Abstract
The glycoprotein Erns of pestiviruses is a virion-associated and -secreted RNase that is involved in virulence. The requirements at the cleavage site in heteropolymeric RNA substrates were studied for Erns. Limited digestion of heteropolymeric RNA substrates indicated a cleavage 5′ of uridine residues irrespective of the preceding nucleotide (Np/U). To further study specificity radiolabeled RNA, molecules of 45 to 56 nucleotides in length were synthesized that contained no or a single Np/U cleavage site. Cleavage was only observed in substrates containing an ApU, CpU, GpU, or UpU dinucleotide and occurred in two steps, an initial NpU-specific and a consecutive unspecific degradation. The NpU-specific cleavage was resistant to 7 M urea while the second-order cleavage was sensitive to denaturation. Kinetic analyses revealed that Erns is a highly active endoribonuclease (kcat/Km = 2 × 106 to 10 × 106 M−1 s−1) with a strong affinity to NpU containing single-stranded RNA substrates (Km = 85 to 260 nM).
Pestiviruses are small enveloped viruses with a single positive-stranded RNA genome and belong to the Flaviviridae (40). Presently the Pestivirus genus contains five species (3), four of which are important pathogens in farm animals, namely, classical swine fever virus (CSFV), bovine viral diarrhea virus types 1 and 2 (BVDV-1 and BVDV-2), and border disease virus (BDV) (40). One of the most remarkable features of pestiviruses is the virus-encoded glycoprotein Erns, which has ribonucleolytic activity. CSFV Erns was shown to be an essential structural component of the virion (41), and the RNase activity has been shown to be a determinant of the virulence of CSFV and BVDV-2 (25, 26). In addition to being a virus-associated protein, Erns is secreted from CSFV-infected cells as soluble protein (32). Erns is a highly N-glycosylated dimeric protein of 227 amino acids, which was typed as a member of the RNase T2 family based on a narrow sequence homology in the active-site domains [cassette 1, XXXHGL(I)WP; cassette 2, F(L)XXH(YD)EXXK(TR)HGT(AW)C] (12, 14, 15, 33, 42). The RNase T2 family encompasses a variety of secreted endoribonucleases with a distribution in all kingdoms (14, 15). RNases of the T2 type are usually larger than 200 amino acids and have no absolute base specificity but a preferred cleavage site. Most fungal T2 RNases cleave at Ap/N bonds, which is in accordance with the finding that in most ribonucleases the base preference is due to interactions with the base located at the 5′-terminal end (B1 site) (36). RNase MC1, which was isolated from bitter gourd (Momordica charantia) and which only encompasses 191 amino acids, also belongs to the T2 RNase family. It can be distinguished from fungal T2 RNases as well as from smaller RNases in RNase A and RNase T1 families because its base specificity is based on interactions with uridine residues at the 3′-terminal site (B2 site) (16, 29, 38).
Thus far Erns has been shown to degrade polyuridylic acid (42), rRNA (12), and viral genomic RNA (12, 42) in vitro. It has been shown that Erns binds to a variety of cells (13), and a C-terminal transport peptide was identified which translocates the protein through the plasma membrane (20). In order to define potential substrates of Erns we have determined the requirements at the cleavage site in heteropolymeric substrates and the kinetic parameters of the RNase.
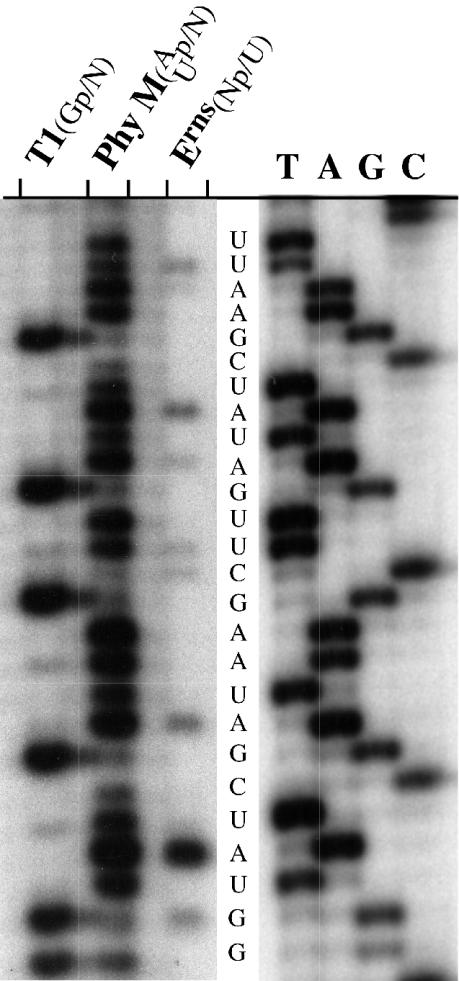
Recombinant CSFV Erns, which was expressed in insect cells and purified by immunoaffinity chromatography, was used for limited digestion of a 32P-labeled RNA substrate (4, 34). As substrate the polylinker sequence of pBS SKII was transcribed with T7 RNA polymerase and was radiolabeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase. Digests using 20 ng of Erns, 2 U of RNase T1, or 2 U of RNase PhyM and 105 cpm (5′-32P) of RNA substrate were performed under denaturing conditions (7 M urea, 1 mM EDTA, and 20 mM sodium-citrate, pH 6.0) for 15 min at 55°C in order to avoid secondary structure formation of the RNA substrate. The RNase digestions were subjected to denaturing gel electrophoresis in an analytical polyacrylamide gel (8% polyacrylamide [PAA], 7 M urea) side by side with a radiolabeled sequencing reaction mixture of the analogous cDNA region (Fig. 1). Comparison of the patterns generated by Erns and the reference endoribonucleases RNase T1 (Gp/N) and PhyM (Up/N, Ap/N) as well as the sequencing reaction revealed that cleavage by Erns occurred 5′ of uridine residues. Cleavage products were identified at all Np/U dinucleotide sequences (ApU, CpU, GpU, and UpU). Erns retains activity in the presence of 7 M urea even at elevated temperatures (55°C), thus the enzyme is remarkably stable. To further determine the requirements at the NpU cleavage site, experiments with synthetic RNA oligonucleotides (5′GCCGACUUC3′) were undertaken. Unfortunately these molecules were not cleaved by Erns, because they were either too small or were uncleavable for other reasons. Therefore, larger single-stranded RNA substrates were designed as runoff transcripts from cDNA. To avoid the formation of secondary structures the substrate molecules consisted mainly of homopolymeric A. For this purpose oligo(dT/dA) adaptors were cloned into the HindIII/SacI sites of pBS SKII to form a cDNA sequence, which was devoid of uridine residues (substrate ΔU [Sub-ΔU]: 5[prime]GAACAAAAGCCGG-A49-GC3′). The four possible NpU dinucleotides were introduced by site-directed mutagenesis 14 nucleotides (nt) upstream of the 3′ end into Sub-ΔU cDNA. Due to variable binding of the mutagenesis primers within the poly(A) sequence, the DNA templates led to synthesis of transcripts with different sizes (Sub-ΔU, 64 nucleotides; Sub-ApU, 57 nucleotides; Sub-CpU, 46 nucleotides; Sub-GpU, 57 nucleotides; Sub-UpU, 53 nucleotides). RNA substrates were generated by using T3 RNA polymerase and were labeled either at the 5′ end with [γ-32P]ATP by T4-polynucleotide kinase (Sub-ΔU [Fig. 2B]; Sub-ApU, not shown) or during transcription with [α-32P]UTP (Sub-ApU [Fig. 2A and C]). Probably due to the addition of nucleotides at the 3′ end (5, 17, 28), the labeled in vitro transcripts resulted in products of different sizes and were therefore purified by denaturing gel electrophoresis. Molecules of appropriate sizes were excised from the gel and eluted in a buffer containing 10 mM Tris-HCl, 300 mM NaCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate (pH 7.5). Despite this purification, a mixture of substrate molecules was obtained which usually differed from each other by a single nucleotide (Fig. 2). For each reaction, 5 × 104 cpm of the purified substrate was subjected to Erns cleavage in 40 mM Tris-acetate, 0.5 mM EDTA (pH 6.0) (Fig. 2A and B). These buffer conditions have previously been described as optimal for Erns activity (42). The uridine-containing substrate Sub-ApU was cleaved by Erns in a concentration-dependent manner (Fig. 2A). This also applied to the substrates Sub-CpU, Sub-GpU, and Sub-UpU (data not shown). In contrast, Sub-ΔU was resistant to cleavage by Erns (Fig. 2B) but was degraded by RNase A (data not shown). The same cleavage pattern was observed when degradation was studied with lysates from PK15 cells which were infected with CSFV at a multiplicity of infection of 1 (Fig. 2C). For this purpose cells were lysed in 1% Triton X-100 24 h postinfection, and the equivalent of 104 cells was used for the degradation of Sub-ApU in 40 mM Tris-acetate, 0.5 mM EDTA (pH 6.0) for the indicated times (Fig. 2C). No degradation of Sub-ApU was observed after incubation with lysates from PK15 cells infected with a recombinant CSFV, which encoded an enzymatically inactive Erns (CSFV-ErnsH79D), or noninfected PK15 cells (Fig. 2C). Independent of the type of 32P-labeling, the cleavage of Sub-ApU resulted in a set of 4 to 5 predominant cleavage products. The size of the largest cleavage product correlated to the position of the NpU bond in the substrate, irrespective of the position of the 32P-label.
FIG. 1.
Erns cleaves 5′ of uridine residues. A T7 transcript encompassing the polylinker sequence of pBS SKII was labeled with [γ-32P]ATP at the 5′ end and was incubated under conditions that prevented the degradation to go on to completion (Erns [20 ng], RNase T1 [2 U], or Phy M [2 U] in a solution of 7 M urea, 1 mM EDTA, 20 mM sodium-citrate [pH 6.0] at 55°C for 15 min). The reactions were subjected to high-voltage polyacrylamide gel electrophoresis in a denaturing 8% PAA sequencing gel side by side with a [α-32P]ATP-labeled dideoxy sequencing reaction of the respective region of the plasmid (lanes are termed according to the dideoxy nucleotides T, A, G, and C). RNase T1 cleaves Gp/N, and RNase Phy M cuts preferentially at Ap/N and Up/N bonds. Cleavage by Erns occurs 5′ of uridine residues.
FIG. 2.
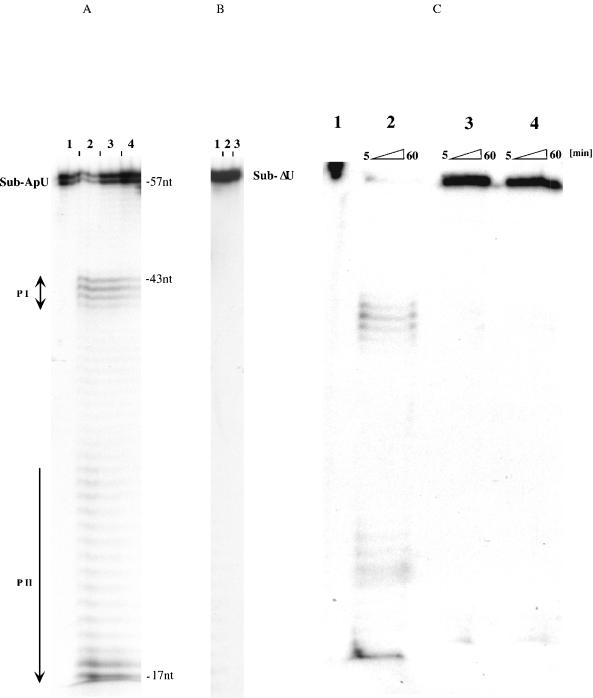
Erns endoribonuclease requires uridine residues in position B2. (A) [32P]UTP labeled Sub-ApU (5 × 104 cpm) was incubated without (lane 1) or with 100 ng (lane 2), 50 ng (lane 3), or 25 ng (lane 4) of purified Erns in 40 mM Tris-acetate, 0.5 mM EDTA (pH 6.0) at 37°C for 20 min. The reactions were subjected to high-voltage polyacrylamide gel electrophoresis in a denaturing 8% PAA sequencing gel. Sub-ApU appears as a doublet of 57 and 58 nucleotides due to purification of the RNA transcript. The predominant cleavage results in 32P-labeled products (PI) that are at least 14 nucleotides shorter than Sub-ApU, which corresponds to the position of the uridine residue. Cleavage products are unstable and are further degraded to oligonucleotides (PII). (B) 32P-labeled Sub-ΔU (5 × 104 cpm) was incubated without (lane 1) or with 100 ng (lane 2) or 50 ng (lane 3) of purified Erns as described above and was analyzed on the same gel. RNA without a uridine residue does not serve as substrate for Erns. (C) 32P-labeled Sub-ApU (5 × 104 cpm) was incubated for 5, 15, 30, and 60 min without (lane 1) or with Triton X-100 lysates (1%) of 104 PK15 cells that were not infected (panel 3) or that were infected with CSFV (panel 2) or recombinant CSFV-ErnsH79D (panel 4) at a multiplicity of infection of 1. nt, nucleotides.
Although the cleavage occurs 5′ of uridine, the labeled phosphate remains attached to the N base of the 5′ cleavage product. RNase T2 and other RNases transfer the phosphate from the 5′ position to the 2′ hydroxyl group of the 5′ product via the formation of a 2′,3′ cyclophosphate intermediate (14, 15, 30). Thus, the labeled phosphate remains linked to the 3′ position of the ribose in the B1 site of the enzyme.
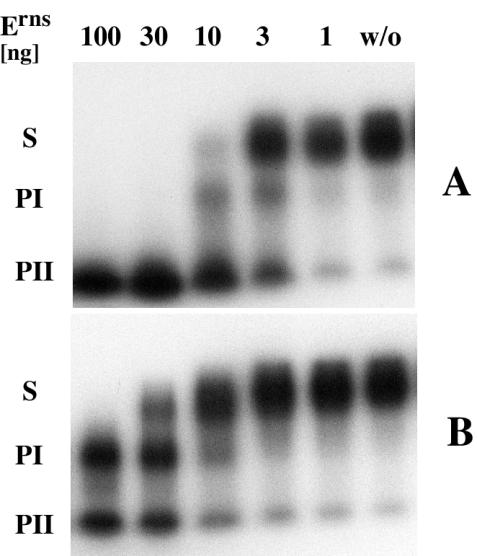
Errors which occur during transcription with T3 or T7 RNA polymerase are a probable reason for the origin of the heterogeneity of the labeled primary cleavage product (PI) product (Fig. 2). Addition of nontemplate nucleotides frequently occurs at the 3′ end as well as at the 5′ end (11, 31). The degradation assays revealed that the labeled 5′ cleavage products (PI) were unstable and further degraded to oligonucleotides (PII). This degradation occurred in the absence of NpU because PI does not contain any uridine residue. That in fact two distinct cleavage events are catalyzed by Erns became apparent by performing the degradation assay under denaturing conditions. Degradation of Sub-ApU with different amounts of Erns (100, 30, 10, 3, and 1 ng) in a solution of 7 M urea, 1 mM EDTA, 20 mM sodium-citrate (pH 6.0) leads to an accumulation of PI (Fig. 3) while the same amount of enzyme in 40 mM Tris-acetate, 0.5 mM EDTA (pH 6.0) produced mainly PII. For these experiments the degradation reaction was separated on denaturing 8% PAA gels in a Mini Protean II apparatus (Bio-Rad). Because the uridine-free substrate Sub-ΔU is not attacked by Erns, there apparently is a difference in the nature of the substrate molecules. Possibly, the initial NpU specific cleavage modifies the PI product (i.e., by generating a 3′ phosphate) so that it is recognized as a substrate. As an alternative it is conceivable that Erns remains attached to PI after completion of the first cleavage and continues to cleave PI at random positions. This model could explain the stabilizing effect of urea on PI, because it likely reduces the affinity of Erns to the NpU-free substrate more efficiently than the affinity to the NpU site. Determination of the precise mechanism of the second cleavage is the subject of future analysis.
FIG. 3.
Separation of primary and secondary cleavage by urea. 32P-labeled Sub-ApU (5 × 104 cpm) was incubated with or without (w/o) Erns (100, 30, 10, 3, or 1 ng) for 15 min in nondenaturing (40 mM Tris-acetate, 0.5 mM EDTA [pH 6.0]) (A) or denaturing buffer (7 M urea, 1 mM EDTA, 20 mM sodium-citrate [pH 6.0]) (B) at 37°C. The reactions were electrophoretically separated in small denaturing polyacryamide gels. Due to the mini-gel format, the cleavage fragments PI and PII form discrete bands. The overall activity of Erns is threefold lower in urea than in Tris-acetate buffer. While in panel A the substrate is rapidly degraded into PII, panel B shows that the presence of 7 M urea stabilizes PI. Also shown is steady-state analysis of the endoribonucleolytic activity of Erns.
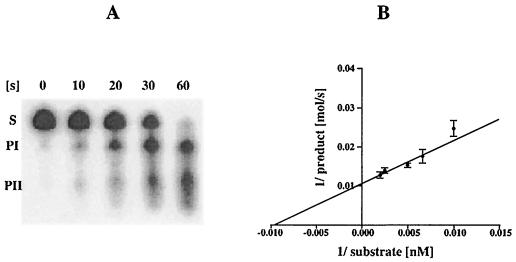
For the functional characterization of an enzyme and its substrate it is important to know the kinetic properties involved. Thus far, kinetic data for Erns are only available for an UpU dinucleotide as substrate (42). To analyze whether the ribonucleolytic activity of Erns in a heteropolymeric substrate depends on the nucleotide in the 5′ position of NpU, the cleavage kinetics for the radiolabeled Sub-ApU, Sub-CpU, Sub-GpU, and Sub-UpU were determined. An Erns degradation assay was developed in which 20, 40, 60, 80, and 100 ng of the unlabeled RNA substrates containing trace amounts (0.2%) of the same 32P-labeled molecule were incubated with 10 ng of purified recombinant Erns at 37°C for 10 to 60 s in 40 mM Tris-acetate, 0.5 mM EDTA (pH 6.0). The degradation reactions were stopped by addition of an equal volume of formamide, heated to 95°C and separated by electrophoresis in 8% PAA gels containing 7 M urea in a Mini Protean II apparatus (Bio-Rad). Each analysis was repeated at least three times. The amount of radioactivity per band was analyzed in a Fuji BAS1000 PhosphorImager. As an example, the degradation of 40 ng of Sub-GpU is shown in Fig. 4A. Because the amount of radioactivity (in counts per minute per nanogram) was known, the turnover of RNA substrate could be measured at all time points (T0 to T60). The kinetic parameters Km and Vmax of Sub-ApU, Sub-CpU, Sub-GpU, and Sub-UpU were calculated from the Lineweaver-Burk plots: 1/v versus 1/[s] (22), by using Sigmablot (SPSS-Science) (shown for GpU in Fig. 4B).
FIG. 4.
(A) Forty nanograms of Sub-GpU, including trace amounts of 32P-labeled Sub-CpU, was incubated with 10 ng of Erns for 10, 20, 30, or 60 s at 37°C and was analyzed by electrophoresis in a small denaturing 8% PAA gel. The radioactivities of substrate and cleavage products were determined by phosphorimaging, and the counts per minute at t = 0 were equated with the amount of RNA substrate which allowed calculation of the turnover of RNA substrate at t = 10, 20, 30, and 60 s. The same experimental procedure was applied for 20, 60, 80, and 100 ng of Sub-GpU and the other substrates: Sub-ApU, -GpU, and -UpU. (B) Lineweaver-Burk analysis of values determined for the degradation of different concentrations of Sub-GpU Erns.
Determinations of the Km values indicate that Erns has a very high affinity (Km, 84 × 10−9 to 259 × 10−9 M) for the tested substrates in the order UpU>GpU>CpU>ApU (Table 1). For Sub-UpU the affinity of Erns is 1,000-fold higher than that for the dinucleotide UpU (Km, 872.5 × 10−6 ± 125.9 × 10−6 M) (42). In contrast, the turnover of the substrates was fastest for Sub-ApU (151.2 fmol/s), which is three times faster than that determined for Sub-CpU. For UpU the results are ambiguous, because the cleavage site (ApUpU) actually allows two cleavages to occur. The catalytic activity of Erns indicated by the kcat/Km values is considerably high (2 × 106 to 1 × 107 M−1 s−1) and operates near the limits of diffusion (10−8 to 10−9 M−1 s−1) (8).
TABLE 1.
Steady-state kinetic parameters for cleavage of Sub-ApU, Sub-CpU, Sub-GpU, and Sub-UpU by Erns
| Substrate | Kinetics parametera
|
|||
|---|---|---|---|---|
| Km (10−9 M) | Vmax (fmol/s) | kcat (s−1) | kcat/Km (M−1 s−1) | |
| Sub-ApU | 258.7 | 151.2 | 1.33 | 5 × 106 |
| Sub-UpU | 83.8 | 104.8 | 0.9 | 10 × 106 |
| Sub-GpU | 103.3 | 95.2 | 0.84 | 8 × 106 |
| Sub-CpU | 220.1 | 53.7 | 0.47 | 2 × 106 |
Erns is a highly active RNase of the T2 RNase family with an unusual cleavage site preference. Interestingly, the same cleavage preference (Np/U) was recently determined for RNase MC1, although there is very little sequence homology outside the active center. RNase MC1 cleaves phosphodiester bonds of A, C, G, or UpU, the most favorable substrate being CpU (16). Kinetic parameters of RNase MC1, which have been determined for CpU (Km, 1.38 × 103 ± 0.2 × 103 M; kcat, 746 ± 95 min−1), and the kcat/Km value (542 × 10−3 ± 50 × 10−3 M−1 min−1) (29), reveal that Erns has a 107-fold higher activity at least with respect to the oligomeric substrates used in our analysis.
The aim of this study was to determine the substrate specificity of the RNase Erns of CSFV. In particular, the question was addressed whether Erns prefers distinct sequence motifs or substrates (i.e., 28S rRNA) as it is described for several secreted ribonucleases (i.e., onconase [21]; BS-RNase [18]; and α-Sarcin, restrictocin, or mitogillin [24]) or targets a wide range of single-stranded RNA molecules. The kinetic data revealed that the substrate recognition of Erns itself is not very discriminating, because the enzyme degraded the test substrates near its theoretical maximum. In other words, the affinity of Erns for substrates containing the ubiquitous NpU cleavage is already so high that an even higher specificity for other (unknown) substrates is unlikely. Although the substrate specificity of Erns was unaltererd by using PK15 cell lysates, it is possible that other factors define the actual targets of Erns as it is reported for vhs RNase of alphaherpesviruses (35). vhs RNase, which is located in the tegument of the virion (19), is released during fusion at the plasma membrane and acts as an unspecific modulator of host cell translation by cleaving mRNAs (7, 37). Degradation of mRNAs by vhs RNase is initiated near the 5′ end of target mRNAs (6) and requires a cellular factor (23).
It was recently shown that nonreplicative recombination in poliovirus requires 3′phosphate and 5′hydroxyl modifications of the joining RNA fragments (9, 10). Strikingly, these ends are produced by many ribonucleases, including RNases of the T2 family, and it is also evident that Erns transfers the 32P-labeled α-phosphate of the uridine residue to the 3′ end of the 5′ fragment. While it is not shown whether nonreplicative recombination also applies for pestiviruses, recombinations of virus genomes with host cellular mRNAs (e.g., ubiquitin) occur with an unprecedented high frequency (1, 2, 27, 39). It is tempting to speculate that Erns may even be advantageous for the virus survival and evolution by being mechanistically involved in the recombination of pestiviruses, but extensive future analyses are needed to test this hypothesis.
Acknowledgments
This work was funded by Deutsche Forschungsgemeinschaft, SFB 535. Y.H. was and G.R.-S. is a fellow of the Graduiertenkolleg “Biochemistry of Nucleoprotein-Complexes.”
REFERENCES
- 1.Baroth, M., M. Orlich, H. J. Thiel, and P. Becher. 2000. Insertion of cellular NEDD8 coding sequences in a pestivirus. Virology 278:456-466. [DOI] [PubMed] [Google Scholar]
- 2.Becher, P., G. Meyers, A. D. Shannon, and H. J. Thiel. 1996. Cytopathogenicity of border disease virus is correlated with integration of cellular sequences into the viral genome. J. Virol. 70:2992-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becher, P., R. A. Ramirez, M. Orlich, S. C. Rosales, M. König, M. Schweizer, H. Stadler, H. Schirrmeier, and H. J. Thiel. 2003. Genetic and antigenic characterization of novel pestivirus genotypes: implications for classification. Virology 311:96-104. [DOI] [PubMed] [Google Scholar]
- 4.Donis-Keller, H., A. M. Maxam, and W. Gilbert. 1977. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 4:2527-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Draper, D. E., S. A. White, and J. M. Kean. 1988. Preparation of specific ribosomal RNA fragments. Methods Enzymol. 164:221-237. [DOI] [PubMed] [Google Scholar]
- 6.Elgadi, M. M., C. E. Hayes, and J. R. Smiley. 1999. The herpes simplex virus vhs protein induces endoribocleolytic cleavage of target RNAs in cell extracts. J. Virol. 73:7153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everly, D. N., P. Feng, S. Mian, and G. S. Read. 2002. mRNA degradation by the virion host shutoff (vhs) protein of herpes simplex virus: genetic and biochemical evidence that vhs is a nuclease. J. Virol. 76:8560-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fersht, A. 1999. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. W. H. Freeman & Co., New York, N.Y.
- 9.Gmyl, A. P., E. V. Belousov, S. V. Maslova, E. V. Khitrina, A. B. Chetverin, and V. I. Agol. 1999. Nonreplicative RNA recombination in poliovirus. J. Virol. 73:8958-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gmyl, A. P., S. A. Korshenko, E. V. Belousov, E. V. Khitrina, and V. I. Agol. 2003. Nonreplicative homologous RNA recombination: promiscuous joining of RNA pieces? RNA 9:1221-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helm, M., H. Brulé, R. Giegé, and C. Florentz. 1999. More mistakes by T7 RNA polymerase at the 5′ ends of in vitro-transcribed RNAs. RNA 5:618-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hulst, M. M., G. Himes, E. Newbigin, and R. J. Moormann. 1994. Glycoprotein E2 of classical swine fever virus: expression in insect cells and identification as a ribonuclease. Virology 200:558-565. [DOI] [PubMed] [Google Scholar]
- 13.Hulst, M. M., and R. J. Moormann. 1997. Inhibition of pestivirus infection in cell culture by envelope proteins E(rns) and E2 of classical swine fever virus: E(rns) and E2 interact with different receptors. J. Gen. Virol. 78:2779-2787. [DOI] [PubMed] [Google Scholar]
- 14.Irie, M. 1997. RNase T1/RNase T2 family RNases, p. 102-130. In G. D'Alessio and J. F. Riordan (ed.), Ribonucleases: structures and functions. Academic Press, New York, N.Y.
- 15.Irie, M., and K. Ohgi. 2001. Ribonuclease T2. Methods Enzymol. 341:42-56. [DOI] [PubMed] [Google Scholar]
- 16.Irie, M., H. Watanabe, K. Ohgi, Y. Minami, H. Yamada, and G. Funatsu. 1993. Base specificity of two plant ribonucleases from Momordica charantia and Luffa cylindrica. Biosci. Biotechnol. Biochem. 57:497-498. [Google Scholar]
- 17.Kholod, N., K. Vassilenko, M. Shlyapnikov, V. Ksenzenko, and L. Kisselev. 1998. Preparation of active tRNA gene transcripts devoid of 3′-extended products and dimers. Nucleic Acids Res. 26:2500-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, J. S., J. Soucek, J. Matousek, and R. T. Raines. 1995. Mechanism of ribonuclease cytotoxicity. J. Biol. Chem. 270:31097-31102. [DOI] [PubMed] [Google Scholar]
- 19.Lam, Q., C. A. Smibert, K. E. Koop, C. Lavery, J. P. Capone, S. P. Weinheimer, and J. R. Smiley. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575-2581. [PMC free article] [PubMed] [Google Scholar]
- 20.Langedijk, J. P. 2002. Translocation activity of C-terminal domain of pestivirus E(rns) and ribotoxin L3 loop. J. Biol. Chem. 277:5308-5314. [DOI] [PubMed] [Google Scholar]
- 21.Leland, P. A., and R. T. Raines. 2001. Review: cancer chemotherapy - ribonucleases to the rescue. Chem. Biol. 8:405-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lineweaver, H., and D. Burk. 1934. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 76:658-666. [Google Scholar]
- 23.Lu, P., H. A. Saffran, and J. R. Smiley. 2001. The vhs1 mutant form of herpes simplex virus virion host shutoff protein retains significant internal ribosome entry site-directed RNA cleavage activity. J. Virol. 75:1072-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarov, A. A., and O. N. Ilinskaya. 2003. Minireview: cytotoxic ribonucleases: molecular weapons and their targets. FEBS Lett. 540:15-20. [DOI] [PubMed] [Google Scholar]
- 25.Meyer, C., M. von Freyburg, K. Elbers, and G. Meyers. 2002. Recovery of virulent and RNase-negative attenuated type 2 bovine viral diarrhea viruses from infectious cDNA clones. J. Virol. 76:8494-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyers, G., A. Saalmuller, and M. Buttner. 1999. Mutations abrogating the RNase activity in glycoprotein E(rns) of the pestivirus classical swine fever virus lead to virus attenuation. J. Virol. 73:10224-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyers, G., N. Tautz, P. Becher, H. J. Thiel, and B. M. Kümmerer. 1996. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J. Virol. 70:8606-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milligan, J. F., D. R. Groebe, G. W. Witherell, and O. C. Uhlenbeck. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15:8783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Numata, T., A. Suzuki, M. Yao, I. Tanaka, and M. Kimura. 2001. Amino acid residues in ribonuclease MC1 from bitter gourd seeds which are essential for uridine specificity. Biochemistry 40:524-530. [DOI] [PubMed] [Google Scholar]
- 30.Ohgi, K., M. Iwama, K. Tada, R. Takizawa, and M. Irie. 1995. Role of Lys108 in the enzymatic activity of RNase Rh from Rhizopus niveus. J. Biochem. (Tokyo) 117:27-33. [DOI] [PubMed] [Google Scholar]
- 31.Pleiss, J. A., M. L. Derrick, and O. C. Uhlenbeck. 1998. T7 RNA polymerase produces 5′ end heterogeneity during in vitro transcription from certain templates. RNA 4:1313-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rümenapf, T., G. Unger, J. H. Strauss, and H. J. Thiel. 1993. Processing of the envelope glycoproteins of pestiviruses. J. Virol. 67:3288-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider, R., G. Unger, R. Stark, E. Schneider-Scherzer, and H. J. Thiel. 1993. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science 261:1169-1171. [DOI] [PubMed] [Google Scholar]
- 34.Simoncsits, A., G. G. Brownlee, R. S. Brown, J. R. Rubin, and H. Guilley. 1977. New rapid gel sequencing method for RNA. Nature 269:833-836. [DOI] [PubMed] [Google Scholar]
- 35.Smiley, J. R., M. M. Elgadi, and H. A. Saffran. 2001. Herpes simplex virus vhs protein. Methods Enzymol. 342:440-451. [DOI] [PubMed] [Google Scholar]
- 36.Steyaert, J. 1997. A decade of protein engineering on ribonuclease T1-atomatic dissection of the enzyme-substrate interactions. Eur. J. Biochem. 247:1-11. [DOI] [PubMed] [Google Scholar]
- 37.Strom, T., and N. Frenkel. 1987. Effects of herpes simplex virus on mRNA stability. J. Virol. 61:2198-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, A., M. Yao, I. Tanaka, T. Numata, S. Kikukawa, N. Yamasaki, and M. Kimura. 2000. Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2′-UMP or 3′-UMP, reveal structural basis for uridine specificity. Biochem. Biophys. Res. Commun. 275:572-576. [DOI] [PubMed] [Google Scholar]
- 39.Tautz, N., G. Meyers, and H. J. Thiel. 1993. Processing of poly-ubiquitin in the polyprotein of an RNA virus. Virology 197:74-85. [DOI] [PubMed] [Google Scholar]
- 40.Thiel, H. J., P. G. W. Plagemann, and V. Moennig. 1996. Pestiviruses, p. 1059-1073. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields Virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 41.Widjojoatmodjo, M. N., H. G. van Gennip, A. Bouma, P. A. van Rijn, and R. J. Moormann. 2000. Classical swine fever virus E(rns) deletion mutants: trans-complementation and potential use as nontransmissible, modified, live-attenuated marker vaccines. J. Virol. 74:2973-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Windisch, J. M., R. Schneider, R. Stark, E. Weiland, G. Meyers, and H. J. Thiel. 1996. RNase of classical swine fever virus: biochemical characterization and inhibition by virus-neutralizing monoclonal antibodies. J. Virol. 70:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]