Abstract
African trypanosomes differentiate between various developmental stages both in mammalian hosts and their tsetse vector to adapt to and survive in the different environments they encounter. In the bloodstream, trypanosomes naturally exist as either proliferative slender-forms or non-proliferative stumpy-forms, the latter being responsible for both prolonged infection and transmission. However, most trypanosome studies are carried out on laboratory-adapted monomorphic cell lines, incapable of differentiating to stumpy-forms or completing the life cycle through the tsetse fly. Partly, this has been due to the inefficiency of transfection of pleomorphic strains which have retained the ability to generate stumpy-forms. Recently, Amaxa Nucleofector® technology was shown to increase transfection efficiency for monomorphic bloodstream forms. Using this technology we have optimised a similar method for pleomorphic bloodstream form transfection, generating transfection efficiencies of 10−7–10−6. This permits routine genetic manipulation of pleomorphic lines, which have the most biological relevance for trypanosomes in the field.
Keywords: Trypanosome, Transfection, Pleomorphic, Stumpy, Trypanosoma brucei
A wide range of genetic manipulations, including gene knock-outs, endogenous-tagging, inducible RNA interference and over-expression are all commonly used in the study of monomorphic bloodstream form and procyclic form Trypanosoma brucei, made possible by the ability to efficiently and reliably transfect these cells. Transfection of procyclic form trypanosomes has been carried out with high efficiency for many years [1], but, historically, bloodstream form trypanosomes have been more difficult to transfect. Recent advances have allowed a marked increase in transfection efficiency for monomorphic bloodstream forms, particularly through the use of the Amaxa Nucleofector® system which increases transfection efficiency 1000-fold compared to traditional electroporation methods [2]. The introduction of site-specific double strand breaks at targeted sites for vector integration also increases transfection efficiency a further 250-fold [3]. These developments have allowed the production of monomorphic bloodstream form RNAi libraries, which have been successfully utilised for whole-genome RNAi screens [4–6].
Laboratory-adapted monomorphic bloodstream forms are proliferative cells which grow uncontrolled in vivo, quickly overwhelming the host. Naturally occurring populations of bloodstream form trypanosomes, in contrast, are pleomorphic; that is they differentiate in a density-dependent manner from the proliferative slender form, through an intermediate stage, to the non-proliferative, transmission-competent stumpy form, limiting parasitaemia and prolonging infection [7,8]. Hence, the stumpy form is a key developmental life stage that plays an important role in disease transmission and within-host infection dynamics [9]. Although monomorphic cell lines are a vital resource for trypanosome research, their inability to generate stumpy forms in vivo limits their utility for the study of genes or pathways involved in infection dynamics, bloodstream form differentiation, or analyses where progression through the complete life cycle is desired. Conversely, pleomorphic cell lines have retained the ability to generate stumpy forms and so can be used for such analyses, yet these lines are less amenable to genetic manipulation. Indeed, the generation of transgenic pleomorphic bloodstream forms, although possible [1], is still considered problematic. In consequence, even common genetic approaches carried out in monomorphic cell lines, such as RNAi, are rarely carried out in pleomorphic cell lines. Based on the success of the Amaxa Nucleofector® system for the transfection of monomorphic bloodstream forms [2] we report here an optimised method for the transfection of pleomorphic cell lines. This method is used routinely in our laboratory for the stable transfection of pleomorphic bloodstream form trypanosomes.
The pleomorphic cell line T. brucei brucei AnTat1.1 90:13 [10], (containing pLew90 and pLew13 [10,11]) has proven particularly amenable to transfection, likely due to a degree of culture adaptation in this cell line. Nonetheless, these cells differentiate to stumpy forms in vivo: stumpy forms are visible from day 4–6 post infection and the parasitaemia plateaus around day 5–7 post infection whereupon the population comprises >80% morphological stumpy forms. The AnTat1.1 90:13 cell line also grows well in vitro when maintained at low parasite density (below 106 cells/ml) and passaged regularly (at least every 2 days) in HMI-9 [12] supplemented with 20% FCS. Although it is possible to transfect such pleomorphic slender grown in vitro that maintain pleomorphism, we found that it is preferable to harvest slender form cells from an in vivo infection, this providing the most healthy starting population for transfection and thereby increasing the likelihood of successfully isolating transfectants. Their limited passage history in vitro could also reduce the risk of a selection for monomorphism. Importantly, the cells were harvested while the population was overwhelmingly slender in morphology (<1 × 108 parasites/ml of blood) since intermediate and stumpy cells are committed to irreversible cell cycle arrest and therefore would not proliferate in vitro. Approximately 1–1.5 ml of blood was harvested using 200 μl of 2% citrate as an anticoagulant from an infected mouse by cardiac puncture. The blood was maintained at 37 °C throughout isolation and was then added as promptly as possible to 25 ml HMI-9 (20% FCS; 100 U/ml penicillin and streptomycin) in vented tissue culture flasks which had been pre-warmed and pre-equilibrated at 37 °C with 5% CO2. To separate the trypanosomes from the mouse blood cells, the flask was positioned upright in the incubator allowing the blood cells to settle to the bottom of the flask. After 2–3 h the supernatant was carefully pipetted (avoiding clumps of red blood cells) into a fresh flask containing pre-warmed and pre-equilibrated HMI-9 to produce a final volume of 60 ml. Further settling of the remaining blood cells was then allowed to occur overnight and the following day the trypanosome containing medium was again transferred to a fresh flask avoiding settled blood cells. The resulting medium was enriched for trypanosome cells but not completely free of blood cells such that the trypanosome density needed to be determined via a haemocytometer rather than, for example, a particle counter. The quantity of trypanosomes generated by this method varied due to variation in the initial parasitaemia and the volume of blood isolated but was typically in the order of 2–4 × 107 cells per mouse.
For the transfection (Fig. 1A), cells were pelleted at 800 g for 8 min in a clinical centrifuge and the supernatant removed. The cell pellet was then resuspended in 100 μl Amaxa transfection buffer (basic parasite nucleofector solution 2) with 10 μg linearised DNA in 5 μl 1 mM Tris-HCl, pH8, 0.1 mM EDTA. The trypanosomes were then transferred to an Amaxa cuvette and transformed in an Amaxa Nucleofector® II using the CD4+ T cells X-001 program. Following transfection, the cells were immediately transferred into 25 ml of pre-warmed and pre-equilibrated HMI-9 and incubated for 6 h. After 6 h, selective drug (0.5 μg/ml puromycin; 10 μg/ml blasticidin; 1.5–3 μg/ml phleomycin, where effective doses required titration) was added and cells were transferred into 24 well plates, diluted 1:2, 1:5, 1:25 and 1:125 using HMI9. Approximately 5–8 days post-transfection resistant clones became detectable and were transferred to fresh selective media with care being taken to avoid cell density from exceeding 106 cells/ml. After expansion in vitro, transfectant clones were cryopreserved in HMI-9 with 7–10% glycerol and genomic DNA harvested, if required. Additionally, to generate blood stocks of the resulting transfectants, cells were infected into a mouse and blood stabilates prepared after harvest when the parasitaemia was slender in morphology.
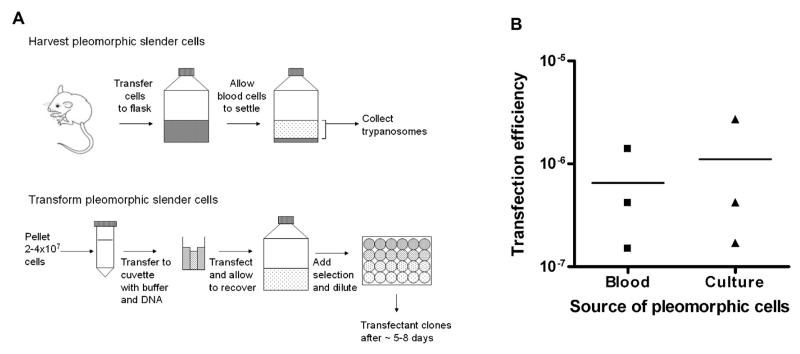
Fig. 1.
Transfection of pleomorphic transfections from cells harvested from blood or from culture. (A) Diagram outlining the method for the harvest and transfection of pleomorphic slender forms (details described in main text). (B) Pleomorphic bloodstream form AnTat1.1 90:13 cells were harvested from mouse blood 3 days post infection or after 7 days in vitro. Between 2.8 and 4.06 × 107 cells were transfected with 10 μg of pALC14 vector in the “Amaxa basic parasite nucleofector solution 2” transfection buffer and selected in 24 well plates at 1:2, 1:5, 1:25 and 1:125 dilutions with 0.5 μg/ml puromycin. Transfection efficiencies were calculated from the number of positive wells per dilution (excluding any dilution where >50% of wells were positive) and extrapolating for the total number of cells transfected. Replicates were carried out from three independent mouse infections and three independent in vitro cultures.
To quantify the transfection efficiency obtained using this method, replicate stable transfections of AnTat1.1 90:13 pleomorph cells [10] were carried out using the ‘empty’ pALC14 RNAi vector (a modified version of pZJM, created from pLew100, which is targeted to insert between ribosomal RNA genes [11,13,14]). Using cells harvested from three independent in vivo infections (as described above) or from three independent cultures (after 7 days growth in vitro) we obtained transfection efficiencies with 10 μg of DNA of 1.5 × 10−7–1.4 × 10−6 and 1.7 × 10−7–2.7 × 10−6, respectively, such that multiple independent transfectant lines were isolated for each transfection (Fig. 1B). In our laboratory over 30 independently transfected lines have been generated by this approach and in no case has a loss of pleomorphism been observed. Moreover, T. brucei brucei AnTat.1.1 cells as well as T. brucei brucei AnTat1.1 90:13 cells have been successfully transfected.
This method enables the routine stable transformation of pleomorphic bloodstream form T. brucei at efficiencies in the order of ~10−7–10−6. This makes the genetic manipulation of differentiation competent pleomorphic trypanosome lines readily achievable using simple methodology and materials and equipment available in most trypanosome research laboratories. This facilitates medium-throughput gene function analyses for trypanosome lines most relevant to parasites in the field, enabling phenotypic analysis throughout the life cycle of the parasite.
Acknowledgements
We thank Julie Wilson for excellent technical assistance. This work was funded by a Wellcome Trust Programme grant (088293MA) to K.M. and by a Wellcome Trust strategic award (095831MA) to the Centre for Immunity, Infection and Evolution. F.R. was supported by SysMO/BBSRC (the Silicotryp).
References
- [1].McCulloch R, Vassella E, Burton P, Boshart M, Barry JD. Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol Biol. 2004;262:53–86. doi: 10.1385/1-59259-761-0:053. [DOI] [PubMed] [Google Scholar]
- [2].Burkard G, Fragoso CM, Roditi I. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol Biochem Parasitol. 2007;153:220–3. doi: 10.1016/j.molbiopara.2007.02.008. [DOI] [PubMed] [Google Scholar]
- [3].Glover L, Horn D. Site-specific DNA double-strand breaks greatly increase stable transformation efficiency in Trypanosoma brucei. Mol Biochem Parasitol. 2009;166:194–7. doi: 10.1016/j.molbiopara.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, et al. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21:915–24. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Baker N, Alsford S, Horn D. Genome-wide RNAi screens in African trypanosomes identify the nifurtimox activator NTR and the eflornithine transporter AAT6. Mol Biochem Parasitol. 2011;176:55–7. doi: 10.1016/j.molbiopara.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Schumann Burkard G, Jutzi P, Roditi I. Genome-wide RNAi screens in bloodstream form trypanosomes identify drug transporters. Mol Biochem Parasitol. 2011;175:91–4. doi: 10.1016/j.molbiopara.2010.09.002. [DOI] [PubMed] [Google Scholar]
- [7].Vickerman K. Polymorphism and mitochondrial activity in sleeping sickness trypanosomes. Nature. 1965;208:762–6. doi: 10.1038/208762a0. [DOI] [PubMed] [Google Scholar]
- [8].Vickerman K. Developmental cycles and biology of pathogenic trypanosomes. Br Med Bull. 1985;41:105–14. doi: 10.1093/oxfordjournals.bmb.a072036. [DOI] [PubMed] [Google Scholar]
- [9].MacGregor P, Szoor B, Savill NJ, Matthews KR. Trypanosomal immune evasion, chronicity and transmission: an elegant balancing act. Nat Rev Microbiol. 2012;10:431–8. doi: 10.1038/nrmicro2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Engstler M, Boshart M. Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev. 2004;18:2798–811. doi: 10.1101/gad.323404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- [12].Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75:985–9. [PubMed] [Google Scholar]
- [13].Bochud-Allemann N, Schneider A. Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J Biol Chem. 2002;277:32849–54. doi: 10.1074/jbc.M205776200. [DOI] [PubMed] [Google Scholar]
- [14].Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem. 2000;275:40174–9. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]