Abstract
Identifying adaptive genetic variation is a challenging task, in particular in non-model species for which genomic information is still limited or absent. Here, we studied distribution patterns of amplified fragment length polymorphisms (AFLPs) in response to environmental variation, in 13 alpine plant species consistently sampled across the entire European Alps. Multiple linear regressions were performed between AFLP allele frequencies per site as dependent variables and two categories of independent variables, namely Moran’s eigenvector map MEM variables (to account for spatial and unaccounted environmental variation, and historical demographic processes) and environmental variables. These associations allowed the identification of 153 loci of ecological relevance. Univariate regressions between allele frequency and each environmental factor further showed that loci of ecological relevance were mainly correlated with MEM variables. We found that precipitation and temperature were the best environmental predictors, whereas topographic factors were rarely involved in environmental associations. Climatic factors, subject to rapid variation as a result of the current global warming, are known to strongly influence the fate of alpine plants. Our study shows, for the first time for a large number of species, that the same environmental variables are drivers of plant adaptation at the scale of a whole biome, here the European Alps.
Keywords: adaptation, climate change, environmental association analysis, genome scan, landscape genetics, loci of ecological relevance
Introduction
Detecting adaptive genetic variation in response to environmental variation helps to better understand the potential of organisms for rapid evolutionary adaptation (Hoffmann & Willi 2008). However, identifying adaptive genetic variation is challenging, in particular in non-model species for which genomic information is still limited or absent (Stinchcombe & Hoekstra 2008). Nevertheless, technical advances in genomic screening for numerous molecular loci spread over the genomes of many individuals allow us to identify genetic variation linked to the environment even in non-model organisms. Such environmental association studies are based on the relationship of the frequencies of alleles at particular loci with the variation in particular environmental variables (Manel et al. 2010a). The underlying assumption is that natural selection along environmental gradients generates gradual changes in allele frequencies at loci physically linked to adaptive genes (Haldane 1948; Endler 1986; Schmidt et al. 2008). The distribution of alleles at loci of ecological relevance is thus different from the distribution of alleles at neutral loci (Holderegger et al. 2010).
Patterns of genetic variation that seem to be caused by natural selection may in fact also result from historical demographic processes (Excoffier et al. 2009; Siol et al. 2010). First, isolation by distance may limit gene flow among populations over a large scale, and the frequency of neutral alleles will change simply as a result of genetic drift (Wright 1938). Second, contact and admixture zones, where populations that diverged in spatial isolation (e.g. glacial survival in different refugia) come into secondary contact, could confound adaptive genetic patterns (Endler 1977). Third, bottlenecks and inbreeding create patterns that mimic those of selective sweeps (Storz 2005). Searching for congruent patterns of adaptive loci across replicated regions represents one way of limiting the confounding effects of historical demographic and spatial processes, as it is unlikely that the latter produced similar genetic patterns at a given locus across independent environmental gradients (e.g. Poncet et al. 2010; Buehler et al. unpublished data). In this study, we argue that finding similar patterns of correlation between loci and ecological factors in different species within the same vast study area constitutes another way of avoiding confounding effects, as it is unlikely that different species are subject to the same demographical processes.
Previous studies in plant species found distinct patterns of genetic variation in allozyme frequencies along environmental gradients (Allard et al. 1993; Linhart & Grant 1996; Prentice et al. 2000). For example Hirao & Kudo (2004) found a correlation between allozyme frequencies and flowering time in Primula cuneifolia along a snowmelt gradient. Shimono et al. (2009) detected morphological traits co-varying with allozyme frequencies in fellfield and snowbed populations of Potentilla matsumurae. Their results suggest that the timing of snowmelt causes a selective pressure that drives local adaptation in this alpine plant species (but see Stanton & Galen 1997). Recent technological advances in molecular markers have made larger genome scans feasible, more powerful and with high genomic resolution, facilitating the identification of loci that are potentially of ecological relevance (Manel et al. 2010a). For example Poncet et al. (2010) studied 825 amplified fragment length polymorphism (AFLP) loci at 208 locations across the European Alps in Arabis alpina. They detected four AFLP loci, common in two independent regions of the French and Swiss Alps, which are linked to mean annual temperature and/or precipitation. Recent studies at the whole genome level also confirmed the existence of loci under selection by climatic factors in the model plant Arabidopsis thaliana (Fournier-Level et al. 2011; Hancock et al. 2011a).
Here, we present patterns of AFLP allele distributions in response to environmental variation in 13 alpine plant species collected across the entire range of the European Alps following a stratified sampling design (Gugerli et al. 2008). Our objective was to (i) identify topographic and climatic factors potentially involved in the adaptation of alpine plants to their environment, to derive testable hypotheses about the mechanisms underlying patterns of adaptive genetic variation (Feder & Mitchell-Olds 2003). Once the important environmental factors were identified per species, we addressed the question of (ii) whether there are common patterns of genetic adaption to environmental variables across alpine plants. To date, published studies have largely focused on single species, preventing the detection of general patterns of adaptation to environmental factors (e.g. Jump & Penuelas 2005; Joost et al. 2007; Segelbacher et al. 2010; Cox et al. 2011). In contrast, multi-species studies, such as the one we provide here, make it possible to draw general conclusions about the environmental factors involved in the adaptive response of species.
Material and method
Plant collection and genotyping
The sampling and genotyping of the 13 alpine plant species studied were carried out according to the methods described in detail in Gugerli et al. 2008; (Table 1, Fig. 1). All samples were collected within a single growing season (June–September 2004). Sampling was standardized across species by using a regular grid with a cell size of 20′ longitude by 12′ latitude across the whole area of the European Alps. Sampling was limited to those cells comprising area above 1500 m a.s.l., and only every second cell was considered. Within each grid cell considered, we arbitrarily chose one location per species based on known occurrences or as expected from species-specific habitat requirements, optimizing field work by searching for areas containing many species across just a few locations within each cell (Gugerli et al. 2008). The number of locations ranged from 74 locations for Gentiana nivalis to 137 for Carex sempervirens. Leaves were collected from three plants per sampling location. Despite the small number of specimens sampled per location, data were considered informative given the main objectives of our study, i.e. identifying broad-scale patterns of general adaptation. At the same time, the large number of sampling locations distributed over a large ecological and altitudinal range per species resulted in a broad variety of environmental conditions (Thiel-Egenter et al. 2009). All samples were genotyped using AFLPs (Vos et al. 1995) with three primer–enzyme combinations (for details, seeGugerli et al. 2008). The primer–enzyme combinations were species specific. Thus, the AFLP loci amplified were clearly independent across species.
Table 1. Alpine plant species (with family) analysed at amplified fragment length polymorphism (AFLP) loci and tested for relations between AFLP allele frequencies and environmental variation.
For each species, the laboratory responsible for AFLP genotyping is indicated. Number of sampling sites and individuals genotyped, Nei’s (1973) gene diversity averaged over all sampling sites (± standard error, SE) and overall genetic differentiation (ΦST) are given for each study species. Ntot, total number of loci; Nler, number of loci of ecological relevance; NMEM, number of broad-scale Moran’s eigenvector map (MEM) variables
| Species | Sampling sites |
Individuals genotyped |
Gene diversity | Φ ST | N tot | N ler | N MEM |
|---|---|---|---|---|---|---|---|
| (1) Arabis alpina L. (UJF)* [Brassicaceae] | 129 | 385 | 0.061 ± 0.004 | 0.664 | 140 | 20 | 22 |
| (2) Campanula barbata L. (UNE)† [Campanulaceae] | 104 | 307 | 0.107 ± 0.004 | 0.385 | 114 | 13 | 16 |
| (3) Carex sempervirens Vill. (WSL)‡ [Cyperaceae] | 137 | 408 | 0.083 ± 0.002 | 0.328 | 122 | 3 | 31 |
| (4) Dryas octopetala L. (UJF) [Rosaceae] | 124 | 370 | 0.123 ± 0.003 | 0.197 | 99 | 4 | 24 |
| (5) Gentiana nivalis L. (UNE) [Gentianaceae] | 74 | 218 | 0.079 ± 0.006 | 0.600 | 157 | 16 | 10 |
| (6) Geum montanum L. (WSL) [Rosaceae] | 122 | 363 | 0.091 ± 0.003 | 0.313 | 85 | 12 | 24 |
| (7) Gypsophila repens L. (WSL) [Caryophyllaceae] | 107 | 319 | 0.110 ± 0.003 | 0.238 | 94 | 2 | 22 |
| (8) Juncus trifidus L. (WSL) [Juncaceae] | 91 | 269 | 0.126 ± 0.005 | 0.289 | 86 | 8 | 14 |
| (9) Loiseleuria procumbens (L.) Desv. (UJF) [Ericaceae] | 90 | 270 | 0.188 ± 0.004 | 0.293 | 116 | 22 | 16 |
| (10) Phyteuma hemisphaericum L. (UV)§ [Campanulaceae] | 76 | 225 | 0.112 ± 0.004 | 0.342 | 234 | 24 | 15 |
| (11) Rhododendron ferrugineum L. (UJF) [Ericaceae] | 126 | 377 | 0.135 ± 0.004 | 0.375 | 111 | 24 | 12 |
| (12) Saxifraga stellaris L. (UV) [Saxifragaceae] | 100 | 283 | 0.074 ± 0.003 | 0.428 | 70 | 2 | 22 |
| (13) Sesleria caerulea (L.) Ard. (UCSB)¶ [Poaceae] | 113 | 265 | 0.201 ± 0.009 | 0.196 | 187 | 19 | 21 |
University Joseph Fourier, France.
University de Neuchâtel, Switzerland.
WSL Birmensdorf, Switzerland.
University of Vienna, Austria.
UCSB Piacenza, Italy.
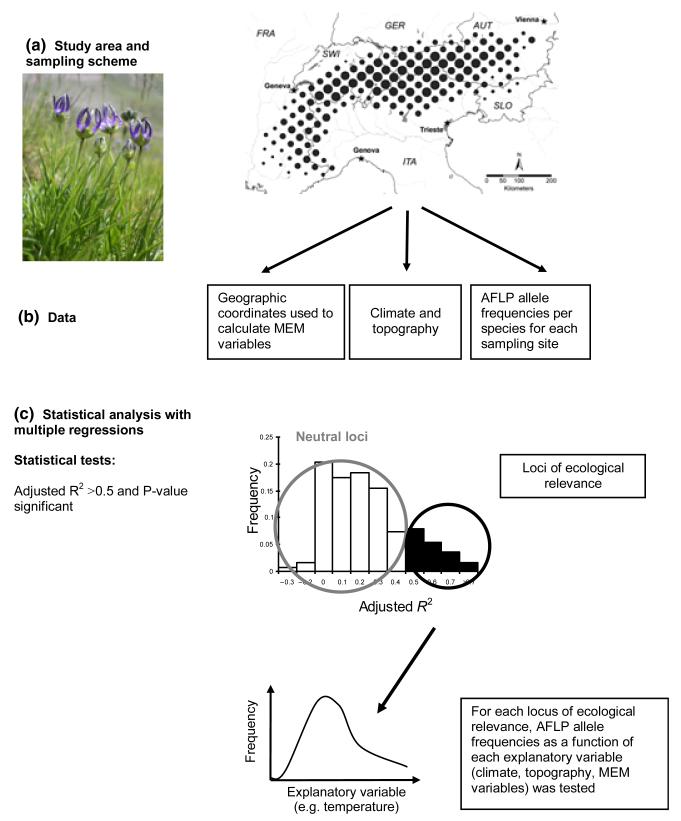
Fig. 1. Approach used in this study to detect loci of ecological relevance in 13 alpine plant species.
(a) Map of the European Alps showing the study range and sampling intensity (circle area proportional to number of species genotyped per grid cell). (AUT, Austria; FRA, France; GER, Germany; ITA, Italy; SLO, Slovenia; SWI, Switzerland). The number of sampling sites per species is given in Table 1. (b) Type of genetic, environmental and spatial data (MEM variables: Moran’s eigenvector maps) used. (c) Schematic of the statistical analyses. The example given in (a) is Phyteuma hemisphaericum, one of the alpine plants studied.
Nei’s gene diversity (Nei 1973), which corresponds to the mean number of pairwise differences between individuals across AFLP loci per sampling site (Kosman 2003), was averaged across sampling sites per species using program ARLEQUIN 3.5.1.3 (Excoffier & Lischer 2010). Overall genetic differentiation per species was estimated with ΦST (Excoffier et al. 1992), assuming Hardy–Weinberg equilibrium at AFLP loci (Bonin et al. 2007) and testing significance based on 1000 bootstrap replicates in ARLEQUIN 3.5.1.3.
Environmental variables
Fourteen monthly and annual environmental variables related to temperature, precipitation, radiation and topography at 200 m resolution were extracted for each sampling location from published GIS topo-climatic layers from 1980 to 1989 (Zimmermann & Kienast 1999; Table 2, Fig. 1). These environmental variables, excluding topographic aspect, were transformed into quadratic orthogonal polynomials to account for non-linear relationships between AFLP allele frequencies and these variables (Legendre & Legendre 2012). Aspect was transformed into sin(aspect) and cos(aspect), rendering this variable appropriate for use in linear regressions.
Table 2. Topo-climatic variables used in this study.
| Yearly or seasonal climate layers (period 1980-1989) |
(1) Annual mean of daily global radiation (horizon/terrain-corrected) [kJ/m2/day] |
| (2) Mean annual precipitation sum [cm] | |
| (3) Summer seasonal precipitation: number of rain days from June to August [mm] | |
| (4) Spring seasonal precipitation: number of rain days from March to May [mm] | |
| (5) Annual degree days above 0 °C from daily climate maps [°C × days] | |
| (6) Number of days with maximum temperature below 0 °C | |
| (7) Annual mean of maximum daily temperatures [°C] | |
| (8) Number of days with minimum temperatures below 0 °C | |
| (9) Annual mean of minimum daily temperatures [°C] | |
| Topography | (10) Slope [%] |
| (11) Integrated topographic exposure map | |
| (12) Potential soil humidity* | |
| (13) Aspect [degree] | |
| (14) Altitude [m] |
Ratio of the upslope contributing region on the tangent of the slope angle (Beven & Kirby 1979).
Moran’s eigenvector map variables
Moran’s eigenvector map (MEM) variables were used as explanatory variables in the regressions, to account for purely spatial and for unmeasured environmental variables. They make it possible to separate and model the spatial patterns comprised in the variations in response data, which in this case are AFLP allele frequencies. Moran’s eigenvector map variables are the eigenvectors of a spatial weighting matrix calculated from the sites’ geographical coordinates (Borcard & Legendre 2002; Dray et al. 2006). Moran’s eigenvector map analysis produces uncorrelated spatial eigenfunctions used to dissect the spatial patterns of the studied variation across a range of spatial scales. The first few MEM variables with large Moran’s I coefficients model broad-scale processes (e.g. genetic variation at large spatial scales such as phylogeographic patterns), whereas subsequent MEM variables with smaller Moran’s I coefficients refer to the spatial autocorrelation generated by processes such as gene flow among sub-populations and genetic drift (Dray et al. 2006). As in our data set the number of sites varied among species, the number of MEM variables was species-specific. For our analysis, we only used the broad-scale MEM variables, i.e. the first half of the MEM eigenfunctions with positive eigenvalues, which model broad-scale spatial variation (Manel et al. 2010b). MEM variables were computed using the ‘PCNM’ R package 2.12.2 (available at http://r-forge.r-project.org/R/?group_id=195). The computation of MEM variables is explained in detail in Borcard et al. (2011; Chapter 7) and Legendre & Legendre (2012; Chapter 14).
Identification of loci of ecological relevance using multiple regression
Our study aimed to find the general responses of AFLP allele frequencies to environmental variation across several species. However, the correlations among environmental variables were species-specific. This meant we could not select only those environmental variables that were uncorrelated before performing single-species multiple regressions. We, therefore, used a two steps-approach:
In the first step, (i), multiple linear regressions were performed between AFLP allele frequencies per site as dependent variables and MEM variables and all environmental variables as independent variables for each of the 13 studied species. This allowed us to identify loci of ecological relevance. In the second step, (ii), these loci putatively under selection were regressed against each of the environmental and MEM variables separately to identify particular variables in the environmental association. By doing so, we were able to assess the relative effects of spatial structure, climate and topography (Fig. 1), although it was not possible to separate these partly interdependent factors fully. This approach is described in more detail below.
-
For each species and each AFLP locus, the allele frequency of band presence (i.e. the frequency of a particular AFLP fragment) per site was regressed simultaneously on 14 environmental factors (Table 2), either untransformed or transformed, for a total of 28 environmental variables tested (i.e. including both linear and quadratic effects, cos and sin aspect), and on MEM variables. Part of the variation accounted for by MEM variables in regression analysis was also explained by the above 28 environmental factors, as the latter could be correlated with MEM variables. The remaining variation explained by MEM variables represented environmental variation that was not modelled by the environmental variables included in the study. We assumed that this broad-scale spatial variation could be partly related to environmental variables that had not been measured in our study, and partly to the historical dynamics of alpine plants (e.g. colonization routes during range contraction and expansion, survival in refugial areas, secondary contact after re-immigration, etc.; Alvarez et al. 2009). Note that in the multiple linear regressions, allele frequencies are discrete rather than continuous variables, with only four possible states (0, 0.33, 0.66 and 1) because only three individuals per site were taken into account.
The significance of the multiple regressions per locus against the environmental and MEM variables was corrected for multiple tests by the Holm correction (Wright 1992). Although the Holm procedure produces a correct experiment-wise error rate, it may still be seen as a liberal criterion because many loci with significant, but small amounts of explained variation will be retained. To make a conservative judgement on the importance of loci of ecological relevance, we therefore also required that a fixed proportion of the variation per locus, measured by (i.e. 50%), was explained by the environmental variables and the broad-scaled MEM variables. The adjusted coefficient of determination, provides unbiased estimates of a response variable’s variation accounted for in a linear model (i.e. explanatory power of variables in multiple regression) (Ohtani 2000). We only considered loci fulfilling both criteria (i.e. significance after accounting for multiple testing and ) as being of ecological relevance.
Because the explanatory variables were not uncorrelated, but species-dependent (Fig. S1, Supporting information), allele frequencies at the identified loci of ecological relevance were then related with each environmental variable and each MEM variables separately in univariate regressions, to estimate the explanatory power provided by each predictor variable.
Results
For the present analysis, a total of 3963 individuals were genotyped for AFLP loci varying in number from 70 in Saxifraga stellaris to 234 in Phyteuma hemisphaericum (Table 1). This produced a total of 1615 AFLP loci analysed across 13 species. The reproducibility of the markers identified was checked with positive controls and 5–10% replicates from DNA isolation to selective polymerase chain reaction. We obtained mismatch error rates of < 5% for all species (Gugerli et al. 2008), which represents a fairly low value for AFLPs (Bonin et al. 2007).
Nei’s gene diversity across AFLP loci averaged across all sampling sites per species ranged between 0.061 for Arabis alpina and 0.201 for Sesleria caerulea (Table 1). All studied species showed significant overall genetic differentiation (ΦST, P < 0.05). Inversely to Nei’s gene diversity, Sesleria caerulea showed the lowest genetic differentiation (0.196), and Arabis alpina revealed the highest genetic differentiation (0.664). These results show that there was ample genetic diversity within and genetic differentiation among the sampling sites in our study species, a prerequisite for the application of environmental association analysis.
The ranges of the environmental variables across the altitudes considered were broad, reflecting the variability of ecological conditions under which populations for all species were sampled in this study (Fig. S2, Supporting information). The number of broad-scaled MEM variables varied from 12 in Geum montanum to 24 in Rhododendron ferrugineum and Phyteuma hemisphaericum (Table 1).
We identified 153 loci (9%) that were significantly related to environmental and MEM variables across the 13 species studied (Table 1). The percentages of loci of ecological relevance varied among species and ranged from 2% in Gypsophila repens to 18% in Rhododendron ferrugineum. Details (i.e. P values and ) of loci identified as being of ecological relevance in multiple regressions are reported in Table S1 (Supporting information).
Univariate regressions showed that the broad-scaled MEM variables were involved in significant relations with allele frequencies in 149 of the 153 AFLP loci identified as being of ecological relevance (Table 3). They were detected as significant predictors for all loci of ecological relevance in six species, and as a major predictor in the seven other species (Tables 1 and 3). After accounting for broad-scale MEM variables, temperature and precipitation were the two major environmental factors related to AFLP allele frequencies at the identified loci of ecological relevance (Table 3). Summer seasonal precipitation was the major environmental factor, involved in 40 significant relations, followed by the number of days with minimum temperatures below 0 °C, involved in 36 significant relations (Table 3). Except for Saxifraga stellaris, for which only altitude was detected as a significant predictor, for all the other species either a precipitation or a temperature variable or both were involved when MEM variables were not the only relevant predictor variables (Table 3). We identified three types of species–environment interaction patterns according to these relations. In three species, precipitation was the major environmental factor affecting AFLP allele frequencies (Campanula barbata, Carex sempervirens and Phyteuma hemisphaericum); in five species, temperature played that role (Arabis alpina, Gypsophila repens, Juncus trifidus, Loiseleuria procumbens and Rhododendron ferrugineum) and, in four species, both precipitation and temperature were involved (Dryas octopetala, Gentiana nivalis, Geum montanum and Sesleria caerulea; Table 3). Topographic variables were only rarely involved in relevant relationships between AFLP allele frequencies and environmental variables (Table 3).
Table 3. Number of significant univariate regressions per species and environmental or MEM variables.
The names of the environmental variables (1–14) are shown in Table 2. Significance values were corrected for multiple tests. If both untransformed and transformed square variables of a particular environmental factor were simultaneously significant in a particular species, we considered only one significant relationship. The same rule was applied to MEM variables. For MEM variables, an AFLP marker associated with any number of the broad-scaled MEM variables was counted as a single relationship. Variables related to precipitations are highlighted by light grey shading and variables related to temperatures by dark grey shading
| Species | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | MEMs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arabis alpina | 0 | 1 | 11 | 0 | 1 | 12 | 12 | 13 | 13 | 0 | 0 | 0 | 0 | 3 | 17 |
| Campanula barbata | 5 | 1 | 1 | 1 | 13 | ||||||||||
| Carex sempervirens | 1 | 3 | |||||||||||||
| Dryas octopetala | 1 | 1 | 1 | 1 | 1 | 1 | 4 | ||||||||
| Gentiana nivalis | 7 | 1 | 0 | 2 | 2 | 1 | 1 | 1 | 1 | 12 | |||||
| Geum montanum | 1 | 3 | 3 | 1 | 2 | 2 | 11 | ||||||||
| Gypsophila repens | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | ||||||
| Juncus trifidus | 3 | 2 | 1 | 3 | 5 | 4 | 8 | ||||||||
| Loiseleuria procumbens | 1 | 6 | 6 | 21 | |||||||||||
| Phyteuma hemisphaericum | 4 | 1 | 16 | 4 | 20 | ||||||||||
| Rhododendron fetrugineum | 7 | 1 | 3 | 6 | 7 | 7 | 1 | 1 | 20 | ||||||
| Saxifraga stellaris | 1 | 2 | |||||||||||||
| Sesleria caerulea | 1 | 2 | 1 | 1 | 1 | 16 | |||||||||
| Total | 0 | 9 | 40 | 27 | 10 | 20 | 24 | 36 | 35 | 1 | 3 | 1 | 0 | 7 | 149 |
Discussion
Considering spatial structure caused by both unaccounted environmental variation and historical demographic processes, we found that environmental factors, mainly temperature and precipitation, influenced allele distributions at nearly 10% of the AFLP loci tested across 13 alpine species from the European Alps. These loci of ecological relevance may be considered as either potentially adaptive or as linked to the genes or genomic regions under selection.
Population-based approaches of Bayesian geographical analysis can be used to test for correlations between allele frequencies and environmental variables, after correcting for background levels of population structure and differences in sample size (Yu et al. 2006; Hancock et al. 2008, 2010, 2011b). However, these approaches can only be applied to cases where a high number of individuals can be sampled in well-defined populations. In addition, they require independent genetic data to estimate population structure. As we only sampled three individuals per species per site, we could not apply these methods and, thus, we used instead the linear regression method described by Manel et al. (2010b). For instance, for most species considered to be relevant in conservation biology regarding climate change, it will neither be possible to sample large numbers of individuals from numerous populations nor to have an independent genetic data set available to test for genetic structure. To consider spatial demographic history, we used MEM variables as complementary explanatory variables. MEMs allowed us to model (i) pure large-scale spatial effects, (ii) environmental effects resulting from unmeasured environmental variables and (iii) spatial effects co-varying with measured environmental variables. However, one cannot fully disentangle these different effects from each other.
After considering spatial effects by MEM variables, the close relationship found between AFLP allele frequencies and temperature and/or precipitation across the 13 species studied (Table 3) strongly suggested that climatic factors are generally involved in affecting (putatively) adaptive genetic variation in alpine plants. This finding confirmed previous single-species studies on various plant species (e.g. Parisod & Bonvin 2008; Richardson et al. 2009; Cox et al. 2011). As we model the spatial effects of the historical dynamics of species using MEM variables and used AFLP loci that are considered being randomly distributed across the genome, it is unlikely that many of the loci identified were false positives (e.g. Richardson et al. 2009; Sork et al. 2010; Bierne et al. 2011). However, the occurrence of false positives cannot be completely ruled out, but should be rather low in this study.
Temperature and precipitation are well known to strongly influence the survival of alpine plants, although their effect may involve complex pathways (Körner 2003). Therefore, these environmental factors are likely to act as major drivers of selective responses in alpine plants (Boyer 1982; Chaves et al. 2003). In concordance with our results that found either or both of these two environmental factors to be important in different species, previous studies have shown the prominent role of temperature and precipitation in the general adaptation of plants (Skot et al. 2002; St Clair et al. 2005; Richardson et al. 2009). The importance of the above two environmental factors in the adaptation of alpine plants has also been found, at different spatial scales, in Arabis alpina (Manel et al. 2010b; Poncet et al. 2010). In Boechera stricta, a wild relative of Arabidopsis thaliana, Lee & Mitchell-Olds (2011) recently emphasized the role of ecological factors versus geographical distance in creating and maintaining adaptive genetic differentiation across a species’ range. However, our study shows, for the first time, and for a large number of species, that the same environmental variables are drivers of plant adaptation at the scale of a whole biome, here the European Alps. One major limitation of this study is that we only tested climatic and topographic factors and that we did not include soil properties, namely calcareous or siliceous bedrock, as environmental factors in this analysis. It is well known that soil properties affect the distribution of alpine plants (Alvarez et al. 2009) and are therefore also likely to play an important role in adaptive evolutionary responses (Körner 2003). However, data for soil variables are currently not available for the entire Alps at the spatial resolution needed for an environmental association analysis.
One contemporary, yet challenging question is whether adaptive evolution can keep pace with the rate and direction of environmental and climate changes induced by human activities (Lavergne et al. 2010). Several studies have shown that species have already shifted their geographic ranges in response to climate change (Walther et al. 2002; Frei et al. 2010), whereas the general potential of species to adapt to rapid environmental change is still being debated (Davis et al. 2005; Reusch & Wood 2007; Jay et al. 2012). Our results identified temperature and precipitation as potential drivers of adaptation. Such information is highly useful in modelling future vegetation dynamics under climate change, which relies on scenarios of ecological change and respective responses of plant communities. As a consequence of the ever-increasing genomic data available as a result of technological progress (Shendure & Ji 2008), it might become possible not only to predict species distributions in response to climate change (Guisan & Thuiller 2005), but also to develop scenarios regarding the spatial distribution of adaptive genetic variation at the whole-genome level in response to changes in temperature and precipitation regimes as proposed by Fournier-Level et al. (2011) and Hancock et al. (2011a). Future steps will be to integrate the results on drivers of genetic adaptation into bioclimatic models and to test the evolutionary and functional relevance of temperature and precipitation in alpine plants under experimental conditions.
The results presented here provide multi-species empirical evidence of genetic variation related to climatic variables. The correlation of temperature and precipitation to signals of adaptation in the alpine species studied here suggested that there should indeed be ample standing genetic variation available in alpine plants based on which adaptation could occur during the course of climate change.
Conclusions
Local adaptation is the only possible response that living organisms have to cope with climate change to avoid extinction, if their dispersal capacity and phenotypic plasticity are insufficient to keep pace with environmental change. So far, the scientific community has largely ignored the potential of adaptive genetic variation as a rapid response to environmental change. By combining genetic and topo-climatic data, we found that loci potentially linked to genes or genomic regions showing adaptive responses to climatic factors are present in alpine plant species. Our approach might allow researchers to assess general patterns of adaptive genetic response to environmental variation at the scale of whole biomes in virtually any group of organisms. This opens new perspectives for understanding the interplay between dispersal and adaptation in the evolutionary response of species to climate change.
Supplementary Material
Acknowledgements
This study was supported by the European Commission (project IntraBioDiv, Framework Programme 6, GOCE-CT-2003-505376) and the Swiss State Secretariat for Education and Research (SER grants 03.0116-1 to F.G. and 03.0116-2 to N.A.). S.M. was supported by the Institut Universitaire de France. F.G. and R.H. received financial support from the CCES Bio-Change project of the ETH domain and the AVE project (Swiss National Science Foundation, CRSI33_127155). N.A. was funded by an Ambizione fellowship from the Swiss National Science Foundation (PZ00P3_126624). W.T. acknowledges support from the European Research Council under the European Community’s Seven Framework Programme FP7/2007-2013 Grant Agreement no. 281422 (TEEMBIO). We thank Sophie Karrenberg, Nigel Yoccoz and three anonymous referees for helpful comments on earlier versions of the manuscript.
Footnotes
Data Accessibility: Sampling locations and AFLP data for all species: DRYAD entry doi:10.5061/dryad.f3rk4 (Meirmans et al. 2011a,b).
References
- Allard RW, Garcia P, Saenz-de-Miera LE, de-la-Vega MP. Evolution of multilocus genetic structure in Avena hirtula and Avena barbata. Genetics. 1993;135:1125–1139. doi: 10.1093/genetics/135.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez N, Thiel-Egenter C, Tribsch A, et al. History or ecology? Substrate type as a major driver of spatial genetic structure in Alpine plants. Ecology Letters. 2009;12:632–640. doi: 10.1111/j.1461-0248.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- Beven K, Kirby M. A physically-based variable contribution area model of catchment hydrology. Hydrology Science Bulletin. 1979;24:43–69. [Google Scholar]
- Bierne N, Welch J, Loire E, Bonhomme F, David P. The coupling hypothesis: why genome scans may fail to map local adaptation genes. Molecular Ecology. 2011;20:2044–2072. doi: 10.1111/j.1365-294X.2011.05080.x. [DOI] [PubMed] [Google Scholar]
- Bonin A, Ehrich D, Manel S. Statistical analysis of amplified fragment length polymorphism data: a toolbox for molecular ecologists and evolutionists. Molecular Ecology. 2007;16:3737–3758. doi: 10.1111/j.1365-294X.2007.03435.x. [DOI] [PubMed] [Google Scholar]
- Borcard D, Gillet F, Legendre P. Numerical Ecology with R. Springer; New York: 2011. [Google Scholar]
- Borcard D, Legendre P. All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecological Modelling. 2002;153:51–68. [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Chaves MM, Maroco JP, Pereira JS. Understanding plant responses to drought – from genes to the whole plant. Functional Plant Biology. 2003;30:239–264. doi: 10.1071/FP02076. [DOI] [PubMed] [Google Scholar]
- Cox K, Vanden Broeck A, Van Calster H, Mergeay J. Temperature-related natural selection in a wind-pollinated tree across regional and continental scales. Molecular Ecology. 2011;20:2724–2738. doi: 10.1111/j.1365-294X.2011.05137.x. [DOI] [PubMed] [Google Scholar]
- Davis MB, Shaw RG, Etterson JR. Evolutionary responses to changing climate. Ecology. 2005;86:1704–1714. [Google Scholar]
- Dray S, Legendre P, Peres-Neto PR. Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM) Ecological Modelling. 2006;196:483–493. [Google Scholar]
- Endler J. Geographic Variation, Speciation, and Clines. Princeton University Press; Princeton: 1977. [PubMed] [Google Scholar]
- Endler J. Natural Selection in the Wild. Princeton University Press; Princeton: 1986. [Google Scholar]
- Excoffier L, Lischer HEL. ARLEQUIN suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes – application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Hofer T, Foll M. Detecting loci under selection in a hierarchically structured population. Heredity. 2009;103:285–298. doi: 10.1038/hdy.2009.74. [DOI] [PubMed] [Google Scholar]
- Feder ME, Mitchell-Olds T. Evolutionary and ecological functional genomics. Nature Reviews Genetics. 2003;4:651–657. doi: 10.1038/nrg1128. [DOI] [PubMed] [Google Scholar]
- Fournier-Level A, Korte A, Cooper MD, et al. A map of local adaptation in Arabidopsis thaliana. Science. 2011;334:86–89. doi: 10.1126/science.1209271. [DOI] [PubMed] [Google Scholar]
- Frei E, Bodin J, Walther G-R. Plant species’ range shifts in mountainous areas – all uphill from here? Botanica Helvetica. 2010;120:117–128. [Google Scholar]
- Gugerli F, Englisch T, Niklfeld H, et al. Relationships among levels of biodiversity and the relevance of intraspecific diversity in conservation – a project synopsis. Perspectives in Plant Ecology, Evolution and Systematics. 2008;10:259–281. [Google Scholar]
- Guisan A, Thuiller W. Predicting species distribution: offering more than simple habitat models. Ecology Letters. 2005;8:993–1009. doi: 10.1111/j.1461-0248.2005.00792.x. [DOI] [PubMed] [Google Scholar]
- Haldane J. The theory of a cline. Journal of Genetics. 1948;48:277–284. doi: 10.1007/BF02986626. [DOI] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Gordon AS, et al. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genetics. 2008;4:e32. doi: 10.1371/journal.pgen.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Ehler E, et al. Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8924–8930. doi: 10.1073/pnas.0914625107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Brachi B, Faure N, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011a;334:83–86. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Alkorta-Aranburu G, et al. Adaptations to climate-mediated selective pressures in humans. PLoS Genetics. 2011b;100137:5. doi: 10.1371/journal.pgen.1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao AS, Kudo G. Landscape genetics of alpine-snowbed plants: comparisons along geographic and snowmelt gradients. Heredity. 2004;93:290–298. doi: 10.1038/sj.hdy.6800503. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Willi Y. Detecting genetic response to environmental change. Nature Reviews Genetics. 2008;9:421–432. doi: 10.1038/nrg2339. [DOI] [PubMed] [Google Scholar]
- Holderegger R, Buehler D, Gugerli F, Manel S. Landscape genetics of plants. Trends in Plant Science. 2010;15:675–683. doi: 10.1016/j.tplants.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Jay F, Manel S, Alvarez N, et al. Forecasting changes in population genetic structure of Alpine plants in response to global warming. Molecular Ecology. 2012;21:2353–2368. doi: 10.1111/j.1365-294X.2012.05541.x. [DOI] [PubMed] [Google Scholar]
- Joost S, Bonin A, Bruford MW, et al. A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Molecular Ecology. 2007;16:3955–3969. doi: 10.1111/j.1365-294X.2007.03442.x. [DOI] [PubMed] [Google Scholar]
- Jump AS, Penuelas J. Running to stand still: adaptation and the response of plants to rapid climate change. Ecology Letters. 2005;8:1010–1020. doi: 10.1111/j.1461-0248.2005.00796.x. [DOI] [PubMed] [Google Scholar]
- Körner C. Alpine Plant Life: Functional Plant Ecology of High Mountain Ecosystems. Springer; New York: 2003. [Google Scholar]
- Kosman E. Nei’s gene diversity and the index of average differences are identical measures of diversity within populations. Plant Pathology. 2003;52:533–535. [Google Scholar]
- Lavergne S, Mouquet N, Thuiller W, Ronce O. Biodiversity and climate change: integrating evolutionary and ecological responses of species and communities. Annual Review of Ecology, Evolution and Systematics. 2010;41:321–350. [Google Scholar]
- Lee C-R, Mitchell-Olds T. Quantifying effects of environmental and geographical factors on patterns of genetic differentiation. Molecular Ecology. 2011;20:4631–4642. doi: 10.1111/j.1365-294X.2011.05310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Legendre L. Numerical Ecology, 3rd English edn. Developments in Environmental Modelling. Vol. 24. Elsevier Science BV; Amsterdam: 2012. [Google Scholar]
- Linhart YB, Grant MC. Evolutionary significance of local genetic differentiation in plants. Annual Review of Ecology and Systematics. 1996;27:237–277. [Google Scholar]
- Manel S, Joost S, Epperson B, et al. Perspectives on the use of landscape genetics to detect genetic adaptive variation in the field. Molecular Ecology. 2010a;19:3760–3772. doi: 10.1111/j.1365-294X.2010.04717.x. [DOI] [PubMed] [Google Scholar]
- Manel S, Poncet B, Legendre P, Gugerli F, Holderegger R. Common factors drive adaptive genetic variation at different spatial scales in Arabis alpina. Molecular Ecology. 2010b;19:3824–3835. doi: 10.1111/j.1365-294X.2010.04716.x. [DOI] [PubMed] [Google Scholar]
- Meirmans PG, Goudet J, IntraBioDiv Consortium. Gaggiotti OE. Ecology and life history affect different aspects of the population structure of 27 high-alpine plants. Molecular Ecology. 2011a;20:3144–3155. doi: 10.1111/j.1365-294X.2011.05164.x. [DOI] [PubMed] [Google Scholar]
- Meirmans PG, Goudet J, IntraBioDiv Consortium. Gaggiotti OE. Data from: Ecology and life history affect different aspects of the population structure of 27 high-alpine plants. Dryad Digital Repository. 2011b doi: 10.1111/j.1365-294X.2011.05164.x. doi:10.5061/dryad.f3rk4. [DOI] [PubMed] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the United States of America. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K. Bootstrapping R2 and adjusted R2 in regression analysis. Economic Modelling. 2000;17:473–483. [Google Scholar]
- Parisod C, Bonvin G. Fine-scale genetic structure and marginal processes in an expanding population of Biscutella laevigata L. (Brassicaceae) Heredity. 2008;101:536–542. doi: 10.1038/hdy.2008.95. [DOI] [PubMed] [Google Scholar]
- Poncet B, Herrmann D, Gugerli F, et al. Tracking genes of ecological relevance using a genome scan: application to Arabis alpina. Molecular Ecolog. 2010;19:2896–2907. doi: 10.1111/j.1365-294X.2010.04696.x. [DOI] [PubMed] [Google Scholar]
- Prentice HC, Lonn M, Lager H, Rosen E, Van Der Maarel E. Changes in allozyme frequencies in Festuca ovina populations after a 9-year nutrient/water experiment. Journal of Ecology. 2000;88:331–347. [Google Scholar]
- Reusch TBH, Wood TE. Molecular ecology of global change. Molecular Ecology. 2007;16:3973–3992. doi: 10.1111/j.1365-294X.2007.03454.x. [DOI] [PubMed] [Google Scholar]
- Richardson BA, Rehfeldt GE, Kim MS. Congruent climate-related genecological responses from molecular markers and quantitative traits for western white pine (Pinus monticola) International Journal of Plant Sciences. 2009;170:1120–1131. [Google Scholar]
- Schmidt PS, Serrao EA, Pearson GA, et al. Ecological genetics in the North Atlantic: environmental gradients and adaptation at specific loci. Ecology. 2008;89:S91–S107. doi: 10.1890/07-1162.1. [DOI] [PubMed] [Google Scholar]
- Segelbacher G, Cushman SA, Epperson BK, et al. Applications of landscape genetics in conservation biology: concepts and challenges. Conservation Genetics. 2010;11:375–385. [Google Scholar]
- Shendure J, Ji HL. Next-generation DNA sequencing. Nature Biotechnology. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Shimono Y, Watanabe M, Hirao AS, Wada N, Kudo G. Morphological and genetic variation of Potentilla matsumurae (Rosaceae) between fellfield and snowbed populations. American Journal of Botany. 2009;96:728–737. doi: 10.3732/ajb.0800242. [DOI] [PubMed] [Google Scholar]
- Siol M, Wright SI, Barrett SCH. The population genomics of plant adaptation. New Phytologist. 2010;188:313–332. doi: 10.1111/j.1469-8137.2010.03401.x. [DOI] [PubMed] [Google Scholar]
- Skot L, Sackville Hamilton NR, Mizen S, Chorlton KH, Thomas ID. Molecular genecology of temperature response in Lolium perenne: 2. Association of AFLP markers with ecogeography. Molecular Ecology. 2002;11:1865–1876. doi: 10.1046/j.1365-294x.2002.01568.x. [DOI] [PubMed] [Google Scholar]
- Sork VL, Davis FW, Westfall R, et al. Gene movement and genetic association with regional climate gradients in California valley oak (Quercus lobata Nee) in the face of climate change. Molecular Ecology. 2010;19:3806–3823. doi: 10.1111/j.1365-294X.2010.04726.x. [DOI] [PubMed] [Google Scholar]
- St Clair JB, Mandel NL, Vance-Boland KW. Genecology of Douglas fir in western Oregon and Washington. Annals of Botany. 2005;96:1199–1214. doi: 10.1093/aob/mci278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton M, Galen C. Life on the edge: adaptation versus environmentally mediated gene flow in the snow buttercup, Ranunculus adoneus. American Naturalist. 1997;150:143–178. doi: 10.1086/286061. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Hoekstra HE. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity. 2008;100:158–170. doi: 10.1038/sj.hdy.6800937. [DOI] [PubMed] [Google Scholar]
- Storz JF. Using genome scans of DNA polymorphism to infer adaptive population divergence. Molecular Ecology. 2005;14:671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Thiel-Egenter C, Gugerli F, Alvarez N, et al. Effects of life history traits on the genetic diversity of high-mountain plants: a multi-species study across the Alps and the Carpathians. Global Ecology and Biogeography. 2009;18:78–87. [Google Scholar]
- Vos P, Hagers R, Bleeker M, et al. AFLP: new technique for DNA fingerprinting. Nucleic Acid Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther G-R, Post E, Convey P, et al. Ecological responses to recent climate change. Nature. 2002;416:389–395. doi: 10.1038/416389a. [DOI] [PubMed] [Google Scholar]
- Wright S. Size of population and breeding structure in relation to evolution. Science. 1938;87:430–431. [Google Scholar]
- Wright SP. Adjusted p-values for simultaneous inference. Biometrics. 1992;48:1005–1013. [Google Scholar]
- Yu JM, Pressoir G, Briggs WH, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Zimmermann NE, Kienast F. Predictive mapping of alpine grasslands in Switzerland: species versus community approach. Journal of Vegetation Science. 1999;10:469–482. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.