Abstract
Immunoglobulins (Igs) are known to be synthesized and secreted only by B lymphocytes. Class switch recombination (CSR) is a key event that enables B cells to express Igs, and one of the crucial steps for CSR initiation is the germline transcription of Ig genes. Surprisingly, recent studies have demonstrated that the Ig genes are also expressed in some epithelial cancer cells; however, the mechanisms underlying how cancer cells initiate CSR and express Igs are still unknown. In this study, we confirmed that the Ig Iα1 promoter in cancer cell lines was activated by the Ets-1 transcription factor, and the activity of the Ig Iα1 promoter and Ig Iα1–Cα1 germline transcription were attenuated after knockdown of Ets-1 by specific small interfering RNAs (siRNA). Furthermore, the expression of Ets-1 and Igα heavy chain in cancer cells was dose dependently upregulated by TGF-β1. These results indicate that activation of the Ig Iα1 promoter by the transcription factor Ets-1 is a critical pathway and provides a novel mechanism for Ig expression in non-B cell cancers.
Keywords: epithelial cancer cells, Ets-1, Ig Iα1 promoter
Introduction
Immunoglobulins (Igs) are generally believed to be produced only by B lymphocytes. Each Ig molecule consists of two identical heavy chains and two identical light chains. Five classes of heavy chain include α, γ, δ, ε and μ, representing five Ig isotypes IgA, IgG, IgD, IgE and IgM, respectively. For the light chain, only two classes, κ and λ, have been discovered. When stimulated by foreign antigens and affected by various regulators, the Ig genes undergo V(D)J recombination and class switch recombination (CSR), and only then can Igs be expressed in B lymphocytes.
Interestingly, recent studies have confirmed that Igs are abnormally synthesized by non-lymphoid cells,1,2 such as epithelial cancer cells and normal cells. In 1991, Cao et al.3 cloned a transforming gene, referred to as Tx (GenBank accession number: AF279037), from the gDNA library of the nasopharyngeal carcinoma cell line CNE2. The Tx gene was then determined to be an aberrant human Igκ gene that lacked variable regions.4 Later, in 1998, using highly sensitive RT-nested PCR, Kimoto5 demonstrated the expression of Ig transcripts in five cancer cell lines, which indicated that Igs were expressed in these cancer cells. Additionally, other research groups have subsequently reported Ig expression in non-lymphoid cells,6,7,8,9,10,11,12,13 especially in epithelial cancer cells. Although the expression of Ig molecules in cancer cells has been confirmed, evidence regarding the biological function of cancerous Ig has not been well documented. The blockade of cancerous IgG can increase programmed cell death and inhibit the growth of cancer cells in vitro.6 Our previous work demonstrated that the Igα heavy chain could increase the percentage of cancer cells in S phase.14 We also showed that Igs produced by cancer cells could specifically reduce antibody-dependent cell-mediated cytotoxicity (ADCC).15 These findings support a positive role for cancer-generated Igs in cancer cell proliferation, and reveal a distinct mechanism for the immune evasion of cancer cells.
Integrated Ig molecules can be expressed and exercise their functions only after V(D)J recombination16 and class switch recombination.17 V(D)J recombination assembles the mature V region of the Ig molecule, and CSR connects the distinct C region to the V region, thus forming an integrated Ig molecule. The mechanism of V(D)J recombination in cancer cells has been reported by our group and others.18,19 The recombination activating gene (RAG), encoding the recombinases RAG1 and RAG2, which are essential enzymes for initiating V(D)J recombination, is also expressed in cancer cells.18 The abnormal expression of RAG proteins in cancer was shown to be regulated by the E2A, FOXO1 and FOXP1 transcription factors and was similar to that observed in B lymphocytes.20 However, the mechanism of CSR in cancer cells has not been elucidated. The process of CSR requires germline (GL) transcription of unrearranged C region genes and is initiated by activation-induced cytidine deaminase (AID), a B cell-specific factor. When GL transcription begins, a short region of ssDNA, which can be targeted by AID at the transcription bubble, will be generated.21,22 Additionally, GL transcription makes the DNA structure more accessible to AID by altering histone modifications in the transcribed region.23 When AID targets the ssDNA of the S region (upstream from the C region), U:G mismatches will be produced in the Ig genes, forming double-strand breaks (DSBs).24,25 Then, two DSBs in different S regions will recombine by performing non-homologous end-joining (NHEJ) to complete CSR.26 Overall, the GL transcript Ig Iα-Cα is a key regulator of Igα heavy chain class switch recombination, which is crucial for the expression of IgA.
Two subclasses of IgA are found in humans and include the heavy chains of IgA1 and IgA2 that are encoded by the two distinct Cα1 and Cα2 genes, respectively. Our previous studies have confirmed the expression of the Ig Iα1–Cα1 transcript in several cancer cell lines.18 Studies showed that TGF-β1 can regulate Ig Iα1–Cα1 GL transcription through the Smad signaling pathway in B cells.27 Moreover, the transcriptional process of the Ig Iα1–Cα1 transcript is mainly regulated by cis-acting elements and their corresponding trans-acting factors. In this study, we focus on the cis-acting elements of the Ig Iα1 promoter that is located upstream of the Cα1 exon. We first determined whether the Ig Iα1 promoter is activated in cancers, and the results showed that the Ig Iα1 promoter was highly activated in nasopharyngeal carcinoma cells. Through bioinformatic analysis, we found several binding sites for various transcription factors, including NF-κB and PU.1, in the Ig Iα1 promoter. Further studies confirmed that the ETS family member, Ets-1, could bind to the PU.1 motif and then transactivate the Ig Iα1 promoter. These results indicate that Ets-1 activates the expression of the Ig Iα1–Cα1 GL transcript, which is critical for class switch recombination.
Materials and Methods
Cell lines and cell culture
Two epithelial cancer cell lines were cultured to study Igα expression. CNE1 cells are a nasopharyngeal carcinoma cell line, and HeLa (ATCC number: CCL-2) cells are a cervical cancer cell line. Our previous studies have demonstrated that CNE1 and HeLa cells can produce and secrete Igs spontaneously.18,28 The Burkitt's lymphoma cell line Raji (ATCC number: CCL-86) was used as a positive control for expression of the Igα heavy chain. All the cell lines were cultured in complete growth medium according to ATCC protocols, and logarithmically growing cells were used in all experiments.
Plasmid constructs
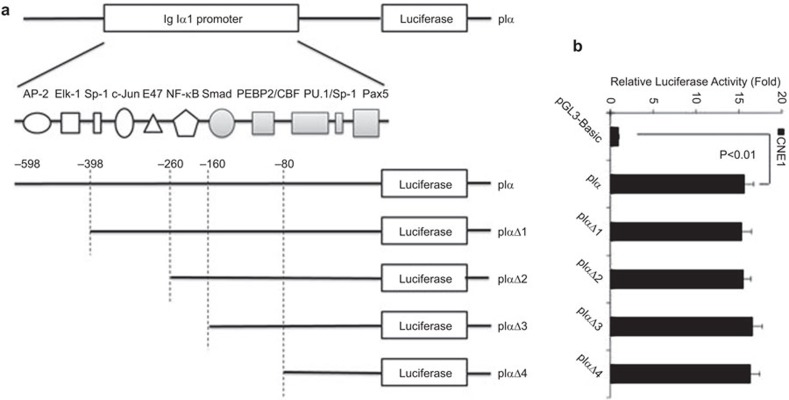
A 674-bp fragment containing the human Ig Iα1 promoter upstream from the TTS site was acquired by PCR amplification of genomic DNA from HeLa cells with the following primers: sense, 5′-cgagctcgtggtgacccacagtaggagt-3′ antisense, 5′-cccaagcttgggtgatggccgtctgtccttag-3′. The fragment was inserted upstream of the luciferase gene of the pGL3-Basic vector (Promega, Madison, WI, USA) and the plasmid was named pIα. The PCR products were confirmed by restriction enzyme digestion and DNA sequencing. To verify the key cis-acting element for Ig Iα1 promoter activation, we first used continuous deletion variation to construct four reporter plasmids: pIαΔ1, pIαΔ2, pIαΔ3 and pIαΔ4 (Figure 1a), and we checked the mutational sites and the integrity of the sequence by DNA sequencing. The internal control plasmid pRL-TK was purchased from Promega.
Figure 1.
The Ig Iα1 promoter is activated in nasopharyngeal carcinoma cells. (a) The intact or truncated sequence of the Ig Iα1 promoter was inserted into the pGL3-Basic vector to construct the luciferase reporter plasmids. (b) Ig Iα1 promoter activity in human nasopharyngeal carcinoma cells (CNE1) transfected with luciferase reporter vectors containing the intact or truncated Ig Iα1 promoter. Luciferase activity was measured after 24 h of culture, and at least three independent transfection experiments were performed in triplicate for each experimental construct. Ig, immunoglobulin.
RNA interference
A specific, anti-Ets-1 small interfering RNA (Ets-1 siRNA) and scrambled oligonucleotide (control siRNA) were both purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). CNE1 or HeLa cells were cultured in 24-well plates and transfected with Ets-1 siRNA (40 pmol/well) or control siRNA (40 pmol/well) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Cells were also grown in six-well plates and the molar masses of the siRNAs were matched to 100 pmol for each well. Proteins from cells transfected with Ets-1 siRNA or control siRNA were harvested for immunoblotting to confirm Ets-1 knockdown.
Dual-luciferase reporter assays
The dual-luciferase reporter assays were performed as previously described.29 Cells were transfected with 1 µg/well of pIα firefly luciferase reporter plasmids or the control pGL3-Basic vector (Promega) using Lipofectamine 2000, and the renilla luciferase vector pRL-TK (10 ng/well) was cotransfected to correct for variations in transfection efficiency. Cells were harvested at 24 h after transfection and lysates were analyzed for firefly and renilla luciferase activity using the Dual-Luciferase Reporter Assay Kit (Promega) with a GloMax 20/20 luminometer (Promega). The final results are represented as the fold luciferase induction compared to that of the pGL3-Basic vector. The data from dual-luciferase reporter assays were calculated using statistical software (SPSS 16.0), and a value of P<0.05 or P<0.01 was considered to be statistically significant.
Reverse transcription and polymerase chain reaction
CNE1 and HeLa cells were treated or not treated with Ets-1 siRNA or control siRNA for 72 h. Total RNA was isolated as described,29 and cDNAs were synthesized using SuperScript II (Invitrogen). The primers for human Ig Iα1–Cα130 were 5′-cagcagccctcttggcaggcagccag-3′ (sense) and 5′-gggtggcggttagcggggtcttgg-3′ (antisense) and yielded a 1194-bp product. The primers for Ets-1 were 5′-acccagcctatccagaatcc-3′ (sense) and 5′-tctgcaaggtgtctgtctgg-3′ (antisense) and yielded a 225-bp product. The products of PCR were detected in 2% agarose gels and stained with SYBR Safe (Invitrogen).
Western blot analysis
Cells in logarithmic-growth phase were washed three times with ice-cold phosphate-buffered saline (PBS), disrupted on ice with IP lysis buffer (Pierce, Rockford, IL, USA) containing a protease inhibitor cocktail tablet (Roche, Basel, Switzerland) for 30 min. Then, after harvesting by scraping, the lysate was centrifuged at 15 000g for 15 min. Western blot analysis was performed as described.29 The following antibodies were used: rabbit anti-human IgA (A0262; DAKO, Glostrup, Denmark), rabbit anti-Ets-1 (sc-350; Santa Cruz Biotechnology) and mouse anti-β-actin (sc-8432; Santa Cruz Biotechnology).
Electrophoretic mobility shift assays (EMSA)
EMSA analysis and nuclear extracts were collected as described previously.29 The reaction mixtures (20 µl) containing 8 µg of nuclear extract were incubated for 20 min at room temperature with 20 fmol of the biotin-labeled double-stranded oligonucleotide probes in reaction buffer (Pierce). For competition experiments, a 400- to 800-fold excess of unlabeled, wildtype or mutant probe was included in the reaction system. For supershift-EMSA assays, the reaction mixtures were pre-incubated at room temperature for 1 h with 2 µg of anti-Ets-1 rabbit IgG (sc-350X; Santa Cruz Biotechnology) or normal rabbit IgG. The wild-type Ets probe sequence was 5′-GCTGGGGCAGGAAGTGGGCGAGT-3′ and 5′-ACTCGCCCACTTCCTGCCCCAGC-3′ and was derived from the binding site of Ets-1 in the human Ig Iα1 promoter. The mutated Ets probe sequence was 5′-GCTGGGGCGAGAAGTGGGCGAGT -3′ and 5′-ACTCGCCCACTTCTCGCCCCAGC-3′.
Chromatin immunoprecipitation assay
Cells were cultured overnight and fixed in 1% formaldehyde, after which the reaction was stopped with glycine. Nuclear protein–DNA complexes were harvested from cells disrupted by SDS lysis buffer with protease inhibitors and processed by sonication to produce DNA fragments with an average length of 100–500 bp. After pre-clearing with proteinG agarose/salmon sperm DNA for 1 h at 4 °C, 20% of each sample was saved as ‘input DNA' for later PCR analysis. Immunoprecipitation was performed with 2 µg of anti-Ets-1 rabbit IgG (sc-350X; Santa Cruz Biotechnology) or normal rabbit IgG. After reversal of crosslinking, PCR amplification assays were used to detect the target DNA using the primers 5′-cagaccacaggccagacat-3′ and 5′-ccgtctgtccttagcagagc-3′ (Ig Iα1 promoter including the PU-binding region; 187 bp).
Results
The Ig Iα1 promoter is activated in nasopharyngeal carcinoma cells
To investigate whether the Ig Iα1 promoter could be activated in cancer cells, we first cloned the Ig Iα1 promoter and inserted it into the pGL3-Basic vector to construct a reporter plasmid, which was referred to as pIα (Figure 1a). We chose a nasopharyngeal carcinoma cell line (CNE1) as our model. When transfected into CNE1 cells, the pIα plasmid showed a high luciferase activity compared to the pGL3-Basic vector (P<0.01) (Figure 1b). This finding indicated that the Ig Iα1 promoter was highly activated in nasopharyngeal carcinoma cells. To further confirm the presence of a cis-acting element in the Ig Iα1 promoter, we constructed continuous deletion mutations to create four reporter plasmids, referred to as pIαΔ1, pIαΔ2, pIαΔ3 or pIαΔ4 (Figure 1a). The binding sites in the Ig Iα1 promoter for AP-2 and Elk-1 were deleted in the pIαΔ1 plasmid, and the binding sites for Sp-1, c-Jun and E47 were deleted in the pIαΔ2 plasmid. The promoter sequence in pIαΔ3 included the binding sites for Smad, PEBP2/CBF, PU.1/Sp-1 and Pax5; and pIαΔ4 only included the binding sites for PU.1/Sp-1 and Pax5. Dual luciferase reporter assays revealed high luciferase activities for all constructs, even though the Ig Iα1 promoter sequence was truncated (Figure 1b). This result indicated that a crucial element for Ig Iα1 promoter activation was located 80 bp upstream of the TSS.
The Ets-1 binding site plays a key role in the activation of the Ig Iα1 promoter
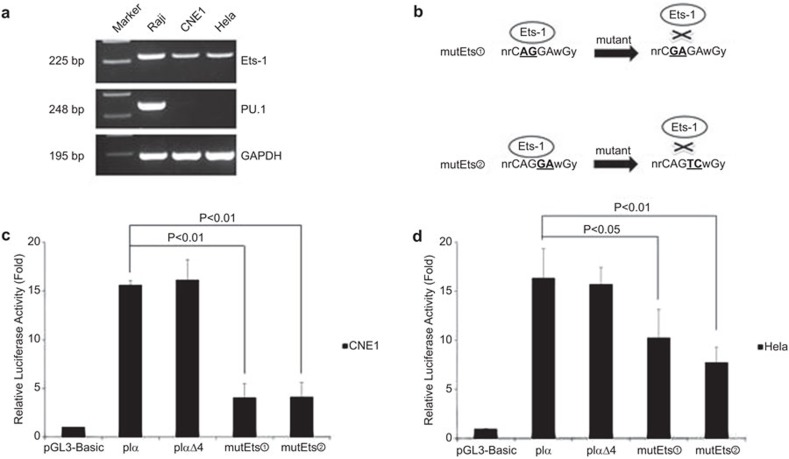
Using two bioinformatic software programs, ‘Genomatix' and ‘TFSEARCH', we discovered a PU.1 binding site in the Ig Iα1 promoter that was included in the pIαΔ4 plasmid. PU.1 is an ETS family transcription factor that plays an important role in hematopoietic cell differentiation, proliferation and apoptosis.31 Because PU.1 is not expressed in epithelial cancer cells, we examined another member of the ETS family, Ets-1, which is expressed in Raji and epithelial cancer cells (Figure 2a). We hypothesized that Ets-1 could bind to the PU.1 motif because they share a highly homologous binding site. To confirm this idea, we constructed two luciferase reporter vectors containing the Ig Iα1 promoter with a mutation in the PU.1/Ets-1 binding site, and the mutation did not yield new binding sites for other transcription factors. The first mutant vector contained a mutation that changed CAGGA to CGAGA, and the second mutant vector contained a mutation that changed CAGGA to CAGTC (Figure 2b). Because our previous data showed that IgA was expressed at high levels in CNE1 and HeLa cells, we conducted dual luciferase reporter assays in the two cell lines. Significant downregulation of Ig Iα1 promoter activity was observed in CNE1 and HeLa cells after the mutation in the PU.1/Ets-1 binding site (P<0.01 or P<0.05; Figure 2c and d). These results indicated that the Ets-1 transcription factor might play an important role in Ig Iα1 promoter activation in epithelial cancer cells.
Figure 2.
The activity of the Ig Iα1 promoter is decreased after mutation of the Ets binding site. (a) The expression of the Ets family members PU.1 and Ets-1 in lymphoma and epithelial cancer cells was assessed by RT-PCR. PU.1 was the only factor that was expressed in the lymphoma cell line. (b) The strategy for mutation of the Ets binding site. Two mutant plasmids were constructed by mutating two base pairs at different sites of the core binding site. (c, d) Dual luciferase reporter assays show significant downregulation of Ig Iα1 promoter activity in CNE1 and HeLa cells after mutation of the Ets binding site. At least three independent transfection experiments were performed in triplicate for each experimental construct. Ig, immunoglobulin.
The Ets-1 transcription factor binds to the Ig Iα1 promoter in vitro
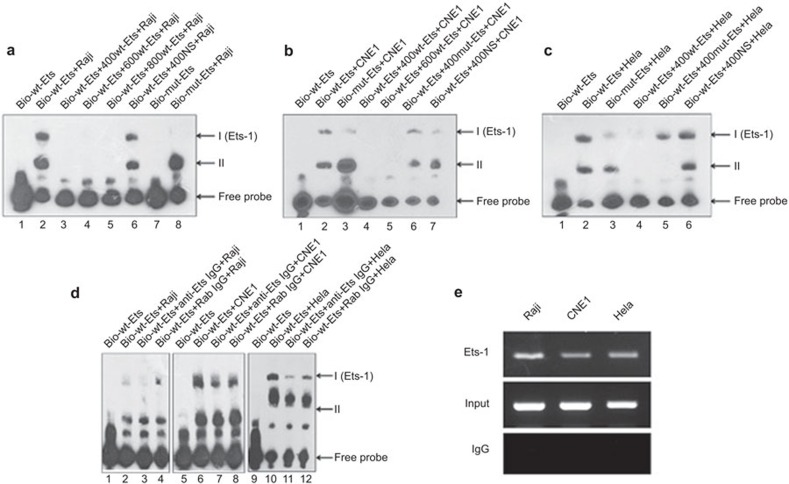
Transcription factors exhibit their biologic functions by binding to a cis-acting element and trans-activating target genes. We therefore performed EMSA assays to determine whether Ets-1 could bind to the Ig Iα1 promoter. Two different shifted bands could be detected when EMSA assays were performed with nuclear extracts from Raji cells (i.e., positive control cells; Figure 3a, lane 2). However, the shifted binding complexes could be inhibited completely by a 400- to 800-fold excess of unlabeled, wild-type Ets probe (Figure 3a, lanes 3–5). The specificity of the formation of complexes was supported by the results of the nonspecific competition assay (Figure 3a, lane 6). Of the two complexes, complex I was not detected, but complex II remained when the EMSA assay was performed using a biotin-labeled, mutant-type Ets oligonucleotide (Figure 3a, lane 8), indicating that complex I was specifically formed by the Ets-related protein. Using nuclear extracts from CNE1 or HeLa cells, the results demonstrated that an Ets family member bound to the Ig Iα1 promoter in vitro (Figure 3b and c). To further confirm that the Ets family member was Ets-1, we pre-incubated the nuclear lysate with a specific Ets-1 antibody, which led to decreased formation of only complex I (Figure 3d, lanes 3, 7 and 11). However, the binding ability was not affected by the addition of a normal rabbit IgG (Figure 3d, lanes 4, 8 and 12). These results suggested that the transcription factor Ets-1 is capable of binding to the Ets site that is found in the Ig Iα1 promoter.
Figure 3.
The Ets-1 transcription factor binds to the Ig Iα1 promoter in vitro and in cells. (a–c) EMSA assays indicate that two different DNA/protein complexes could be observed when incubating the Ets binding site probe with a nuclear extract from Raji, CNE1 or HeLa cells. (d) SuperShift-EMSA assays demonstrate possible Ets-1 transcription factor binding to the Ets site in the Ig Iα1 promoter. (e) ChIP assays confirm that the Ets-1 transcription factor exerts its regulatory function through direct binding to the human Ig Iα1 promoter in cells. ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility shift assay; Ig, immunoglobulin.
The Ets-1 transcription factor can bind to the Ig Iα1 promoter in cells
To verify the binding ability of Ets-1 to the Ig Iα1 promoter in human cancer cells, chromatin from the positive-control Raji, CNE1 and HeLa epithelial cancer cells were used to perform a chromatin immunoprecipitation assay. The short chromatin was subjected to an immunoprecipitation reaction with a specific Ets-1 antibody or normal rabbit IgG, and the precipitated DNA/protein/antibody complex was reverse-crosslinked and amplified by PCR using primers specific for the Ets binding site of the Ig Iα1 promoter. As shown in Figure 3e, immunoprecipitation with an antibody specific for Ets-1 pulled down the Ig Iα1 promoter region, but normal rabbit IgG had no effect on any of the samples. Therefore, the above data indicate that Ets-1 exhibits its regulatory function by directly binding to the human Ig Iα1 promoter in cells.
RNA interference against Ets-1 downregulates the activity of the Ig Iα1 promoter
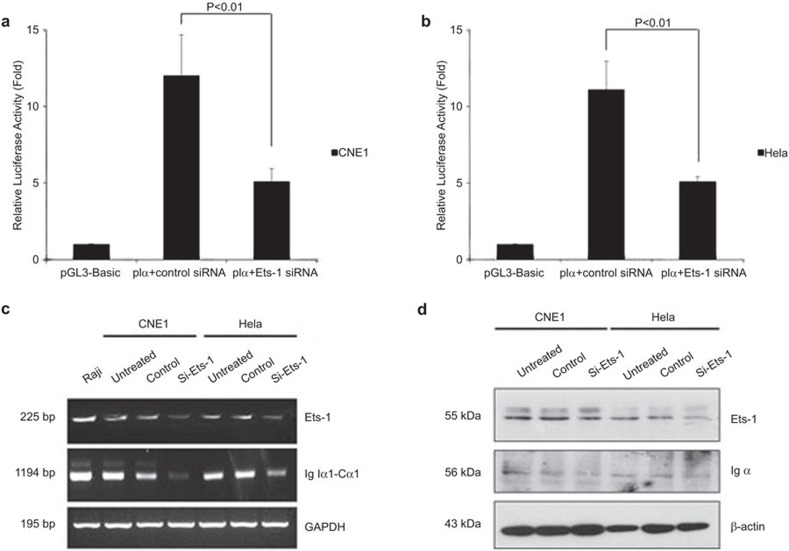
To further study the regulatory function of Ets-1 in the activation of the Ig Iα1 promoter, specific siRNA sequences targeting Ets-1 were transfected into CNE1 and HeLa cells along with the pIα and pRL-TK plasmids, and a nonspecific siRNA was used as a negative control. At 24 h after transfection, cells were collected for dual luciferase reporter assays. Transfection with the siRNA sequence targeting Ets-1 decreased the activity of the Ig Iα1 promoter in CNE1 and HeLa cells compared to the control siRNA (P<0.01; Figure 4a and b), suggesting that silencing the Ets-1 gene downregulates the activation of the human Ig Iα1 promoter in epithelial cancer cells.
Figure 4.
Knockdown of Ets-1 decreases the activity of the Ig Iα1 promoter, Ig Iα1–Cα1 germline transcription and Igα expression. (a, b) Dual luciferase reporter assays indicate that RNA interference of Ets-1 downregulates the activity of the Ig Iα1 promoter. At least three independent transfection experiments were performed in triplicate for each experimental construct. (c, d) RT-PCR and western blot assays indicate that the knockdown of Ets-1 decreases the Ig Iα1–Cα1 germline transcript and Igα expression, respectively. Ig, immunoglobulin.
Knockdown of Ets-1 decreases the Ig Iα1-Cα1 GL transcript and Igα expression
Activation of the Ig Iα1 promoter is critical for Ig Iα1–Cα1 GL transcript expression. We further determined whether knockdown of endogenous Ets-1 could affect Ig Iα1–Cα1 GL transcript and Igα heavy chain expression. At 72 h after transfection with a specific Ets-1 or control siRNA, total RNA and total protein were extracted from CNE1 and HeLa cells. RT-PCR results showed that the Ig Iα1-Cα1 GL transcript level declined after knockdown of Ets-1 (Figure 4c). Moreover, western blot assay results showed that Igα heavy chain expression was reduced after knockdown of Ets-1 compared to the control (Figure 4d). These results suggest that knockdown of endogenous Ets-1 influenced the expression of the Ig Iα1–Cα1 GL transcript and Igα expression.
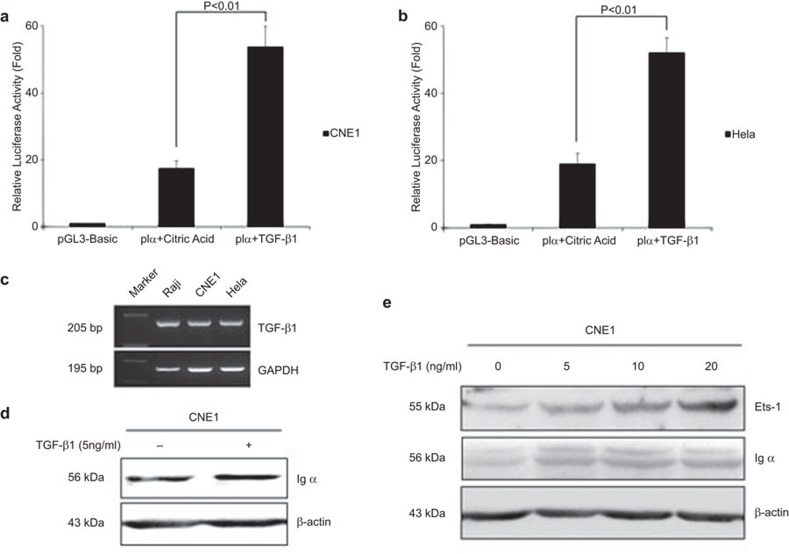
TGF-β1 increases Igα expression by upregulating Ets-1 in epithelial cancer cells
In B lymphocytes, Igα class switch recombination is induced by the TGF-β1 signaling pathway. We found that TGF-β1 could be synthesized by Raji, CNE1 and HeLa cells (Figure 5C). To explore whether TGF-β1 can affect Igα expression in cancer cells, we first performed dual-luciferase reporter assays to evaluate the effect of TGF-β1 on Ig Iα1 promoter activity. The results showed that Ig Iα1 promoter activity was increased when CNE1 and HeLa cells were treated with TGF-β1 (100-21; Peprotech, Rocky Hill, NJ, USA) (P<0.01; Figure 5a and b). Accordingly, Igα expression was also upregulated by TGF-β1 in CNE1 cells (Figure 5d). Furthermore, we found that TGF-β1 could upregulate Igα expression by increasing the level of Ets-1 in a dose-dependent manner (Figure 5e). These data provide evidence that TGF-β1 plays a role in Ig Iα1 promoter activation in cancer cells.
Figure 5.
TGF-β1 upregulates the activity of the Ig Iα1 promoter and Igα expression by increasing the Ets-1 level. (a, b) Dual luciferase reporter assays indicate that the activity of the Ig Iα1 promoter is increased by TGF-β1. After transfection with reporter plasmids for 24 h, the cells were treated with TGF-β1 (10 ng/ml) or citric acid (solvent of TGF-β1, negative control) for 24 h, and luciferase activity was assessed. At least three independent transfection experiments were performed in triplicate for each experimental construct. (c) RT-PCR showed that TGF-β1 could be synthesized by Raji, CNE1 and HeLa cells. (d–e) Western blot analysis indicates that TGF-β1 can upregulate Igα expression by increasing the Ets-1 level in a dose-dependent manner. Proteins were harvested from CNE1 cells treated with TGF-β1 for 24 h. Ig, immunoglobulin.
Discussion
Ig GL transcription is an important regulator of Ig heavy chain class switch recombination. Our research results demonstrated that the Ig Iα1 promoter, which is essential for initiating Ig Iα1–Cα1 GL transcription, was highly activated in cancer cells. In further investigations, we confirmed that Ets-1 (an ETS family member) could bind to the PU.1 motif and transactivate the Ig Iα1 promoter. These results indicate that transcription factor Ets-1 activates the expression of the Ig Iα1–Cα1 GL transcript, which is critical for class switch recombination.
Recent research focusing on transcription factors has become a hot topic in tumor development and progression, and ETS factors represent one of the largest families of transcriptional regulators that exhibit oncogenic and suppressive activity. Some Ets genes are often expressed abnormally in human cancers, and transcription factors of the ETS family, especially Ets-1, have been reported to be associated with tumor progression, including invasion and metastasis, through their transactivation of oncogenes. Therefore, these factors are potential molecular targets for selective cancer therapies.32 The B cell-specific expression of Ig genes is controlled by an orchestrated action of variable (V) region promoters and intronic or 3′ enhancers, all of which are active in a lymphoid-specific manner. Research findings showed that octamer-related proteins (Oct-1 or Oct-2) and PU.1 (a member of the Ets family) could bind to the Igκ V19 promoter region. In transfection experiments with non-B cells, PU.1 was demonstrated to be able to activate this promoter in concert with Oct-2, which suggests an important role for PU.1 or other members of the Ets family in the activation of the Ig promoter.33 Another study indicated that the Ig VH4-59 promoter was highly activated in several non-B-cell cancer lines. Furthermore, the octamer element located in the Ig VH4-59 promoter played an important role in Ig gene activation in non-B cells, and research confirmed that Oct-1 could bind to the octamer element of the Ig VH4-59 promoter and activate Ig gene transcription in epithelial cancer cells. These results reveal a distinct mechanism for Ig gene expression in non-B cancer cells.34
In B lymphocytes, TGF-β signal regulates Ig Iα–Cα GL transcription mainly through the Smad pathway. However, in cancer cells, TGF-β is also known to perform an essential role in cancer progression and metastasis by regulating non-Smad pathways, such as Ras/ERK, PI3-K/Akt and MAPK signaling.35 TGF-β ligands bind to the TGF-β receptor, leading to the phosphorylation of Smad proteins, which recruit cofactors such as AP-1 and Ets to target DNA binding elements and then activate gene transcription.36
We previously found that expression of the Igκ light chain in nasopharyngeal carcinoma cells could be upregulated by the EBV-encoded, latent membrane protein 1 (LMP1) through the NF-κB and AP-1 signaling pathways. Detection of intracellular Igκ by western blot and flow cytometry analysis indicated that Igκ expression could be suppressed by LMP1-targeted DNAzyme and inhibitors of JNKs and NF-κB,37 and further studies suggested that LMP1 promotes the interactions between NF-κB and AP-1 with the human iEκ enhancer, which is crucial for the upregulation of the Igκ light chain in LMP1-positive nasopharyngeal carcinoma cells.38 Furthermore, our previous study provided some evidence showing that the virus-encoded protein LMP1 activated the Igκ 3′ enhancer by activating Ets-1 through the ERKs signaling pathway in non-B epithelial cancer cells. From our results, we inferred that LMP1 could also regulate the activity of the Ig Iα1 promoter by activating Ets-1. This evidence suggests a possible mechanism by which virus-encoded oncoproteins regulate Ig expression through signal transduction pathways in human cancers.
Acknowledgments
This work was supported by the National High Technology Research and Development Program (863) of China (No. 2006AA02A404) and the National Nature Science Foundation of China (Nos. 30772465 and 30973399).
References
- Hu D, Zheng H, Liu H, Li M, Ren W, Liao W, et al. Immunoglobulin expression and its biological significance in cancer cells. Cell Mol Immunol. 2008;5:319–324. doi: 10.1038/cmi.2008.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Qiu X, Gu J. Immunoglobulin expression in non-lymphoid lineage and neoplastic cells. Am J Pathol. 2009;174:1139–1148. doi: 10.2353/ajpath.2009.080879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Sun Y, Poirier S, Winterstein D, Hegamyer G, Seed J, et al. Isolation and partial characterization of a transformation-associated sequence from human nasopharyngeal carcinoma. Mol Carcinog. 1991;4:297–307. doi: 10.1002/mc.2940040408. [DOI] [PubMed] [Google Scholar]
- Li M, Ren W, Weng XX, Liao W, Xia LQ, Deng X, et al. Nucleotide sequence analysis of a transforming gene isolated from nasopharyngeal carcinoma cell line CNE2: an aberrant human immunoglobulin kappa light chain which lacks variable region. DNA Seq. 2001;12:331–335. doi: 10.3109/10425170109084456. [DOI] [PubMed] [Google Scholar]
- Kimoto Y. Expression of heavy-chain constant region of immunoglobulin and T-cell receptor gene transcripts in human non-hematopoietic tumor cell lines. Genes Chromosomes Cancer. 1998;22:83–86. doi: 10.1002/(sici)1098-2264(1998)22:1<83::aid-gcc12>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Qiu X, Zhu X, Zhang L, Mao Y, Zhang J, Hao P, et al. Human epithelial cancers secrete immunoglobulin g with unidentified specificity to promote growth and survival of tumor cells. Cancer Res. 2003;63:6488–6495. [PubMed] [Google Scholar]
- Babbage G, Ottensmeier CH, Blaydes J, Stevenson FK, Sahota SS. Immunoglobulin heavy chain locus events and expression of activation-induced cytidine deaminase in epithelial breast cancer cell lines. Cancer Res. 2006;66:3996–4000. doi: 10.1158/0008-5472.CAN-05-3704. [DOI] [PubMed] [Google Scholar]
- Chen Z, Gu J. Immunoglobulin G expression in carcinomas and cancer cell lines. FASEB J. 2007;21:2931–2938. doi: 10.1096/fj.07-8073com. [DOI] [PubMed] [Google Scholar]
- Geng LY, Shi ZZ, Dong Q, Cai XH, Zhang YM, Cao W, et al. Expression of SNC73, a transcript of the immunoglobulin alpha-1 gene, in human epithelial carcinomas. World J Gastroenterol. 2007;13:2305–2311. doi: 10.3748/wjg.v13.i16.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Sun X, Mao Y, Zhu X, Zhang P, Zhang L, et al. Expression of immunoglobulin gene with classical V–(D)–J rearrangement in mouse brain neurons. Int J Biochem Cell Biol. 2008;40:1604–1615. doi: 10.1016/j.biocel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Zhu X, Li C, Sun X, Mao Y, Li G, Liu X, et al. Immunoglobulin mRNA and protein expression in human oral epithelial tumor cells. Appl Immunohistochem Mol Morphol. 2008;16:232–238. doi: 10.1097/PAI.0b013e31814c915a. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang L, Ma T, Zhang P, Qiu X. Expression of immunoglobulin gene with classical V–(D)–J rearrangement in mouse testis and epididymis. J Histochem Cytochem. 2009;57:339–349. doi: 10.1369/jhc.2008.951434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Mao Y, Huang J, Ma T, Zhang L, Zhu X, et al. Immunoglobulin gene locus events in epithelial cells of lactating mouse mammary glands. Cell Mol Life Sci. 2010;67:985–994. doi: 10.1007/s00018-009-0231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Li M, Liu H, Ren W, Hu DS, Shi Y, et al. Immunoglobulin alpha heavy chain derived from human epithelial cancer cells promotes the access of S phase and growth of cancer cells. Cell Biol Int. 2007;31:82–87. doi: 10.1016/j.cellbi.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Li M, Zheng H, Duan Z, Liu H, Hu D, Bode A, et al. Promotion of cell proliferation and inhibition of ADCC by cancerous immunoglobulin expressed in cancer cell lines. Cell Mol Immunol. 2012;9:54–61. doi: 10.1038/cmi.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Li M, Ren W, Zeng L, Liu HD, Hu D, et al. Expression and secretion of immunoglobulin alpha heavy chain with diverse VDJ recombinations by human epithelial cancer cells. Mol Immunol. 2007;44:2221–2227. doi: 10.1016/j.molimm.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Zheng J, Huang J, Mao Y, Liu S, Sun X, Zhu X, et al. Immunoglobulin gene transcripts have distinct VHDJH recombination characteristics in human epithelial cancer cells. J Biol Chem. 2009;284:13610–13619. doi: 10.1074/jbc.M809524200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Xiao Y, Zhang J, Li J, Liu Y, Zhao Y, et al. Transcription factors E2A, FOXO1 and FOXP1 regulate recombination activating gene expression in cancer cells. PLoS ONE. 2011;6:e20475. doi: 10.1371/journal.pone.0020475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci USA. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Whang N, Wuerffel R, Kenter AL. AID-dependent histone acetylation is detected in immunoglobulin S regions. J Exp Med. 2006;203:215–226. doi: 10.1084/jem.20051774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Schrader CE. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. Trends Genet. 2006;22:23–28. doi: 10.1016/j.tig.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- Cerutti A. The regulation of IgA class switching. Nat Rev Immunol. 2008;8:421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Duan Z, Li M, Jiang Y, Liu H, Zheng H, et al. Heterogeneity of aberrant immunoglobulin expression in cancer cells. Cell Mol Immunol. 2011;8:479–485. doi: 10.1038/cmi.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Duan Z, Zheng H, Hu D, Li M, Tao Y, et al. EBV-encoded LMP1 upregulates Igkappa 3′enhancer activity and Igkappa expression in nasopharyngeal cancer cells by activating the Ets-1 through ERKs signaling. PLoS ONE. 2012;7:e32624. doi: 10.1371/journal.pone.0032624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Zan H, Schaffer A, Bergsagel L, Harindranath N, Max EE, et al. CD40 ligand and appropriate cytokines induce switching to IgG, IgA, and IgE and coordinated germinal center and plasmacytoid phenotypic differentiation in a human monoclonal IgM+IgD+ B cell line. J Immunol. 1998;160:2145–2157. [PMC free article] [PubMed] [Google Scholar]
- Oikawa T, Yamada T, Kihara-Negishi F, Yamamoto H, Kondoh N, Hitomi Y, et al. The role of Ets family transcription factor PU.1 in hematopoietic cell differentiation, proliferation and apoptosis. Cell Death Differ. 1999;6:599–608. doi: 10.1038/sj.cdd.4400534. [DOI] [PubMed] [Google Scholar]
- Hahne JC, Okuducu AF, Sahin A, Fafeur V, Kiriakidis S, Wernert N. The transcription factor ETS-1: its role in tumour development and strategies for its inhibition. Mini Rev Med Chem. 2008;8:1095–1105. doi: 10.2174/138955708785909934. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H, Newell JW, Matthias P. Involvement of the Ets family factor PU.1 in the activation of immunoglobulin promoters. J Biol Chem. 1995;270:898–907. doi: 10.1074/jbc.270.2.898. [DOI] [PubMed] [Google Scholar]
- Zhu X, Wu L, Zhang L, Hao P, Zhang S, Huang J, et al. Distinct regulatory mechanism of immunoglobulin gene transcription in epithelial cancer cells. Cell Mol Immunol. 2010;7:279–286. doi: 10.1038/cmi.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- Sundqvist A, Zieba A, Vasilaki E, Herrera Hidalgo C, Soderberg O, Koinuma D, et al. Specific interactions between Smad proteins and AP-1 components determine TGFbeta-induced breast cancer cell invasion. Oncogene. 2013;32:3606–3615. doi: 10.1038/onc.2012.370. [DOI] [PubMed] [Google Scholar]
- Liu HD, Zheng H, Li M, Hu DS, Tang M, Cao Y. Upregulated expression of kappa light chain by Epstein-Barr virus encoded latent membrane protein 1 in nasopharyngeal carcinoma cells via NF-kappaB and AP-1 pathways. Cell Signal. 2007;19:419–427. doi: 10.1016/j.cellsig.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Liu H, Zheng H, Duan Z, Hu D, Li M, Liu S, et al. LMP1-augmented kappa intron enhancer activity contributes to upregulation expression of Ig kappa light chain via NF-kappaB and AP-1 pathways in nasopharyngeal carcinoma cells. Mol Cancer. 2009;8:92. doi: 10.1186/1476-4598-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]