Abstract
The duck (Anas platyrhynchos) is one of the principal natural hosts of influenza A viruses. We present the duck genome sequence and perform deep transcriptome analyses to investigate immune-related genes. Our data indicate that the duck possesses a contractive immune gene repertoire, as in chicken and zebra finch, and this repertoire has been shaped through lineage-specific duplications. We identify genes that are responsive to influenza A viruses using the lung transcriptomes of control ducks and ones that were infected with either a highly pathogenic (A/duck/Hubei/49/05) or a weakly pathogenic (A/goose/Hubei/65/05) H5N1 virus. Further, we show how the duck’s defense mechanisms against influenza infection have been optimized through the diversification of its β-defensin and butyrophilin-like repertoires. These analyses, in combination with the genomic and transcriptomic data, provide a resource for characterizing the interaction between host and influenza viruses.
Birds are distinct from other vertebrates in many characteristics, both morphological (for example, feathers and eggs) and physiological (for example, their lightweight skeletons and high metabolic rates). Birds occupy habitats from the Arctic to the Antarctic, and they have body size ranging from 5 cm to 2.8 m. The diversity found in birds inspires us to investigate the genomic differences underlying their phenotypic differences from mammals and the variation within the avian class. The sequencing of the chicken genome1 provided the first insight into the evolutionary events between birds and other vertebrates. Avian evolutionary events have subsequently been elucidated with the recent availability of the zebra finch and turkey genomes2,3. Additional avian genomes, however, are needed to provide more detailed evolutionary information and insight into adaptation mechanisms. The duck (A. platyrhynchos) is particularly well suited for further exploration in these areas. Ducks diverged from the related chicken and turkey and zebra finch approximately 90–100 million years ago4. The duck is also one of the most economically important waterfowl as a source of meat, eggs and feathers.
Of special interest to medicine and agriculture is the fact that ducks serve as the principal natural reservoir for influenza A viruses and harbor all 16 hemagglutinin (HA) and 9 neuraminidase (NA) subtypes that are currently known5,6, with the exception of the H13 and H16 subtypes7. Often, influenza strains cause the duck no harm. However, the long-standing equilibrium between influenza A viruses and the duck has been disrupted with the emergence of H5N1 viruses8. H5N1 strains have caused unprecedented outbreaks in poultry in more than 60 countries and have caused 622 human infections (as of March 2013), with an overall fatality rate of 59% in humans. Recently, it was reported that an engineered influenza virus encoding hemagglutinin from the highly pathogenic avian H5N1 influenza A strains can be transmitted between ferrets9, emphasizing the potential for a human pandemic to emerge from birds. Furthermore, weakly pathogenic avian influenza A viruses, such as H9N2 (ref. 10) and H7N2 (ref. 11) strains, have caused transient infections in humans12. The exceptional virulence of avian influenza viruses in humans, in combination with their ongoing evolution in birds, motivates us to better understand host immune responses to avian influenza viruses. We report here the duck whole-genome sequence and compared it to the genomes of mammals and other birds. We performed deep transcriptome analyses of lungs from control ducks and ones that were infected with either a highly pathogenic (A/duck/Hubei/49/05, DK/49) or a weakly pathogenic (A/goose/Hubei/65/05, GS/65) H5N1 virus13.
RESULTS
The genome landscape
We sequenced the genome of a 10-week-old female Beijing duck using methods similar to those applied to sequence the giant panda genome14. In total, we generated 77 Gb of paired-end reads (approximately 64-fold coverage of the whole genome) with an average length of 50 bp (Supplementary Figs. 1–3 and Supplementary Table 1). Using SOAPdenovo (Supplementary Note), we combined short reads to generate a draft assembly, which consisted of 78,487 scaffolds and covered 1.1 Gb. The contig N50 and scaffold N50 values of this draft assembly were 26 kb and 1.2 Mb, respectively (Supplementary Table 2). We then constructed superscaffolds and created chromosomal sequences according to the duck genetic map15 and the comparative physical map16. This effort resulted in the construction of a total of 47 superscaffolds, which contained 225 scaffolds and spanned 289 Mb (Supplementary Table 3).
We generated transcriptomes from several different tissues (Supplementary Note). These transcriptomes comprised 1.87 million ESTs and approximately 121 million 75-bp and approximately 917 million 90-bp paired-end reads, which were generated using either the 454/Roche Life Sciences Analyzer or Illumina Genome sequencing technology (Table 1 and Supplementary Tables 4 and 5). Next, we estimated the coverage of the duck assembly with alignments to seven finished BACs (completed independently using Sanger sequencing technology17), 240 microsatellite markers15 and the 319,996 ESTs assembled in this project (Supplementary Note) using BLASTN (E value < 1 × 10−5). These analyses suggested that 7 BACs covering 640 kb on chromosomes 1, 3 and 4 were aligned over more than 95% of their lengths (Supplementary Fig. 4). Similarly, greater than 97% of 240 microsatellite markers and 96% of the 319,996 ESTs were aligned to the duck assembly. We further aligned the duck and chicken assemblies to the human using Narcisse. This effort showed that the coverage of the two avian assemblies of the human genome (GRCh37) was similar, indicating that the quality of the duck and chicken assemblies was comparable. In addition, in aligning the duck assembly onto the chicken genome sequence using GLINT software with its default parameters18, only 41 of the 78,847 duck scaffolds were identified as possible chimeras when applying the criterion that they contain sequences that partially map to two different chicken chromosomes. These data indicate that the duck assembly has good coverage and generally provides a reasonable substrate for the analysis presented in this study.
Table 1. Number and length of reads and number of genes detected using RNA sequencing in control and H5N1 virus–infected ducks.
| Number of uniquely mapped reads |
Number of DEGs and PSGsa (versus control) |
Number of DEGs and PSGsa (versus GS/65-infected ducks) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | Total reads | Total length of reads (bp) | Number of expressed genes | Genome | Gene | DEGs | Frequency of PSGs | DEG set | DEGs | Frequency of PSGs | DEG set |
| Control | 146,618,064 | 13,195,625,760 | 18,181 | 84,895,562 | 72,455,573 | ||||||
| DK/49 (day 1) | 116,561,634 | 10,490,547,060 | 18,276 | 60,873,691 | 49,811,986 | 2,257 | 63/957 | 1,506 | 38/624 | ||
| DK/49 (day 2) | 133,943,194 | 12,054,887,460 | 18,233 | 73,475,680 | 59,066,187 | 3,101 | 89/1,413 | 1,436 | 44/654 | ||
| DK/49 (day 3) | 136,923,500 | 12,323,115,000 | 17,951 | 81,116,464 | 65,172,557 | 3,066 | 86/1,383 | 5,038 | 1,396 | 44/625 | 3,232 |
| GS/65 (day 1) | 138,642,320 | 12,477,808,800 | 18,135 | 81,176,979 | 66,830,938 | 916 | 20/367 | ||||
| GS/65 (day 2) | 127,576,536 | 11,481,888,240 | 18,140 | 72,750,242 | 58,940,732 | 2,060 | 58/918 | ||||
| GS/65 (day 3) | 116,267,820 | 10,464,103,800 | 17,983 | 69,337,186 | 56,072,340 | 1,251 | 29/538 | 2,741 | |||
DEGs are genes that showed significantly different expression with FDR ≤0.001 and fold change ≥2. PSGs are genes that are predicted to be under positive selection with FDR <0.05. The numerator and denominator for the frequencies of PSGs are the number of positively selected genes and genes where a likelihood ratio test was performed for PSGs, respectively.
We defined reference gene sets from the duck assembly using the BGI and Ensembl pipelines (Supplementary Note). For the BGI reference set, we aligned the duck transcriptome data and human (22,232) and chicken (16,701) genes to the duck genome assembly, yielding 15,065 predicted protein-coding duck genes. We applied GENSCAN19 and Augustus20 with the default parameters and predicted 32,383 and 22,739 protein-coding genes in the duck draft genome for the respective reference sets (Supplementary Table 6). Finally, we integrated all gene sources and created a reference set containing 19,144 genes, constituting approximately 2.3% of the total duck assembly. Of these 19,144 genes, 9,678 were mapped to categories established by the Gene Ontology (GO) Project, 14,725 had orthologs in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, and 18,012 were supported by the duck ESTs (Supplementary Figs. 5 and 6). Comparing the BGI reference gene set to the Ensembl reference gene set, which contained 15,634 protein-coding genes (Supplementary Note), we found that 14,049 of the BGI reference genes and 13,811 of the Ensembl reference genes were predicted by both pipelines, whereas the remaining 5,095 and 1,823 genes from the two sets, respectively, were annotated with the BGI and Ensembl pipelines alone. We predicted 817 noncoding RNA genes in the duck assembly with a homology-based annotation method (Supplementary Tables 7 and 8 and Supplementary Note). In addition, we identified 29 families of DNA transposons, 61 families of retrotransposons and 414 families of microsatellites, comprising 5.9% of the duck genome (Supplementary Table 9 and Supplementary Note).
Changes in gene family size
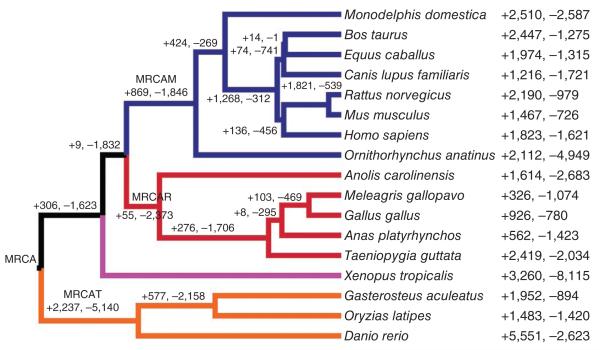
We examined large-scale differences in gene complements within birds and between birds and either mammals or fish using the duck reference gene set and combined gene sets from three birds, one reptile-amphibian, three fish and eight mammals (Fig. 1). Using a likelihood model21, we estimated at least 23,044 genes distributed in 14,466 gene families in the most recent common ancestor (MRCA) of 17 vertebrates (Fig. 1). We estimated the average rate of change across all 14,466 gene families in the MRCA via maximum likelihood under a single- or multiple-rate model for each clade21. A comparison of likelihood values showed that the best-fit model (P < 0.01) estimated the average rates of genomic turnover (represented by the λ value, given per gene per million years) to be 0.0012 for teleosts, 0.0019 for Xenopus tropicalis, 0.0011 for reptiles (including birds) and 0.0017 for mammals. These estimates are similar to previous values for yeast (0.0020)21, Drosophila melanogaster (0.0023)22 and mammals (0.0017)23, supporting the theory that rates of gene duplication and deletion across eukaryotes are comparable.
Figure 1.
Numbers of gene losses and gains across 17 vertebrates. Data are shown for 17 vertebrates, 3 teleosts, 5 reptilians and 8 mammals. The numbers of gene gains (+) and losses (−) are given on branches or to the right of the taxa. The rates of gene gain and loss for the clades derived from the MRCAR (MRCA of reptiles), MRCAT (MRCA of teleosts) and MRCAM (MRCA of mammals) and for Xenopus tropicalis are 0.0011, 0.0012, 0.0017 and 0.0019 per gene per million years, respectively.
We compared gene family sizes at parent and daughter nodes along the vertebrate tree (Fig. 1), finding that the total numbers of contractions outnumbered expansions in the MRCAs of teleosts, mammals and reptiles. This tendency, where contractions outnumbered expansions, was continued into the duck and turkey. However, the reverse case, where expansions outnumbered contractions, seemed to be true in the chicken and zebra finch. From the terminal branch leading to the duck and chicken, we inferred a gain of 562 out of 1,029 genes and a loss of 1,423 out of 1,249 genes in ~90 million years (Fig. 1).
Expansion and contraction of immune gene families
We identified the predicted duck, chicken and zebra finch genes that were homologs of 3,542 human and 1,415 mouse immune genes (comprising 4,344 unique immune genes), which were derived from analyses of Import, IRIS, Septic Shock Group, MAPK–NF-κB network and immunome databases using TreeFam24. In total, 6,044 human and 5,715 mouse genes were clustered into 3,726 immune-related gene families, all of which included at least one of the 4,344 unique immune genes. However, only 3,116 duck genes, 3,294 chicken genes and 3,355 zebra finch genes were clustered into the immune-related gene families, indicating that avian immune gene repertoires were contractive.
We used cytokines as an example to compare immune gene repertoires in mammals and birds. After detecting cytokines in the above five species with TreeFam24, we manually queried their repertoires against the non-redundant database in NCBI and examined their assemblies in Ensembl (version 57). Using the combined duck transcriptome and assembly data, we identified 150 duck cytokine genes; although this number resembles the numbers of such genes identified in chicken (149 genes) and zebra finch (150 genes), it is substantially lower than the numbers of mammalian cytokine genes (230 in humans and 218 in mice) (Table 2).
Table 2. Comparison of cytokines in the duck, chicken, zebra finch, human and mouse genomes.
| Number of genes |
||||||
|---|---|---|---|---|---|---|
| Class | Family | Duck | Chicken | Zebra finch | Human | Mouse |
| Class I cytokines | IL-2 receptor | 7 | 7 | 7 | 8 | 8 |
| IL-3 receptor | 2 | 2 | 1 | 3 | 3 | |
| IL-6 receptor | 8 | 7 | 8 | 12 | 12 | |
| Single-chain family | 4 | 3 | 6 | 6 | 4 | |
| Class II cytokines | Type I interferons | 4 | 6 | 4 | 20 | 17 |
| Type II interferons | 1 | 1 | 1 | 1 | 1 | |
| IL-10 family | 4 | 4 | 4 | 6 | 5 | |
| PDGF family | Cysteine-knot growth factors | 8 | 8 | 8 | 9 | 9 |
| 4-helix bundle growth factors | 0 | 0 | 0 | 2 | 2 | |
| β-trefoil growth factors | 19 | 19 | 21 | 22 | 21 | |
| Other growth factors | 15 | 17 | 18 | 20 | 20 | |
| TNF family | 11 | 11 | 10 | 18 | 18 | |
| IL-1 family | 2 | 3 | 2 | 10 | 9 | |
| IL-17 family | 5 | 5 | 6 | 6 | 6 | |
| TGF-β family | BMP2 subfamily | 2 | 2 | 2 | 2 | 2 |
| BMP5 subfamily | 4 | 4 | 2 | 5 | 5 | |
| GDF5 subfamily | 3 | 2 | 2 | 3 | 3 | |
| VGL subfamily | 1 | 1 | 1 | 2 | 2 | |
| BMP3 subfamily | 2 | 2 | 3 | 2 | 2 | |
| ADMP | 2 | 2 | 2 | 0 | 0 | |
| Intermediate members | 5 | 4 | 5 | 5 | 5 | |
| Activin subfamily | 4 | 2 | 2 | 4 | 4 | |
| TGF subfamily | 3 | 3 | 2 | 3 | 3 | |
| Distant members | 6 | 6 | 6 | 8 | 8 | |
| Chemokines | CC chemokines | 13 | 14 | 12 | 28 | 27 |
| CXC chemokines | 8 | 8 | 8 | 17 | 14 | |
| CXC3C chemokines | 2 | 1 | 2 | 1 | 1 | |
| C chemokines | 1 | 1 | 1 | 2 | 1 | |
| Total | 150 | 149 | 150 | 230 | 218 | |
We found that the duck genome contains 16 defensins distributed over 3 scaffolds, a number that was slightly higher than that of the 14 defensins found in chicken25. Closing the sequence gaps within the three scaffolds via Sanger sequencing, we identified three additional duck defensin genes (including one pseudogene). Structural and phylogenetic analyses of avian defensins and mammalian β-defensins showed that all duck defensins are β-defensins (Supplementary Fig. 7a), supporting the hypothesis that birds lack α- and θ-defensins25,26. Molecular phylogenetic analysis indicated that the avian defensin genes were divided into 12 subfamilies: 1 subfamily (AvDB14) lost its member in the zebra finch, 2 subfamilies (AvDB1-AvDB3 and AvDB6-AvDB7) contained lineage-specific duplications (LSDs), and 9 subfamilies (AvDB2, AvDB4, AvDB5 and AvDB8–AvDB13) remained one-to-one orthologs in three birds (Supplementary Fig. 7a). Evolutionary comparison of the avian defensin genes suggested that single clusters of these genes in duck, chicken and zebra finch were collinear, with the exception of AvDB1, AvDB3 and AvDB14 (Supplementary Fig. 7b). These observations suggest that the ancient Neognathae had 13 avian defensin genes, including AvDB1–AvDB5 and AvDB7–AvDB14. Gene duplications along with the pseudogenization of defensin genes further increased the repertoire of these genes in both duck and zebra finch. Two gene duplication events of AvDB1 have been described in the zebra finch; however, duplication of the ancient AvDB7 gene seems to have led to the introduction of the AvDB6 gene in chicken.
LSDs in birds
Using a cutoff of 2 duplication events, we identified 5, 76, 577 and 1,752 LSDs in turkey, duck, chicken and zebra finch, respectively (Supplementary Note). Of the 14 gene families that contained 76 duck LSDs, we found that 3 were significantly expanded in this lineage (family-wide P value < 0.0005). One family is a BTNL (butyrophilin-like) family, which includes the mammalian BTNL genes, with the exception of BTNL9 (Supplementary Fig. 7c). Domain prediction using SMART software27 suggested that 6 out of 17 duck gene fragments encoded a structure that was typical of BTNLs28. The high prevalence of these genes in duck is in sharp contrast to their frequency in chicken and turkey, where only 1 or 2 out of 4 genes encoded this structure (Supplementary Fig. 7d). The second family significantly expanded in the duck lineage was an olfactory receptor gene family, and the third family was a novel gene family that included only five duck epidermal growth factor (EGF)-like genes (Supplementary Table 10).
Evidence for positive selection
We performed likelihood ratio tests (LRTs) for positive selection with the codeml program under a branch-site model29 using 8,409 avian quaternions (Supplementary Note). These LRTs (false discovery rate (FDR) <0.05) predicted that 2.7%, 5.2%, 6.2% and 10.9% of genes showed evidence of positive selection in chicken, duck, zebra finch and turkey, respectively. These proportions are significantly lower than the previously reported values (10.7% in chicken and 11.3% in zebra finch)30. This large variation in the proportion of genes showing evidence of positive selection within a particular lineage is partly attributed to alignment problems and poor sequence quality31,32. A comparison across species suggested that the proportion of positively selected genes in duck was in the range of the proportions detected in high-quality genomes, such as those of the chicken (2.7%) and zebra finch (6.2%). This observation, along with reports from the assessment of the quality of the duck draft genome (Supplementary Note), encouraged us to further investigate the biological functions of positively selected genes in the duck. Ingenuity Systems Pathway Analysis (IPA) showed that positively selected genes in duck were enriched in cellular assembly and organization, cellular function and maintenance, and cell signaling. In addition, genes related to amino acid metabolism and small-molecule biochemistry tended to be under positive selection in duck (Supplementary Table 11).
Gene profiles after avian influenza virus infections
We examined global gene expression profiles using seven lung transcriptomes of ducks infected with H5N1 viruses and control individuals (Table 1). Alignment of approximately 916 million Illumina paired-end reads with the merged reference gene set suggested that between 17,951 and 18,276 genes were expressed in these lung tissues, and 16,404 genes were transcribed in all 7 lung tissues. In general, the overall gene expression patterns of DK/49-infected animals 1–2 d after inoculation were similar to those of GS/65-infected animals 1 d after inoculation, whereas the gene expression profiles of DK/49-infected animals 3 d after inoculation were similar to those of GS/65-infected animals 2–3 d after inoculation (Fig. 2a). Compared to control animals, DK/49-infected ducks had 2,257, 3,101 and 3,066 genes with significantly altered expression (FDR ≤0.001, fold change ≥2) 1–3 d after inoculation, and GS/65-infected ducks had 916, 2,060 and 1,251 genes with significantly altered expression (FDR ≤0.001, fold change ≥2) 1–3 d after inoculation (Table 1). These findings, together with hierarchical clustering analysis, showed an appreciably more dynamic response during infection with GS/65 virus compared to DK/49 virus. Further comparison suggested that 1,506, 1,436 and 1,396 genes had significantly different expression levels (FDR ≤0.001, fold change ≥2) in DK/49-infected ducks and GS/65-infected ducks 1–3 d after inoculation, respectively (Table 1 and Supplementary Fig. 8). Compared to the duck reference gene set (with 5.2% predicted to be under positive selection), these sets of differentially expressed genes were predicted to have a slightly higher proportion of genes (5.4–7.0%) under positive selection (Table 1), supporting the hypothesis that ducks might alter their sensitivity to avian influenza viruses through the positive selection of many genes evolving in the host response to these viruses33,34.
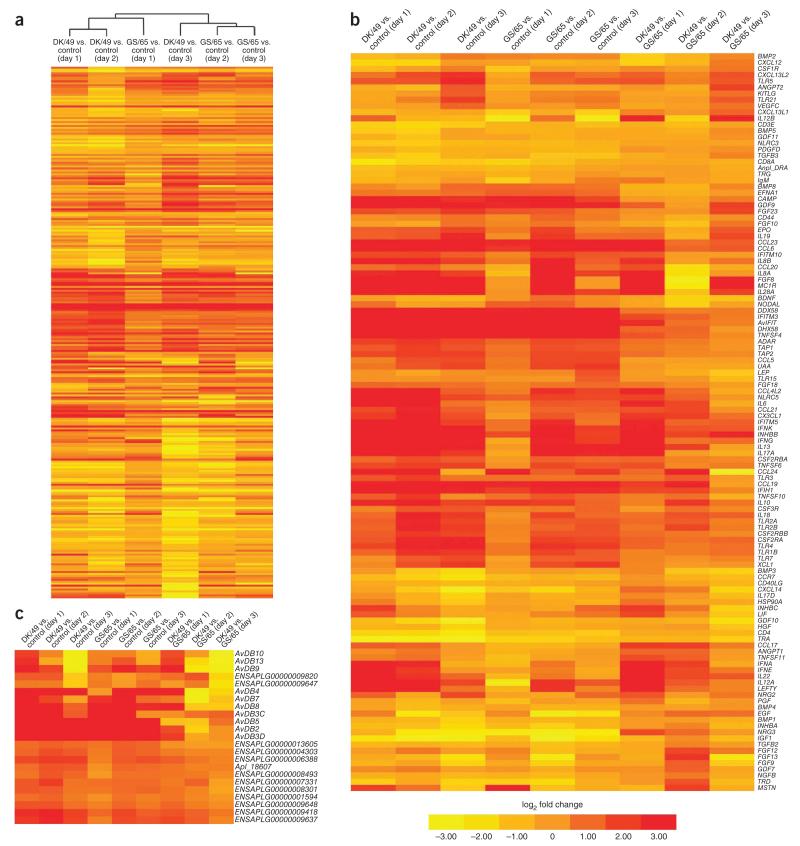
Figure 2.
Identification of genes responsive to influenza A viruses in the lungs of ducks infected with one of two H5N1 viruses on days 1, 2 and 3 after inoculation. The genes included here showed significant differences in gene expression (FDR ≤0.001, fold change ≥2) in at least one experiment. Genes shown in red had upregulated expression, and those shown in yellow had downregulated expression in infected ducks relative to controls or in DK/49-infected relative to GS/65-infected ducks. (Full gene names are given in supplementary table 13.) Hierarchical clusters of genes and samples were based on Pearson’s correlation and Spearman’s rank correlation analyses, respectively. (a) Overall gene expression profiles in DK/49- or GS/65-infected ducks compared to control animals. The heatmap was generated from hierarchical cluster analyses of both genes and samples. (b) Expression of 119 innate immune genes in DK/49- or GS/65-infected ducks. The heatmap was generated from hierarchical analysis of genes, showing significant changes in gene expression for 119 innate immune genes in DK/49- or GS/65-infected ducks 1–3 d after inoculation. (c) Expression of two significantly expanded gene families (β-defensins and BTNLs) in DK/49- or GS/65-infected ducks. The heatmap was generated from hierarchical analysis of genes, showing that most of the avian defensin and BTNL genes, including two LSDs of AvDB3 and eight LSDs of BTNL genes, have significantly altered gene expression in DK/49- or GS/65-infected ducks 1–3 d after inoculation.
We merged the sets of genes determined to have differential expression in DK/49- or GS/65-infected ducks compared with control animals into set 1 (5,038) and set 2 (2,741), respectively, and we combined the genes with differential expression in DK/49- and GS/65-infected ducks into set 3 (3,232) (Table 1). IPA of sets 1 and 2 identified four enriched categories related to cell activation and one enriched category associated with antigen presentation (P < 0.0005; Supplementary Table 12), suggesting that ducks infected with DK/49 or GS/65 virus both have severe disruption of cellular functions. IPA of set 3 also identified three enriched categories related to cell activation (cellular movement, cellular growth and proliferation, and cellular development), as well as one associated with molecular transport and one related to lipid metabolism (Supplementary Table 12).
Innate immune response in avian influenza virus infections
Transcriptome analysis of 150 cytokines (listed in Table 2) showed that, compared to control ducks, ones infected with DK/49 or GS/65 had 74 cytokines with expression levels that were significantly changed (FDR ≤ 0.001, fold change ≥ 2) 1–3 d after inoculation (Fig. 2b; full gene names are given in Supplementary Table 13). Of these cytokines, 20 growth factor genes (BMP1–BMP5, EGF, FGF9, FGF12, FGF13, GDF10, GDF11, HGF, IGF1, INHBA, NGFB, NRG3, PDGFD, PGF, TGFB2 and TGFB3) had expression that was significantly decreased by 2.0- to 9.8-fold, and 13 growth factor genes (BMP8, EFNA1, FGF8, FGF18, FGF23, GDF9, INHBB, INHBC, KITLG, MSTN, NODAL, NRG2 and VEGFC) had expression that was significantly increased by 2.1- to 379-fold with DK/49 or GS/65 infection. Similarly, one tumor necrosis factor (TNF) gene (TNFSF11), one interleukin (IL)-17 gene (IL17D) and three CXC chemokine genes (CXCL12, CXCL13L2 and CXCL14) had expression that was markedly decreased by 2.2- to 5.8-fold, whereas one IL-17 gene (IL17A), three TNF genes (TNFSF4, TNFSF6 and TNFSF10) and four CXC chemokine genes (CX3CL1, CXCL13, IL8A and IL8B) had expression that was markedly increased by 2.3- to 34-fold after infection with DK/49 and GS/65. However, all five interferon genes (IFNA, IFNE, IFNG, IFNK and IL28A), one C chemokine gene (XL1), nine CC chemokine genes (CCL4L2, CCL5, CCL6, CCL17, CCL19, CCL20, CCL21, CCL23 and CCL24) and ten interleukin or interleukin receptor genes (IL1, IL6, IL10, IL13, IL12A, IL12B, IL19, IL22, LEP and LIF) had expression that was markedly upregulated by 2.1- to 1,414-fold in DK/49- or GS/65-infected ducks compared with control ducks. Compared with GS/65-infected ducks, DK/49-infected ducks had 7 cytokine genes with expression that was elevated by 2.4- to 46-fold 1–2 d after inoculation that then had expression decreased by 2.3- to 7-fold 2–3 d after inoculation, 12 cytokine genes with expression that was significantly downregulated (by 2.0- to 4.2-fold) and 38 cytokine genes with expression that were significantly upregulated (by 2.0- to 1,414-fold) 1–3 d after inoculation (FDR ≤0.001, fold change ≥2; Fig. 2b).
DDX58, IFITM3 and IFIT1–IFIT3 have key roles in the antiviral response to avian influenza virus infection35–37 in mammals. Transcriptome analysis showed that the expression levels of the DDX58, IFITM3 and AvIFIT genes (the gene name of AvIFIT is given in Supplementary Fig. 9) in both DK/49- and GS/65-infected ducks were markedly increased by 6.9- to 440-fold 1–3 d after inoculation, with peak expression (increased by 12.5- to 440-fold) at 2 d, compared with control ducks (Fig. 2b). Similar to DDX58, two additional RNA helicases (ADAR and DHX58) showed expression that was significantly elevated by 2.1- to 30-fold 1–3 d after inoculation, with peak expression (increased by 3.2- to 30-fold) at 2 d, in both DK/49- and GS/65-infected ducks compared with control ducks, indicating that these three RNA helicases have key roles in the host response to avian influenza viruses in duck. Moreover, similar to AvIFIT, which had altered gene expression in infected ducks, the genes for three additional interferon-induced proteins (IFIH1, IFITM5 and IFITM10) showed significantly different expression levels during infection with DK/49 virus 1–3 d after inoculation, with peak expression at 2 d. Pronounced changes in gene expression for IFIH1, IFITM5 and IFITM10 were observed in the GS/65-infected ducks 2 d after inoculation, whereas only minor changes in IFITM5 and IFITM10 expression were detected in GS/65-infected ducks at 1 and 3 d after inoculation (Fig. 2b). Toll-like receptors (TLRs) are involved in host-virus interactions and lead to the secretion of interferons by infected cells. Consistent with the changes observed in the expression of the five interferon genes, nine TLR genes, including two members not found in mammals (TLR15 and TLR21), had expression that was significantly increased by 2.0- to 7.5-fold in DK/49- or GS/65-infected ducks 1–3 d after inoculation compared with control ducks (Fig. 2b). However, the immunoglobulin M (IgM) locus, three T cell receptors genes (TRA, TRD and TRG) and four genes encoding CD molecules (CD3E, CD4, CD8A and CD40LG) showed significantly decreased expression in DK/49- or GS/65-infected ducks compared with control ducks (FDR ≤0.001, fold change ≥2). In addition, three major histocompatibility complex (MHC) genes (Anpl-DRA, TAP1 and TAP2) and four colony-stimulation factor receptor genes (CSF2RA, CSF2RBA, CSF2RBB and CSF3R) had significantly elevated expression in DK/49- or GS/65-infected ducks compared with control ducks (FDR ≤0.001, fold change ≥2; Fig. 2b).
LSDs of immune genes responsive to avian influenza viruses
Mammalian defensins have been proposed to contribute to the immune response to avian influenza virus infection. In mice, the expression of β-defensins 3 and 4 is significantly higher in avian influenza virus–infected airways38, and, in humans, β-defensins inhibit avian influenza virus replication and increase the uptake of these viruses by neutrophils39. Similarly, key roles for BTNLs in immune responses have been extensively reported in mammals: four BTNLs (BTNL1, BTNL2, BTNL4 and BTNL6) can attenuate T cell activation and antagonize pathological inflammatory T cell infiltrates28,40. However, the functions of avian defensins and BTNLs in immune responses to influenza viruses in birds are uncertain.
Transcriptome analysis indicated that eight avian defensin genes (AvDB2, AvDB3C, AvDB3D, AvDB4, AvDB5, AvDB7, AvDB8 and AvDB9), including two LSDs of these genes (AvDB3C and AvDB3D), had expression that was markedly increased by 7.6- to 1,551-fold (FDR ≤0.001, fold change ≥2) in DK/49- or GS/65-infected ducks compared with controls. Unexpectedly, AvDB10 and AvDB13 showed expression that was significantly elevated by 3.9- to 5.1-fold 1 d after inoculation that returned to normal, basal levels at 2 d and was downregulated by 8.6- to 12-fold at 3 d in DK/49-infected ducks. However, such pronounced changes in AvDB10 and AvDB13 expression were not detected 1–3 d after inoculation in GS/65-infected ducks, where significant change in gene expression was only observed for AvDB13 2 d after inoculation (Fig. 2c). Compared with GS/65-infected ducks, DK/49-infected ducks had three avian defensin genes (AvDB2, AvDB4 and AvDB9) with expression that was significantly increased by 3.1- to 971-fold 1 d after inoculation, and the reverse was true (with expression significantly decreased by 3.1- to 971-fold) 2 d after inoculation (Fig. 2c). Moreover, two avian defensin genes (AvDB3D and AvDB8) had expression that was significantly elevated by 4.1- to 8.9-fold 1 d after inoculation, and one defensing gene (AvDB7) had expression that was significantly decreased by 9.8-fold at 2 d in DK/49-infected ducks compared with GS/65-infected ducks (Fig. 2c). Of the 17 BTNL genes, 11 genes, including 8 LSDs, showed expression that was markedly elevated by 2.0- to 7.5-fold 1–3 d after inoculation in DK/49- or GS/65-infected ducks compared with control individuals. In comparison with GS/65-infected ducks, DK/49-infected ducks had one BTNL gene with expression that was markedly decreased by 4.5-fold and five BTNL genes, including three LSDs, with expression that was substantially increased by 2.1- to 3.2-fold (Fig. 2c).
DISCUSSION
We draw four noteworthy conclusions from our results. First, using next-generation sequencing technology, we generated the first draft sequence for a waterfowl, one which is a natural host of avian influenza viruses. Second, we showed that immune-related gene repertoires in three bird lineages (including 3,116–3,355 genes) seemed to be contractive compared to equivalent repertories in mammals (with 5,715–6,044 genes). Further efforts identified about 150 cytokines in three bird species using assemblies in Ensembl57 and the NCBI non-redundant database (as of November 2012), whereas there were about 220 cytokines in 2 mammal species (Table 2). Third, we performed deep transcriptome analysis to characterize gene expression profiles and to identify genes that are responsive to avian influence viruses (for example, AvIFT, AvDB7–AvDB10, IFITM5 and IFITM10). Fourth, we found that some LSDs of avian defensin and BTNL genes might be involved in host immune response to both of the two H5N1 viruses whose infection was examined in ducks.
The duck genome possesses a contractive immune-related gene repertoire similar to those of the chicken and zebra finch, and it includes genes that are not present in the other three species whose genomes have been sequenced (Fig. 1). In the analyses presented here, we found that many genes (for example, BTNLs and defensins) were independently duplicated in the duck but not in the chicken genome. These results suggest that gene gain and loss have influenced the divergence of the four avian genomes and the evolution of their respective immune systems. β-defensins are induced in response to influenza virus infection38, and BTNLs are involved in T cell activation and infiltration in mammals28,40. Notably, most of the defensin and BTNL genes, including two LSDs of AvDB3 and eight LSDs of BTNL genes, may be implicated in the host immune response to influenza in duck; therefore, a functional analysis of the defensins and BTNLs of birds will be of interest to the study of avian influenza virus infection. Moreover, the duck seems to benefit from positive selection on specific genes functioning in the host-virus interaction. This presence of this benefit is supported by the detection of slightly higher frequencies of positively selected genes in the sets of differentially expressed genes identified following infection with H5N1 viruses compared to the duck reference gene set reported in this study. The protein sequences of the two H5N1 viruses investigated in this study are highly conserved, with the exception of 20 amino acid alterations distributed over 7 genes; however, one virus is highly pathogenic (DK/49), and the other is weakly pathogenic (GS/65) in ducks13. Notably, the optimized immune system of ducks can be overcome by the highly pathogenic H5N1 virus but not by the weakly pathogenic H5N1 virus. This distinction identifies disruptions in the long-standing equilibrium between ducks and avian influenza viruses. Our future ability to assess the functions of genes showing significantly different expression induced by highly pathogenic H5N1 viruses compared with weakly pathogenic H5N1 viruses, using genetic manipulations and co-evolutionary analyses, will certainly extend knowledge of the avian genes related to influenza in birds.
ONLINE METHODS
Sequence assembly
We constructed paired-end DNA libraries with insert size smaller and larger than 2 kb according to the manuals for the standard DNA and mate-pair libraries, respectively (Illumina). The sequencing process followed the manufacturer’s recommendations. After removing duplicate reads introduced by PCR, base calling and adaptor sequence contained in the raw reads, we assembled the duck genome with SOAPdenovo14.
Gene evolution
We created a reference gene set by merging the homology set, the de novo set and the GLEAN set. We built gene families using the Ensembl pipeline41 with genomes in Ensembl59 in addition to the duck and turkey genomes, and we subsequently estimated the change of gene complements using the CAFÉ (computational analysis of gene family evolution) tool21. We predicted the positively selected genes using the codeml program under the branch-site model29. We constructed maximum-likelihood trees with protein sequences using PHYML version 2.4.4 under the JTT model with four substitution rate classes42.
Facility
Studies of the H5N1 viruses (DK/49 and GS/65) were conducted in a biosecurity level 3+ laboratory that was approved by the Chinese Ministry of Agriculture. All animal studies were approved by the Review Board of the Harbin Veterinary Research Institute, Chinese Academy of Agriculture Sciences.
Viruses, duck infection and RNA extraction
The DK/49 and GS/65 H5N1 viruses were isolated from a duck and a goose, respectively, during the avian influenza outbreak of 2005 in China43. The viruses were propagated in 10-d-old fertilized chicken eggs. Two groups of 4-week-old specific pathogen–free (SPF) Shaoxin ducks from the Harbin Veterinary Research Institute, China Academy of Agricultural Science, were inoculated intranasally with 103 of 50% egg infectious doses (EID50) of the DK/49 and GS/65 viruses after adapted to a biosecurity level 3+ environment for 4 d. The lungs were collected from the above H5N1 virus–infected ducks at days 1, 2 and 3 after inoculation and from uninfected 4-week-old SPF ducks (n = 3, except n = 2 for the GS/65-infected ducks 2 d after inoculation, and n = 1 for the DK/49-infected ducks 3 d after inoculation).
Total RNA was extracted from approximately 100 mg of each lung tissue sample using the Qiagen RNeasy kit. RNA concentration and quality were measured using an Agilent 2100 Bioanalyzer, showing that all RNA samples had an RNA integrity number (RIN) of >7.3 and a ratio of 28S:18S rRNA of >1.0. Subsequently, each pooled lung RNA sample for DK/49-infected ducks 1–2 d after inoculation, GS/65-infected ducks 1–3 d after inoculation and control ducks was separately prepared from equal masses of two or three individuals.
RNA sequencing
cDNA libraries were prepared according to the manufacturer’s instructions (Illumina). One mRNA sample of the lung collected from a duck infected with the DK/49 virus 3 d after inoculation and six pooled lung RNA samples were purified from total RNA using Dynal Oligo(dT) beads and fragmented into small pieces of approximately 200 nt using RNA Fragmentation Reagents (Ambion). Cleaved mRNA fragments were reverse transcribed into single cDNAs using SuperScript II (Invitrogen) and were primed with random primers; double-stranded cDNA was then synthesized using RNase H (Invitrogen) and DNA Pol I (Invitrogen). Subsequently, cDNA was subjected to end repair and phosphorylation using Klenow polymerase (Enzymatics), T4 DNA polymerase (Enzymatics) and T4 polynucleotide kinase (to blunt end the DNA fragments) (Enzymatics). End-repaired cDNA fragments were 3′ adenylated using Klenow (exo–) DNA polymerase (Enzymatics). Then, Illumina paired-end adapters were ligated to the ends of these 3′-adenylated cDNA fragments. Gel electrophoresis was used to separate the cDNA fragments from any unligated adapters. cDNA fragments from 180–220 bp in size were selected. cDNA libraries were amplified using 12 cycles of PCR with Phusion polymerase (NEB), and 75-cycle paired-end sequencing was performed on the Illumina Genome Analyzer.
Transcriptome analysis
We clustered the BGI and Ensembl reference gene sets (Supplementary Note) and the duck transcripts deposited in the NCBI database to create a merged reference gene set consisting of 20,647 protein-coding genes. We aligned the high-quality reads to the genome and the merged reference gene set using SOAPaligner with a threshold of five mismatches. For multiposition hits, one of the best matching loci was chosen randomly. Only uniquely mapped reads were used for the analysis of gene expression levels. Differentially expressed genes were identified using Fisher’s exact test44 and Bonferroni-Hochberg correction (FDR ≤0.001, fold change ≥2)45. Differentially expressed genes identified in DK/49- or GS/65-infected ducks compared with control ducks were merged to create sets 1 and 2. Differentially expressed genes identified in DK/49-infected ducks compared with GS/65-infected ducks were combined into set 3 (Table 1). The three sets of differentially expressed genes were used to investigate biological processes using IPA.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank S. Hu and Q. Li for duck sample collection. We also thank S. Zou and X. Liu for assistance in mRNA purification. The sequencing of the duck genome was funded by the National High Technology Research and Development Program of China (2010AA10A109), the National Key Technology Support Program of China (2008BADB2B08) and the Recommend International Advanced Agricultural Science and Technology 948 Program (2012-G1 and 2013-G1(2)) of the Ministry of Agriculture of China.
Footnotes
URLs. Cumulative number of confirmed human cases of avian influenza A (H5N1) infection reported to the World Health Organization,http://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/index.html; influenza activity in the United States and worldwide, 2004–2005 season, http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5425a3.htm; Narcisse, http://narcisse.toulouse.inra.fr/multi/cgi-bin/narcisse.cgi; InnateDB, http://www.innatedb.com/index.jsp; Ingenuity Systems Pathway Analysis (IPA), http://www.ingenuity.com/.
Accession codes. Duck genome assembly and reads have been deposited under GenBank accession PRJNA46621. Transcript sequencing data have been deposited under GenBank Gene Expression Omnibus (GEO) accession GSE22967 and Short Read Archive (SRA) accession PRJNA194464.
Note: Supplementary information is available in the online version of the paper.
COMPETING FINANCIAL INTERESTS The authors declare no completing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 2.Warren WC, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalloul RA, et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 2010;8:e1000475. doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hackett SJ, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- 5.Olsen B, et al. Global patterns of influenza A virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 6.Wilcox BR, et al. Influenza-A viruses in ducks in northwestern Minnesota: fine scale spatial and temporal variation in prevalence and subtype diversity. PLoS ONE. 2011;6:e24010. doi: 10.1371/journal.pone.0024010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munster VJ, et al. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 2007;3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sturm-Ramirez KM, et al. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J. Virol. 2005;79:11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imai M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmissio to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin YP, et al. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA. 2000;97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belser JA, et al. Contemporary North American influenza H7 viruses possess human receptor specificity: implications for virus transmissibility. Proc. Natl. Acad. Sci. USA. 2008;105:7558–7563. doi: 10.1073/pnas.0801259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butt KM, et al. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J. Clin. Microbiol. 2005;43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song J, et al. The PA protein directly contribute to the virulence of H5N1 avian influenza viruses in domestic ducks. J. Virol. 2011;85:2180–2188. doi: 10.1128/JVI.01975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li R, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, et al. A genetic and cytogenetic map for the duck (Anas platyrhynchos) Genetics. 2006;173:287–296. doi: 10.1534/genetics.105.053256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skinner BM, et al. Comparative genomics in chicken and Pekin duck using FISH mapping and microarray analysis. BMC Genomics. 2009;10:357. doi: 10.1186/1471-2164-10-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, et al. Molecular evolution of the vertebrate TLR1 gene family—a complex history of gene duplication, gene conversion, positive selection and co-evolution. BMC Evol. Biol. 2011;11:149. doi: 10.1186/1471-2148-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Courcelle E, et al. Narcisse: a mirror view of conserved syntenies. Nucleic Acids Res. 2008;36:D485–D490. doi: 10.1093/nar/gkm805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(suppl. 2):ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 21.Hahn MW, De Bie T, Stajich JE, Nguyen C, Cristianini N. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 2005;15:1153–1160. doi: 10.1101/gr.3567505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn MW, Han MV, Han SG. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 2007;3:e197. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahn MW, Demuth JP, Han SG. Accelerated rate of gene gain and loss in primates. Genetics. 2007;177:1941–1949. doi: 10.1534/genetics.107.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao Y, et al. A genome-wide screen identifies a single β-defensin gene cluster in the chicken: implications for the origin and evolution of mammalian defensins. BMC Genomics. 2004;5:56. doi: 10.1186/1471-2164-5-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellgren O, Ekblom R. Evolution of a cluster of innate immune genes (β-defensins) along the ancestral lines of chicken and zebra finch. Immunome Res. 2010;6:3. doi: 10.1186/1745-7580-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bas A, et al. Butyrophilin-like 1 encodes an enterocyte protein that selectively regulates functional interactions with T lymphocytes. Proc. Natl. Acad. Sci. USA. 2011;108:4376–4381. doi: 10.1073/pnas.1010647108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 30.Nam K, et al. Molecular evolution of genes in avian genomes. Genome Biol. 2010;11:R68. doi: 10.1186/gb-2010-11-6-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider A, et al. Estimates of positive Darwinian selection are inflated by errors in sequencing, annotation, and alignment. Genome Biol. Evol. 2009;1:114–118. doi: 10.1093/gbe/evp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fletcher W, Yang Z. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol. Biol. Evol. 2010;27:2257–2267. doi: 10.1093/molbev/msq115. [DOI] [PubMed] [Google Scholar]
- 33.Rothenburg S, Seo EJ, Gibbs JS, Dever TE, Dittmar K. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat. Struct. Mol. Biol. 2009;16:63–70. doi: 10.1038/nsmb.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barber MR, Aldridge JR, Jr., Webster RG, Magor KE. Association of RIG-I with innate immunity of ducks to influenza. Proc. Natl. Acad. Sci. USA. 2010;107:5913–5918. doi: 10.1073/pnas.1001755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pichlmair A, et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 37.Everitt AR, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chong KT, Thangavel RR, Tang X. Enhanced expression of murine β-defensins (MBD-1, -2,- 3, and -4) in upper and lower airway mucosa of influenza virus infected mice. Virology. 2008;380:136–143. doi: 10.1016/j.virol.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 39.Doss M, et al. Interactions of α-, β-, and θ-defensins with influenza A virus and surfactant protein D. J. Immunol. 2009;182:7878–7887. doi: 10.4049/jimmunol.0804049. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen T, Liu XK, Zhang Y, Dong C. BTNL2, a butyrophilin-like molecule that functions to inhibit T cell activation. J. Immunol. 2006;176:7354–7360. doi: 10.4049/jimmunol.176.12.7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilella AJ, et al. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 43.Chen H. H5N1 avian influenza in China. Sci. China C Life Sci. 2009;52:419–427. doi: 10.1007/s11427-009-0068-6. [DOI] [PubMed] [Google Scholar]
- 44.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 45.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.