 6F).
FIG. 6.
Immunoreactivity of wild type (wt) and mutant HTNV glycoproteins with individual anti-Gn and -Gc MAbs. Vero E6 cells were infected with vTF7-3 followed by transfection with wt HTNV M cDNA (pGEM-HTNM) or glycosylation site mutant cDNA as indicated. Cells were then labeled with 50 μCi of [35S]methionine for 15 h and immunoprecipitated with a pair of anti-Gn and anti-Gc MAbs. The resulting immunoprecipitates were analyzed by SDS-10% PAGE under reducing conditions.
We also examined the immunoreactivities of four double N glycosylation site mutants (Fig. 6G to J). The patterns of reactivity of the three mutants that contained one change on Gn and one on Gc were similar to those of the glycoproteins containing single mutations (Fig. 6H to J). However, consistent with the Golgi targeting study, mutations of both N235 and N399 seriously affected the conformation of Gn, such that the mutant glycoprotein showed much-reduced reactivity with anti-Gn MAbs 8B6, 3D5, and 6D4 and anti-Gc MAb 11E10 (Fig. 6G). The altered immunoreactivity patterns observed with this panel of anti-Gn and -Gc MAbs strongly suggest that lack of glycans at some glycosylation sites affected protein folding and thus their conformation.
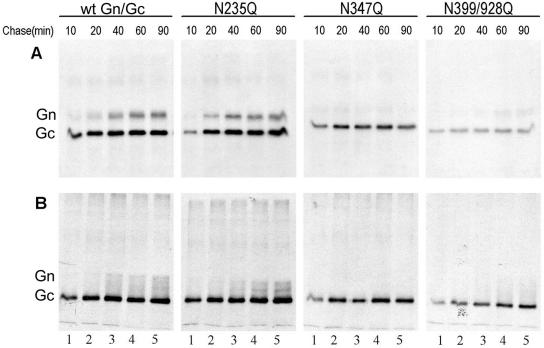
To investigate further the effect of N glycosylation on protein folding of HTNV glycoproteins, pulse-chase experiments were conducted. The wild-type glycoproteins and three N glycosylation mutants (N235Q, N347Q, and N399/928Q) were pulse-labeled with [35S]methionine for 20 min and then chased for up to 90 min in the presence of excess unlabeled methionine (Fig. 7). The labeled proteins were immunoprecipitated with anti-Gn MAb 16D2 and two anti-Gc MAbs (11E10 and HCO2). MAb 16D2 was used because it recognizes only the mature Gn that has properly folded and heterodimerized with Gc (42), whereas MAbs 11E10 and HCO2 react with the Gc protein from the wild type and all N glycosylation mutant clones (Fig. 6). As shown in Fig. 7A, both wild-type Gn and Gc were detectable after 10 min of chase, but the amount of Gc peaked at 20 min while that of Gn did not peak until 60 min (Fig. 7A). This seems analogous to the situation with Uukuniemi phlebovirus glycoproteins, where Gc also folds more rapidly than Gn (30). Under nonreducing conditions, wild-type Gn was apparent as a diffuse band that appeared to contain four forms (Fig. 7B). The pattern seen with mutant N235Q was similar to that of the wild type, but Gn appeared to form only two bands under nonreducing conditions (Fig. 7B). The Gn proteins of mutants N347Q and N399/928Q (Fig. 7) were poorly immunoprecipitated with 16D2 during the 90-min chase, and so we conclude that Gn folding and the heterodimerization with Gc were significantly compromised. Minor bands of higher-molecular-weight material were noted on gels run under both reducing (Fig. 7A) and nonreducing (Fig. 7B) conditions, though their significance requires further investigation.
FIG. 7.
Pulse-chase analysis of wild type (wt) and three N glycosylation site mutant glycoproteins. Vero E6 cells were infected with vTF7-3 followed by transfection with wt or mutant cDNA, labeled for 20 min with 80 μCi of [35S]methionine (200 μCi/ml), and then incubated with excess unlabeled methionine for the indicated times. Cell lysates were immunoprecipitated with anti-Gn MAb 16D2 and two anti-Gc MAbs, 11E10 and HCO2. The resulting immunoprecipitates were analyzed by SDS-10% PAGE under reducing conditions (A) or by SDS-8% PAGE under nonreducing conditions (B).
Association of Gn and Gc with CNX and CRT.
Since the correct folding and maturation of HTNV glycoproteins appears to rely on attachment of N-linked glycan chains, the involvement of molecular chaperones is suggested. Two ER resident chaperones, CNX and CRT, are known to promote protein folding and quality control by binding transiently to many newly synthesized glycoproteins (17, 28, 43). To examine the association of HTNV glycoproteins with these chaperones, transfected cells were pulse-labeled for 30 min and aliquots of radiolabeled cell lysates were reacted with anti-Gn/Gc MAbs (Fig. 8A) or anti-CNX (Fig. 8B) or anti-CRT (Fig. 8C) antibodies. As shown in Fig. 8, radiolabeled bands corresponding in size to HTNV Gn and Gc of the wild type and of two mutants were coimmunoprecipitated by anti-CNX and anti-CRT antibodies, indicating that the two ER chaperones do indeed associate with HTNV glycoproteins. (Other bands seen in all samples in panels B and C, including mock-transfected cells, probably represent the immunoprecipitated chaperones, CNX [65 kDa] and CRT [46 kDa] or possibly coimmunoprecipitated vaccinia virus proteins.) We noted that CNX and CRT appeared to have different affinities for wild-type and mutant glycoproteins. More N134Q Gn protein was precipitated by anti-CNX than wild-type and N347Q Gn (Fig. 8B, lanes 1, 3, and 5), whereas more wild-type and N347Q Gn were precipitated by anti-CRT antibodies (Fig. 8C, lanes 1 to 3). The recognition and binding of CNX and CRT to nascent or misfolded glycoprotein is mediated by protein-linked monoglucosylated glycans (17, 28). Therefore, coimmunoprecipitation with anti-CNX antibodies was performed on lysates of DNJ-treated cells. As seen in Fig. 8B, lanes 2, 4, and 6, very little hantavirus glycoprotein was precipitated. This confirms that the positive interaction seen in untreated samples represents chaperone-glycoprotein association.
FIG. 8.
Association of HTNV glycoproteins with CNX and CRT. Vero E6 cells were infected with vTF7-3 followed by transfection with wild-type or N glycosylation site mutant HTNV M cDNAs and then labeled for 30 min with 50 μCi of [35S]methionine in the absence (A and B, lanes 1, 3, 5, and 7; C, lanes 1 to 4) or presence (A and B, lanes 2, 4, 6, and 8) of DNJ. Cell lysates were divided into three aliquots for immunoprecipitation with anti-Gn/Gc (A), anti-CNX (B), or anti-CRT (C) antibodies. The resulting immunoprecipitates were analyzed by SDS-10% PAGE under reducing conditions.
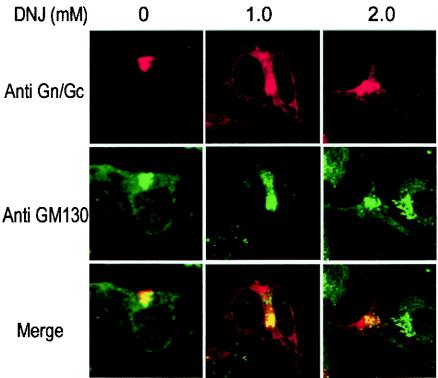
To further study the effect of N-glycan trimming on folding and intracellular trafficking of HTNV glycoproteins, we examined the localization Gn and Gc in HeLaT4+ cells treated with DNJ. Partial colocalization of the glycoproteins with the Golgi marker GM130 was observed in the presence of DNJ, but a significant amount was retained in the ER (Fig. 9). This indicates that HTNV glycoproteins with untrimmed triglucosylated N-linked glycans are still able to fold correctly, form heterodimers, and transport to the Golgi complex, though not with absolute efficiency. In conclusion, our results suggest that the ER chaperones CNX and CRT are involved in the folding process of HTNV glycoproteins by association with N-linked glycan chains and such binding is helpful but not essential for correct folding.
FIG. 9.
Intracellular localization of triglucosylated HTNV glycoproteins. HeLaT4+ cells were transfected with HTNV M cDNA and incubated in the presence of 1 or 2 mM DNJ. Cells were doubly stained with a mixture of anti-Gn and -Gc MAbs and anti-GM130 Golgi marker antibody. Merged confocal microscopic images are also shown, with HTNV glycoproteins stained red, the Golgi complex stained green, and colocalization shown as yellow.
DISCUSSION
HTNV glycoproteins Gn and Gc are both modified by N-linked glycosylation, predominantly of the high-mannose type, though there has been debate whether the N-glycans are completely endo H sensitive (1) or whether some of the oligosaccharides acquire endo H resistance (32, 36). In this study, we analyzed the N-linked oligosaccharides of Gn and Gc, using glycosidase digestion and treatment with the ER α-glucosidase inhibitor DNJ and the N glycosylation inhibitor tunicamycin. Our results indicate that the N-glycans of HTNV glycoproteins are entirely endo H sensitive and of the high-mannose type. Since the conversion of high-mannose type to the complex form occurs in the medial and trans-Golgi compartments (19), our data suggest that the HTNV glycoproteins are just able to transport to the cis-Golgi compartment, consistent with our previous observation that HTNV glycoproteins colocalize with GM130, a cis-Golgi matrix protein (38).
We noted that there was an electrophoretic migration difference between the Gn proteins deglycosylated by endo H and PNGase F. In the presence of DNJ, with the trimming of the N-glycan inhibited, the mobility of Gn protein after endo H and PNGase F treatment was unchanged. DNJ inhibits ER glucosidases I and II and thus prevents trimming of the outmost glucose residues from the 14-saccharide core N-linked glycan GlcNAC2Man9Glc3 (10, 41). Consequently, DNJ would render the glycoproteins uniformly triglucosylated and sensitive to endo H. Gn digested by endo H had a molecular weight approximately 1,000 greater than that digested by PNGase F, which is consistent with the molecular weight of 885 for four N-acetylglucosamine moieties retained on the asparagine residues of the N glycosylation sites after endo H digestion. These results suggest, therefore, that the slightly higher molecular mass of Gn protein was not due to the acquisition of endo H resistance but rather the consequence of the remaining four N-acetylglucosamine residues. The sensitivity of the glycoproteins to endo H was also supported by the results that no radioactive signal was detectable when the [3H]mannose-labeled glycoproteins were subjected to endo H digestion and Gn that was deglycosylated by endo H and PNGase F digestion was the same size as that synthesized in the presence of the N glycosylation inhibitor tunicamycin.
HTNV glycoproteins possess six potential N-linked glycosylation sites, five on Gn and one on Gc (37), of which four sites (at residues N134, N347, N399, and N928) are conserved among all hantaviruses. In this study, we determined that all sites on the ectodomains of Gn (N134, N235, N347, and N399) and Gc (N928) were glycosylated and that the fifth site on Gn (N609) that faces the cytoplasm was not utilized.
It is well accepted that glycosylation plays a pivotal role in the function of a glycoprotein (16, 27), and in this study we demonstrated that N glycosylation is crucial for the proper folding, intracellular transport, and maintenance of the epitope conformation of HTNV glycoproteins. It is evident that N-glycan at the first site is most crucial in directing correct folding of Gn, as the mutant N134Q was retained in the ER and could no longer be recognized by anti-Gn MAbs. It has been reported that glycoproteins are most sensitive to removal of glycosylation sites near their N termini, as these sites first engage the protein folding machinery in the ER to initiate the correct folding process (6, 15, 16). Glycosylation at the third site (N347) in Gn also markedly affected its folding, as the Golgi targeting of N347 was substantially impaired and it was no longer recognized by two neutralizing MAbs, 3D5 and 16D2. Single mutations at sites N235 and N399 on Gn and N928 on Gc were better tolerated, indicating that lack of glycans at one of these sites is not sufficient to affect folding and targeting or that it can be compensated by the neighboring oligosaccharide chains. However, mutation of two N glycosylation sites on Gn (at residues N235 and N399) together resulted in profound effects, reflected by the loss of Golgi targeting and abolition of reactivity with three MAbs (Fig. 6G), indicating that the oligosaccharide chains on Gn can have additive effects. Global effects of N glycosylation on protein folding have also been observed for other virus proteins, such as vesicular stomatitis virus G protein (22), influenza virus hemagglutinin (11), and avian sarcoma/leucosis virus glycoprotein (5).
For other viruses in the family, heterodimerization of Gn and Gc proteins seems to be necessary for efficient Golgi transport and virus morphogenesis (31), and available evidence indicates the same pertains for hantavirus glycoproteins (1, 38, 40). Some MAbs (such as 2D5, 3D5, 16D2, and 3G1) recognize Gn only when heterodimeric (26, 42), suggesting that conformational differences exist between Gn expressed alone and that expressed in association with Gc. It is likely that the mature folding and conformation of HTNV glycoproteins depends on their mutual interaction. Our data suggest that N glycosylation also affects heterodimerization; Gn expressed from three N glycosylation mutant clones (N134Q, N347Q, and N235/399Q) did not react with 3D5 and 16D2. Using reactivity with MAb 16D2 as an indicator for heterodimerization of Gn and Gc, our pulse-chase analysis revealed that the two proteins interacted within 10 min and interaction peaked within 60 min. Certain N-glycan chains had a significant detrimental effect on heterodimerization, but whether this is a consequence of protein misfolding due to a lack of glycan chains or a direct influence on heterodimer formation requires further study.
It is also possible that a conformational change in one protein would influence the other. Mutation of N glycosylation sites on Gn at residues N134, N347, and N399 reduced the reactivity of the mutant glycoproteins with anti-Gc MAbs, and vice versa, lack of N-glycans on Gc also reduced the immunoreactivity of Gn protein (Fig. 6), though to a lesser extent. Gc appears more tolerant to mutation of its N glycosylation site, whereas Gn is more heavily glycosylated than Gc and so is more dependent on N-glycans for proper folding and assembly.
We also showed that two ER resident chaperones, CNX and CRT, associate with HTNV glycoproteins. When binding of the chaperones was blocked in the presence of DNJ, Golgi targeting of the glycoproteins was considerably compromised. This implies that association with CNX and CRT is needed for efficient folding and heterodimerization of the glycoproteins. However, the association of HTNV glycoproteins with these lectins is not absolutely required for their folding. Many glycoproteins, when prevented from binding to CNX and CRT, manage to fold and assemble correctly (12, 13, 23, 24), indicative of the redundancy of folding factors in the ER.
In summary, our studies determined the utilization of five potential N glycosylation sites and confirmed that N-glycans of HTNV glycoproteins are completely of the high-mannose type. Our data demonstrated that certain N-linked glycosylation is crucial for correct folding and intracellular trafficking of the proteins, though the importance of individual N-glycan chains varies. Two ER resident chaperones, CNX and CRT, were found associated with newly synthesized HTNV glycoproteins and play a role in their folding.
Acknowledgments
We express our gratitude to Connie Schmaljohn for providing the HTNV MAbs, to Martin Lowe for anti-GM130, and to Klaus Conzelmann for the BSR-T7 cells.
This work was supported by grants 046745 and 048378 from the Wellcome Trust and grant G9604479 from the Medical Research Council.
REFERENCES
- 1.Antic, D., K. E. Wright, and C. Y. Kang. 1992. Maturation of Hantaan virus glycoproteins G1 and G2. Virology 189:324-328. [DOI] [PubMed] [Google Scholar]
- 2.Arikawa, J., A. L. Schmaljohn, J. M. Dalrymple, and C. S. Schmaljohn. 1989. Characterization of Hantaan virus envelope glycoprotein antigenic determinants defined by monoclonal antibodies. J. Gen. Virol. 70:615-624. [DOI] [PubMed] [Google Scholar]
- 3.Braakman, I., and E. van Anken. 2000. Folding of viral envelope glycoproteins in the endoplasmic reticulum. Traffic 1:533-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchholz, U. J., S. Finke, and K.-K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BSRV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delos, S. E., M. J. Burdick, and J. M. White. 2002. A single glycosylation site within the receptor-binding domain of the avian sarcoma/leukosis virus glycoprotein is critical for receptor binding. Virology 294:354-363. [DOI] [PubMed] [Google Scholar]
- 6.Doms, R. W., A. Helenius, and J. White. 1985. Membrane fusion activity of the influenza virus hemagglutinin. The low pH-induced conformational change. J. Biol. Chem. 260:2973-2981. [PubMed] [Google Scholar]
- 7.Doms, R. W., R. A. Lamb, J. K. Rose, and A. Helenius. 1993. Folding and assembly of viral membrane proteins. Virology 193:545-562. [DOI] [PubMed] [Google Scholar]
- 8.Elbein, A. D. 1991. Glycosidase inhibitors: inhibitors of N-linked oligosaccharide processing. FASEB J. 5:3055-3063. [DOI] [PubMed] [Google Scholar]
- 9.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuhrmann, U., E. Bause, and H. Ploegh. 1985. Inhibitors of oligosaccharide processing. Biochim. Biophys. Acta 825:95-110. [DOI] [PubMed] [Google Scholar]
- 11.Gallagher, P. J., J. M. Henneberry, J. F. Sambrook, and M. J. Gething. 1992. Glycosylation requirements for intracellular transport and function of the hemagglutinin of influenza virus. J. Virol. 66:7136-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudin, Y. 1997. Folding of rabies virus glycoprotein: epitope acquisition and interaction with endoplasmic reticulum chaperones. J. Virol. 71:3742-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond, C., I. Braakman, and A. Helenius. 1994. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. USA 91:913-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hebert, D. N., B. Foellmer, and A. Helenius. 1995. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell 81:425-433. [DOI] [PubMed] [Google Scholar]
- 15.Hebert, D. N., J.-X. Zhang, W. Chen, B. Foellmer, and A. Helenius. 1997. The number and location of glycans on influenza hemagglutinin determine folding and association with xalnexin and calreticulin. J. Cell Biol. 139:613-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helenius, A., and M. Aebi. 2001. Intracellular functions of N-linked glycans. Science 291:2364-2369. [DOI] [PubMed] [Google Scholar]
- 17.Helenius, A., E. S. Trombetta, D. N. Hebert, and J. F. Simons. 1997. Calnexin, calreticulin and the folding of glycoproteins. Trends Cell Biol. 7:193-200. [DOI] [PubMed] [Google Scholar]
- 18.Imperiali, B., and S. E. O'Connor. 1999. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr. Opin. Chem. Biol. 3:643-649. [DOI] [PubMed] [Google Scholar]
- 19.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631-664. [DOI] [PubMed] [Google Scholar]
- 20.Lappin, D. F., G. W. Nakitare, J. W. Palfreyman, and R. M. Elliott. 1994. Localization of Bunyamwera bunyavirus G1 glycoprotein to the Golgi requires association with G2 but not with NSm. J. Gen. Virol. 75:3441-3451. [DOI] [PubMed] [Google Scholar]
- 21.Lober, C., B. Anheier, S. Lindow, H. D. Klenk, and H. Feldmann. 2001. The Hantaan virus glycoprotein precursor is cleaved at the conserved pentapeptide WAASA. Virology 289:224-229. [DOI] [PubMed] [Google Scholar]
- 22.Machamer, C. E., and J. K. Rose. 1988. Vesicular stomatitis virus G proteins with altered glycosylation sites display temperature-sensitive intracellular transport and are subject to aberrant intermolecular disulfide bonding. J. Biol. Chem. 263:5955-5960. [PubMed] [Google Scholar]
- 23.McGinnes, L. W., and T. G. Morrison. 1998. Role of carbohydrate processing and calnexin binding in the folding and activity of the HN protein of Newcastle disease virus. Virus Res. 53:175-185. [DOI] [PubMed] [Google Scholar]
- 24.Molinari, M., and A. Helenius. 1999. Glycoproteins form mixed disulphides with oxidoreductases during folding in living cells. Nature 402:90-93. [DOI] [PubMed] [Google Scholar]
- 25.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura, N., C. Rabouille, R. Watson, T. Nilsson, N. Hui, P. Slusarewicz, T. E. Kreis, and G. Warren. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131:1715-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parodi, A. J. 2000. Protein glucosylation and its role in protein folding. Annu. Rev. Biochem. 69:69-93. [DOI] [PubMed] [Google Scholar]
- 28.Parodi, A. J. 2000. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem. J. 348:1-13. [PMC free article] [PubMed] [Google Scholar]
- 29.Pensiero, M. N., G. B. Jennings, C. S. Schmaljohn, and J. Hay. 1988. Expression of the Hantaan virus M genome segment by using a vaccinia virus recombinant. J. Virol. 62:696-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Persson, R., and R. F. Pettersson. 1991. Formation and intracellular transport of a heterodimeric viral spike protein complex. J. Cell Biol. 112:257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettersson, R. F., and L. Melin. 1996. Snythesis, assembly, and intracellular transport of Bunyaviridae membrane proteins, p. 159-188. In R. M. Elliott (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 32.Ruusala, A., R. Persson, C. S. Schmaljohn, and R. F. Pettersson. 1992. Coexpression of the membrane glycoproteins G1 and G2 of Hantaan virus is required for targeting to the Golgi complex. Virology 186:53-64. [DOI] [PubMed] [Google Scholar]
- 33.Schmaljohn, C. 1996. Molecular biology of hantavirus, p. 63-90. In R. M. Elliott (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 34.Schmaljohn, C. S., S. E. Hasty, J. M. Dalrymple, J. W. LeDuc, H. W. Lee, C. H. von Bonsdorff, M. Brummer-Korvenkontio, A. Vaheri, T. F. Tsai, H. L. Regnery, et al. 1985. Antigenic and genetic properties of viruses linked to hemorrhagic fever with renal syndrome. Science 227:1041-1044. [DOI] [PubMed] [Google Scholar]
- 35.Schmaljohn, C. S., S. E. Hasty, S. A. Harrison, and J. M. Dalrymple. 1983. Characterization of Hantaan virions, the prototype virus of hemorrhagic fever with renal syndrome. J. Infect. Dis. 148:1005-1012. [DOI] [PubMed] [Google Scholar]
- 36.Schmaljohn, C. S., S. E. Hasty, L. Rasmussen, and J. M. Dalrymple. 1986. Hantaan virus replication: effects of monensin, tunicamycin and endoglycosidases on the structural glycoproteins. J. Gen. Virol. 67:707-717. [DOI] [PubMed] [Google Scholar]
- 37.Schmaljohn, C. S., A. L. Schmaljohn, and J. M. Dalrymple. 1987. Hantaan virus M RNA: coding strategy, nucleotide sequence, and gene order. Virology 157:31-39. [DOI] [PubMed] [Google Scholar]
- 38.Shi, X., and R. M. Elliott. 2002. Golgi localization of Hantaan virus glycoproteins requires coexpression of G1 and G2. Virology 300:31-38. [DOI] [PubMed] [Google Scholar]
- 39.Spiropoulou, C. F. 2001. Hantavirus maturation. Curr. Top. Microbiol. Immunol. 256:33-46. [DOI] [PubMed] [Google Scholar]
- 40.Spiropoulou, C. F., C. S. Goldsmith, T. R. Shoemaker, C. J. Peters, and R. W. Compans. 2003. Sin Nombre virus glycoprotein trafficking. Virology 308:48-63. [DOI] [PubMed] [Google Scholar]
- 41.Suh, K., C. Gabel, and J. Bergmann. 1992. Identification of a novel mechanism for the removal of glucose residues from high mannose-type oligosaccharides. J. Biol. Chem. 267:21671-21677. [PubMed] [Google Scholar]
- 42.Wang, M., D. G. Pennock, K. W. Spik, and C. S. Schmaljohn. 1993. Epitope mapping studies with neutralizing and non-neutralizing monoclonal antibodies to the G1 and G2 envelope glycoproteins of Hantaan virus. Virology 197:757-766. [DOI] [PubMed] [Google Scholar]
- 43.Williams, D. B. 1995. Calnexin: a molecular chaperone with a taste for carbohydrate. Biochem. Cell Biol. 73:123-132. [DOI] [PubMed] [Google Scholar]
- 44.Wyss, D. F., and G. Wagner. 1996. The structural role of sugars in glycoproteins. Curr. Opin. Biotechnol. 7:409-416. [DOI] [PubMed] [Google Scholar]