Abstract
A major problem hampering the development of an effective vaccine against human immunodeficiency virus type 1 (HIV-1) is the resistance of many primary viral isolates to antibody-mediated neutralization. To identify factors responsible for this resistance, determinants of the large differences in neutralization sensitivities of HIV-1 pseudotyped with Env proteins derived from two prototypic clade B primary isolates were mapped. SF162 Env pseudotypes were neutralized very potently by a panel of sera from HIV-infected individuals, while JR-FL Env pseudotypes were neutralized by only a small fraction of these sera. This differential sensitivity to neutralization was also observed for a number of monoclonal antibodies (MAbs) directed against sites in the V2, V3, and CD4 binding domains, despite often similar binding affinities of these MAbs towards the two soluble rgp120s. The neutralization phenotypes were switched for chimeric Envs in which the V1/V2 domains of these two sequences were exchanged, indicating that the V1/V2 region regulated the overall neutralization sensitivity of these Envs. These results suggested that the inherent neutralization resistance of JR-FL, and presumably of related primary isolates, is to a great extent mediated by gp120 V1/V2 domain structure rather than by sequence variations at the target sites. Three MAbs (immunoglobulin G-b12, 2G12, and 2F5) previously reported to possess broad neutralizing activity for primary HIV-1 isolates neutralized JR-FL virus at least as well as SF162 virus and were not significantly affected by the V1/V2 domain exchanges. The rare antibodies capable of neutralizing a broad range of primary isolates thus appeared to be targeted to exceptional epitopes that are not sensitive to V1/V2 domain regulation of neutralization sensitivity.
There is a consensus that a broadly neutralizing humoral response is an essential component of a protective human immunodeficiency virus (HIV) vaccine. Unfortunately, current vaccine approaches have not been able to produce such neutralizing responses against primary HIV isolates despite induction of high titers of antibodies, including antibodies capable of neutralizing specific test strains (1, 2, 11, 14, 21, 25, 35, 36). Factors that determine the sensitivity of HIV type 1 (HIV-1) isolates to neutralization have not been clearly defined. Earlier studies indicated that X4-tropic laboratory strains in general were highly sensitive to neutralization and that R5-tropic primary isolates were relatively resistant (35, 38). Later evidence showed that neutralization sensitivities differ even among primary isolates (27) and that neutralization sensitivity does not correlate with coreceptor usage (6, 37).
One of the factors that can contribute to poor neutralization of primary HIV isolates in standard assays is the presence of viral variants whose neutralization epitopes are absent or modified in ways that result in reduced affinity towards the antibodies being tested. This complexity can be avoided by the use of single-cycle viral transduction assays mediated by noninfectious virions pseudotyped with molecularly cloned Env proteins. Such particles contain homogenous Env proteins; thus, differences in the extent of neutralization should reflect inherent differences in the sensitivities of the Env proteins rather than the presence of a resistant fraction of virus.
This assay was used to examine the neutralization sensitivities of SF162 and JR-FL env, two highly related env genes derived from primary, non-syncytium-inducing, macrophagetropic HIV-1 strains that were isolated from brain tissue of patients in the San Francisco area who were infected with clade B viruses (10, 28). The two env genes possess a high level of sequence similarity in both their gp120 and gp41 domains (>89%) but differed greatly in their sensitivity to neutralization by patient sera and the majority of monoclonal antibodies (MAbs) that were examined. The neutralization phenotype of chimeras in which the gp120 V1/V2 domains were exchanged mapped a major determinant of antibody-mediated neutralization sensitivity to this region. These results suggested that modulation of resistance to neutralization via targets in multiple domains of gp120 by determinants in the V1/V2 domain might be an important factor in the inability of the humoral response to control HIV replication.
MATERIALS AND METHODS
Viruses and generation of chimeric viruses.
Infectious viral pseudotypes were generated by transfecting 60-mm-diameter plates of 293 cells with 3 μl of FuGENE 6 transfection reagent (Boehringer Mannheim) combined with 1 μg of total DNA consisting of equal amounts of a plasmid expressing env-defective, luciferase-expressing HIVNL4-3 genomes (7) and a plasmid expressing HIV Env envelope proteins. The cells were refed at 24 h with RPMI 1640-10% fetal bovine serum (FBS), and supernatant medium containing pseudotyped luciferase-expressing viruses was harvested approximately 48 h after refeeding.
Human sera and MAbs.
Sera from HIV-infected individuals were obtained from S. Kreiswirth (Astor Medical Group, New York, N.Y.) and F. Valentine (New York University Medical Center, New York, N.Y.). Env-specific MAbs directed against the specified regions were used. MAbs immunoglobulin G (IgG)-b12 (3), 2F5 (41), and 2G12 (51) were obtained from the AIDS Research and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. MAbs 17b and 48d (49) against CD4-induced epitopes were obtained from J. Robinson (the Tulane University Medical School, New Orleans, La.), and the X5 Fab (40) was obtained from D. Dimitrov at the National Cancer Institute. The following MAbs were produced in our laboratories: for the V3 domain, 4117C and 4148D (44), 447-52D (12), and 2182, 2191, and 2219 (19); for the CD4 binding domain, 5145A (43) and 1125H (50); and for the V2 domain, 8.22.2 (22), 2158, and 830A. MAbs 2158 and 830A were produced according to a method previously described (17, 20). Briefly, peripheral blood mononuclear cells from an HIV-1-infected individual were transformed with Epstein-Barr virus; those cells that were producing antibodies reactive with V1V2Case-A2 fusion protein (26) were fused with the human × mouse heteromyeloma SHM-D33 (48). The resulting hybridomas were cloned at limiting dilutions until monoclonality was achieved.
Expression and purification of recombinant Env proteins.
JR-FL env was expressed from an SspI (5473)-to-XhoI (8216) fragment (numbering according to GenBank accession no. U63632) cloned from pSVJR Δ112-1 (42) (obtained from Irvin Chen) into a derivative of pcDNA3.1zeo(−) (Invitrogen) in which the promoter had been replaced with the intron-containing human cytomegalovirus major immediate-early promoter taken from pEE14 (CellTech). SF162 env was expressed from an EcoRI (1)-to-HindIII (3851) fragment (numbering according to GenBank accession no. M38428) cloned from p162-4.5 (9) (obtained from Cecilia Cheng-Mayer) into pcDNA3.1zeo(−) (Invitrogen).
V1/V2 chimeras were created by exchanging DraIII/StuI fragments between JR-FL and SF162 env-encoding plasmids (positions 5908 to 6139 in JR-FL; positions 849 to 1083 in SF162). These restriction sites are in the conserved stem of the V1/V2 loops. The chimeric env genes were expressed in the same formats as the parental genes.
Stable cell lines expressing SF162 and JFL rgp120 were isolated from subcloned 293(T) cells transfected with the Env-expressing vectors and selected with Zeocin. Secreted rgp120 proteins were purified from supernatant medium harvested from cells grown for 48 to 72 h in RPMI 1640 containing reduced FBS (0.5%) by affinity chromatography on agarose columns containing immobilized snowdrop lectin (Galanthus nivalis) (Sigma) as previously described (16). Methyl α-d-mannopyranoside (Sigma) (1 M) in phosphate-buffered saline (PBS) buffer was used to elute the bound proteins. The soluble gp120s produced by these cell lines possessed the same mobilities in sodium dodecyl sulfate-polyacrylamide electrophoresis gels as those isolated from infectious virions both before and after enzymatic removal of N-linked glycans, indicating similar extents of glycosylation and other posttranslational modification for gp120 produced by the sgp120 and gp160 vectors.
Antibody binding assays with soluble gp120.
Sheep anti-HIV-1 gp120 C5 polyclonal antibody (Cliniqa Corp.) dissolved in bicarbonate buffer (pH 9.8) was adsorbed onto enzyme-linked immunosorbent assay (ELISA) plates (Falcon), blocked with 2% dry milk in PBS (pH 7.4), and used with similar concentrations (2 μg/ml) of the two proteins to capture either SF162 or JRFL gp120. The relative levels of affinity of MAbs to the captured gp120 proteins were then determined by titrations. Binding was performed in the presence of 2% dry milk-PBS buffer, wells were washed with PBS-0.1% Tween 20, and bound MAbs were detected using alkaline phosphatase-conjugated species-specific secondary IgG (Zymed Laboratories) followed by addition of substrate (1 mg of p-nitrophenol/ml in DEA buffer [pH 9.8]). A405 measurements were taken using a Spectra SLT ELISA plate reader (TECAN Instruments) at various time points. Relative affinities were determined by the concentration of MAb required to achieve 50% maximal binding to rgp120.
Virus neutralization assays.
Neutralization activity was determined with a single-cycle infectivity assay using virions generated from the Env-defective luciferase-expressing HIVNL4-3 genome pseudotyped with molecularly cloned HIV Env, as previously described (29). Briefly, pseudotyped virions in culture supernatants from transfected 293T cells were incubated with serial dilutions of MAbs or polyclonal sera from HIV-infected subjects for 1 h at 37°C and were then added in the presence of Polybrene (10 μg/ml) to U87-T4-CCR5 target cells plated out in 96-well plates. After 24 h, cells were refed with RPMI medium containing 10% FBS and Polybrene and incubated for an additional 24 to 48 h. Luciferase activity was determined 48 to 72 h postinfection using assay reagents from Promega and a microtiter plate luminometer (HARTA Inc.). The virus input was routinely adjusted to yield between 50,000 and 100,000 relative light units in a 3-day assay. Under these conditions the luciferase activity obtained was linear over an at least 125-fold range of virus concentrations.
RESULTS
Differential sensitivity of HIV pseudotyped with two related Env proteins to neutralization by human sera.
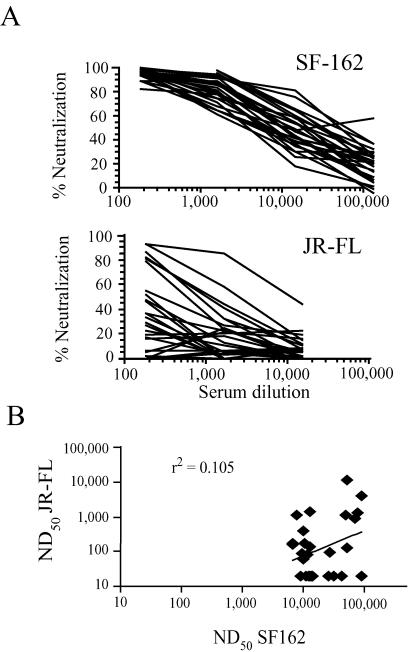
The sensitivity of HIV-1 pseudotyped with either the SF162 or JR-FL envelope proteins to neutralization by a panel of 28 HIV-positive human sera was determined. The sera were all strongly positive against rgp120s by ELISA but were not selected by any other criteria. Despite the high sequence similarity of the SF162 and JR-FL envelope proteins, the two pseudotypes differed greatly in their levels of susceptibility to antibody-mediated neutralization (Fig. 1). All of the analyzed sera strongly neutralized SF162: the serum dilutions required to neutralize 50% of viral activity (ND50s) of all of the sera were above 1:6,000, and 24 of 28 sera exhibited >90% neutralization at a dilution of 1:180. In contrast, only 9 of 28 sera achieved 50% neutralization of the JR-FL pseudotypes at the 1:180 dilution and only two of these gave >90% neutralization at this dilution. A pool of normal human sera did not neutralize either virus at a dilution of 1:20.
FIG. 1.
(A) Neutralization curves for a panel of 28 sera (from HIV-1-infected humans) against SF162 (top panel) and JR-FL (bottom panel) pseudotypes. Sera selected for high-level anti-gp120 titers were serially diluted and preincubated for 30 min with pNL4-3 luc virions pseudotyped with either SF162 or JR-FL Env proteins; the mixture was then added to 96-well plates containing U87/CD4/CCR5 cells. Luciferase activity was measured after 3 days, and percent neutralization was calculated by dividing the difference between that value and luciferase activity for a control sample (virus without antibody) and multiplying by 100. (B) Scatter graph showing relationship between ND50s obtained for SF162 pseudotypes versus those for JR-FL pseudotypes for individual sera analyzed as described for Fig. 1A. The best-fit regression line and r2 value are shown.
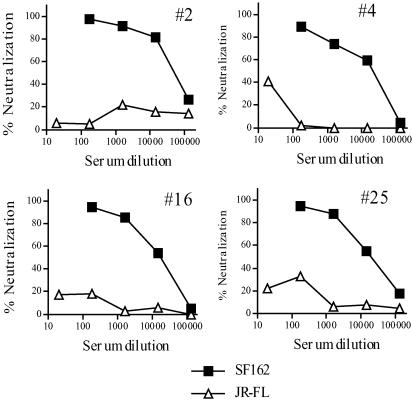
Examination of the neutralization end points of individual sera indicated a lack of correlation between the neutralizing titers of individual sera for the two viruses (Fig. 1B). Eleven sera with no detectable neutralizing activity for JR-FL neutralized SF162 pseudotypes very potently, with ND50s ranging from 1:9,000 to 1:90,000. A total of 13 sera possessed detectable neutralizing activity for JR-FL (ND50, >1:100); of these, only 7 had ND50s above 1:200. All of these also preferentially neutralized the SF162 pseudotype; the average ND50 ratio (SF162/JR-FL) for these 13 sera was 92, and only 3 had ND50 ratios below 10. Representative neutralization curves are shown for four sera that had high-level selectivity for SF162 (Fig. 2).
FIG. 2.
Representative neutralization patterns for four human sera (serum samples 2, 4, 16, and 25) with high-level neutralization selectivity for SF162 against pNL4-3 luc virions pseudotyped with SF162 env or JR-FL env.
Comparative neutralization sensitivities of SF162 and JR-FL pseudotypes to MAbs directed against the V3 domain of HIV Env.
One possible explanation for the discordant results with respect to the neutralizing activity of polyclonal sera for SF162 and JR-FL pseudotypes is that this neutralization was mediated by key epitopes that were present in the SF162 sequence but not in the JR-FL envelope protein. Although the diverse origins of the sera and the high level of sequence similarity between the two env genes make this unlikely, this possibility could not be ruled out a priori. To address this possibility, binding and neutralization assays were performed with purified MAbs directed against previously described neutralization domains.
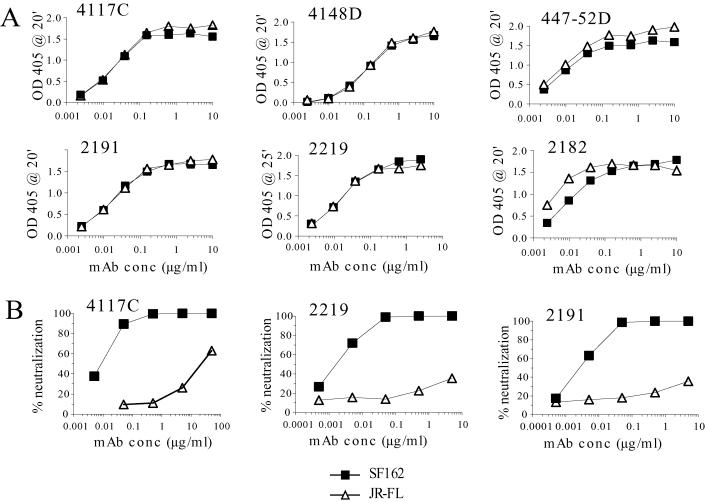
The V3 domain of JR-FL gp120 has the same sequence as the clade B consensus (http://HIV-web.lanl.gov), while that of SF162 gp120 differs at three positions (the HIGPGRAFYTTGE sequence at the center of the JR-FL V3 loop is TIGPGRAFYATGD for SF162). With a few exceptions, these changes have relatively small effects on the apparent affinities of a panel of six V3-specific human MAbs isolated from infected subjects, as evidenced by binding curves against purified SF162 and JR-FL gp120s (Fig. 3A and Table 1). Five of the six MAbs tested produced overlapping binding curves, indicating that they possessed essentially identical affinities for the two rgp120s; MAb 2182 bound the JR-FL gp120 with higher affinity, requiring a fivefold-lower concentration to reach 50% maximal binding.
FIG. 3.
(A) Binding curves of anti-V3 MAbs to rgp120 derived from SF162 and JR-FL env genes. The soluble gp120s were captured with sheep anti-C5 polyclonal antibody; antibody binding was determined at the indicated time points as described in Materials and Methods. (B) Neutralization of SF162 and JR-FL HIV pseudotypes by selected anti-V3 MAbs. Neutralization assays were performed with U87 cells expressing CD4 and CCR5 receptors; percent neutralization was determined as described above. OD 405 @ 20′, optical density at 405 nm after 20 min; conc, concentration.
TABLE 1.
Ratios of 50% maximal binding concentrations and ND50s of MAbs to SF162 and JR-FL pseudotypes
| Epitope | MAb | 50% Maximal binding concna
|
Binding ratio (JR/SF) | ND50b
|
ND50 ratio (JR/SF) | ||
|---|---|---|---|---|---|---|---|
| JR-FL | SF162 | JR-FL | SF162 | ||||
| V3 | 447-52D | 0.009 | 0.009 | 1.0 | 15 | 0.015 | 1,000 |
| 4117C | 0.026 | 0.023 | 1.1 | 35 | 0.015 | 2,333 | |
| 4148D | 0.14 | 0.13 | 1.1 | 100 | 0.042 | 2,381 | |
| 2191 | 0.024 | 0.021 | 1.1 | 18 | 0.004 | 4,500 | |
| 2219 | 0.017 | 0.021 | 0.8 | 24 | 0.003 | 8,000 | |
| 2182 | 0.0024 | 0.01 | 0.2 | 33 | 1.1 | 30 | |
| CD4-bs | 5145A | 0.081 | 0.057 | 1.4 | 50 | 0.040 | 1,250 |
| 1125H | 0.29 | 0.11 | 2.6 | >>100 | 1 | >>100 | |
| IgG-b12 | 0.035 | 0.040 | 0.9 | 0.037 | 0.032 | 1.2 | |
| V2 | 8.22.2 | 0.51 | 0.56 | 0.9 | >>100 | 37 | >>3 |
| 2158 | 0.067 | (1.3)c | (0.05)c | >>50 | 11 | >>5 | |
| 830A | 0.04 | (0.5)c | (0.08)c | >50 | 2 | >25 | |
| CD4-i | X5 (Fab) | NDe | ND | ((20))d | 47 | 0.54 | 87 |
| 17b | ND | ND | ((2.4))d | >>100 | 16 | >>6 | |
| 48d | ND | ND | ND | >>100 | 4.8 | >>20 | |
| 2G12 | 2G12 | 0.12 | 0.42 | 0.3 | 0.38 | 2.4 | 0.16 |
| gp41 | 2F5 | NAf | NA | NA | 0.38 | 1.3 | 0.29 |
Titers were determined for serially diluted MAbs in ELISA plates coated with purified gp120 antigens; antibody concentrations (in micrograms per milliliter) that gave 50% maximal binding are reported.
ND50s were determined for pNL4-3 luc virions pseudotyped with the indicated envelope proteins by interpolation from neutralization curves and are reported in micrograms per milliliter.
The 50% maximal binding concentrations and ratios listed in parentheses represent values estimated by assuming that the maximum binding plateau for SF162 gp120 (value not achieved) was the same as that for JR-FL gp120.
Binding plateaus were not obtained. Values listed in double parentheses represent ratios of concentrations required to achieve an optical density of 0.55 after 25 min.
ND, not determined (binding plateaus not obtained).
NA, not applicable.
Despite their similar affinities for soluble gp120s from the two viruses, these MAbs preferentially neutralized the SF162 pseudotype, in many cases requiring concentrations between 3 and 4 orders of magnitude lower than that required for equivalent neutralization of JR-FL (Fig. 3B, Table 1). Consistent with its lower affinity for SF162 gp120, the potency of MAb 2182 for SF162 was considerably lower than that of the other V3 MAbs (Table 1). Yet despite its greater affinity for JR-FL gp120, 2182 neutralized SF162 pseudotypes with a 30-fold-lower ND50 than JR-FL pseudotypes. These results showed that the V3 domain was a much less effective neutralization target in the JR-FL Env protein than in the SF162 Env protein complex, even for antibodies possessing equivalent or greater affinities for the soluble JR-FL gp120 protein.
Differential neutralizing activity of MAbs directed against other epitopes for SF162 and JR-FL pseudotypes.
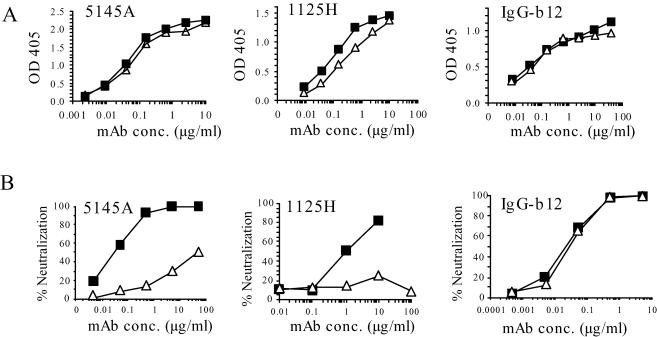
The relative neutralization sensitivities of non-V3 target sites on the SF162 and JR-FL Env proteins were examined by performing similar binding and neutralization assays with MAbs directed against epitopes located in other regions of HIV Env proteins. Three MAbs directed against the CD4 binding site (CD4-bs) were tested: 5145A (43) and 1125H (50), two standard human MAbs isolated from B cells of HIV-infected patients, and IgG-b12, isolated from a phage library expressing human heavy- and light-chain sequences cloned from an HIV-infected individual. As was observed for the anti-V3 MAbs, the affinities of 5145A and IgG-b12 for the two rgp120s were very similar whereas 1125H bound more strongly to SF162 rgp120, with a 2.6-fold-lower 50% maximal binding concentration (Fig. 4A and Table 1). Both 5145A and 1125H preferentially neutralized SF162 pseudotypes (Fig. 4B and Table 1). For 5145A, the ND50 ratio was more than 3 orders of magnitude; this ratio could not be determined for 1125H because no neutralization was detected for JR-FL pseudotypes even at the highest concentration tested, but the shapes of the curves suggested that the preference of 1125H for SF162 was at least as great. In contrast to this result, IgG-b12 possessed essentially equivalent neutralization potency levels for the two viral pseudotypes.
FIG. 4.
(A) Binding curves of anti-CD4-bs MAbs to SF162 (squares) and JR-FL (triangles) rgp120s. Binding assays were performed as described for the anti-V3 MAbs. (B) Neutralization curves of three anti-CD4-bs MAbs against SF162 (squares) and JR-FL (triangles) pseudotypes of pNL4-3 luc. OD 405, optical density at 405 nm; conc, concentration.
Similar comparisons were performed for three MAbs directed to the V2 domain: 8.22.2, directed against a linear epitope located at the beginning of the V2 crown (22), and 2158 and 830A, directed against conserved V2-specific disulfide-dependent epitopes similar to that recognized by the previously described human MAb 697D (18). 8.22.2 bound equally well to the two gp120s, but while it weakly neutralized SF162 (ND50 = 37 μg/ml) it did not neutralize the JR-FL pseudotypes at 100 μg/ml (Table 1). MAbs 2158 and 830A bound considerably more strongly to JR-FL gp120 than to SF162 gp120 and yet also neutralized SF162 but not JR-FL pseudotypes. These results indicated that sites in the V2 domain were also more potent neutralization targets in SF162 pseudotypes than in JR-FL pseudotypes.
MAbs with reported neutralizing activity directed against additional target sites were also tested in similar assays. Two MAbs (17b and 48d) (49) and one Fab (X5) (40) directed against “CD4-induced” epitopes in gp120 whose expression is enhanced upon binding of CD4 were examined. Because of their relatively low affinity levels and the limited amounts of material available, binding plateaus were not obtained for these antibodies. However, the binding curves indicated that these antibodies recognized SF162 gp120 more strongly than JR-FL gp120 (Table 1). MAbs 17b and 48d had weak neutralizing activities for SF162 and no detectable activity for JR-FL pseudotypes. The X5 Fab had stronger neutralizing activity for the SF162 pseudotype (ND50 = 0.5 μg/ml); the higher activity of this reagent was consistent with a recent report indicating that antibodies to CD4-induced epitopes had limited access to CD4-engaged gp120 complexes and that this can be overcome by the reduced size of Fab fragments (33). The X5 Fab fragment was 87-fold less potent against the JR-FL pseudotype (ND50 = 47 μg/ml). These data showed that the CD4-induced epitopes were also preferential neutralization targets in the SF162 envelope protein. For these antibodies, however, increased neutralization potencies may be directly due to increased binding affinities.
A strikingly different pattern was obtained for MAb 2G12 (Table 1). This antibody is directed against a glycan-dependent epitope in the C-terminal half of gp120 (4, 45, 46) and has been reported to possess potent neutralizing activity against a large number of primary HIV isolates (52). 2G12 possessed a higher level of neutralizing activity for the JR-FL pseudotype than for the SF162 pseudotype, with a sixfold-lower ND50 for JR-FL. This enhanced neutralization of JR-FL may be accounted for in part by the preferential affinity of 2G12 for JR-FL gp120, as reflected by a 3.5-fold-lower 50% maximum binding concentration.
Preferential neutralizing activity for the JR-FL pseudotype was also obtained for MAb 2F5, directed against a site in gp41 C terminal to the second heptad repeat region (13, 56). The ND50 for this MAb was 3.4-fold lower for JR-FL pseudotypes than for SF162 pseudotypes. Although the binding efficiency of 2F5 for JR-FL and SF162 gp41 was not tested, the extended sequence encoding this epitope (IEESQNQQEKNEQELLELDKWASLW) (56) is completely conserved between the SF162 and JR-FL gp41 sequences, suggesting that this difference in neutralization sensitivity to 2F5 is not due to a sequence change in the epitope itself but is instead due to changes elsewhere in Env.
Determinants of the difference in levels of neutralization sensitivity between SF162 and JR-FL Env map to the V1/V2 domain.
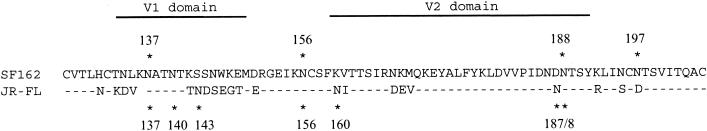
A comparison of the SF162 and JR-FL gp120 sequences showed a high level (89%) of conservation, with changes concentrated in the V1/V2, V4, and V5 domains. Previous studies have shown that introducing mutations or deletions into the V1/V2 domain of HIV-1 gp120s can have a dramatic effect on the sensitivity of viral isolates to neutralization by human antisera, sCD4, and MAbs to the V3 region and CD4-bs (5, 8, 34, 39, 47). To examine the contribution of the V1/V2 domain to the differential levels of neutralization sensitivity of the SF162 and JR-FL viral pseudotypes, chimeric env genes were prepared in which the entire V1/V2 domain was switched between the two sequences. A total of 21 out of 79 amino acids in the V1/V2 regions differed between the two sequences, and the JR-FL sequence contained three more sites for N-linked glycosylation than the SF162 sequence (7 sites versus 4) (Fig. 5). Viruses pseudotyped with the chimeric Env proteins yielded high levels of luciferase activity (data not shown), indicating that the two chimeric env genes were both functional.
FIG. 5.
Sequence alignment of the V1/V2 domains of SF162 and JR-FL gp120 proteins. Homologous residues are indicated by hyphens; modified asparagines of N-linked glycosylation sites are marked with asterisks. Numbering is based on the relative positions in the HXB2 sequence.
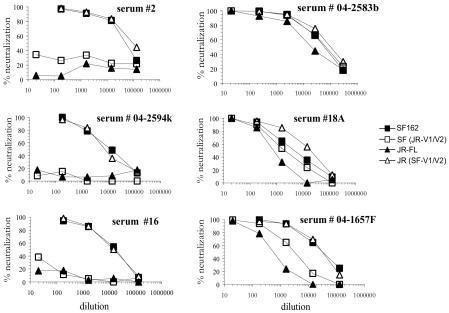
Upon comparing the neutralization sensitivities of the chimeric Envs with those of both human immune sera and MAbs, it was clear that the neutralization phenotype was determined by the V1/V2 domain and not by the sequences at the target epitopes (Fig. 6 and Table 2). In general, the neutralization activity of human immune sera for the JR(SF V1/V2) chimera was similar to that of SF162. All of the sera, including those with no neutralizing activity for the JR-FL parent, potently neutralized the JR(SF V1/V2) chimera. For most sera, ND50s for this chimera differed less than twofold from that for SF162, while five sera neutralized the JR(SF V1/V2) chimera more potently (two- to eightfold) than SF162. Thus, the JR-FL backbone was not intrinsically more resistant; resistance was conferred by its V1/V2 domain. Similarly, the SF(JR V1/V2) chimera more closely resembled the resistant JR-FL parent than the sensitive SF162 parent. For almost half of the sera (13 of 28) this chimera was more than 100-fold (129- to 4,500-fold) more resistant than the SF162 parent, while for 9 sera it was 10- to 80-fold more resistant. For five sera the resistance was increased only two- to ninefold, and one serum sample neutralized this chimera as strongly as it did SF162. Thus, with but a few exceptions, the JR-FL V1/V2 domain conferred significant neutralization resistance to the SF162 backbone.
FIG. 6.
Representative neutralization curves for human sera against SF162 and JR-FL parental and JR(SF-V1/V2) and SF(JR-V1/V2) Env pseudotypes. The three left panels show patterns for sera that preferentially neutralize SF162 pseudotypes; the three right panels show patterns for the sera with the greatest cross-neutralizing activities.
TABLE 2.
Relative ND50s of human sera for parental and chimeric SF162/JR-FL Env pseudotypes
| Human serum | ND50 for pseudotypea:
|
|||
|---|---|---|---|---|
| SF162 | JR(SF V1/V2) | JR-FL | SF(JR V1/V2) | |
| 2 | 90,000 | 115,000 | <20 | <20 |
| 4 | 43,000 | 75,000 | <20 | 120 |
| 25 | 31,000 | 13,000 | <20 | <20 |
| 16 | 25,000 | 16,000 | <20 | <20 |
| 04-2594k | 14,000 | 10,800 | <20 | <20 |
| 26 | 14,000 | 10,500 | <20 | 40 |
| 04-2435j | 13,500 | 8,500 | <20 | <20 |
| 04-1642F | 12,800 | 32,000 | <20 | <20 |
| 04-8942K | 12,200 | 13,500 | <20 | 20 |
| 9 | 11,000 | 11,800 | <20 | 20 |
| 04-2416d | 9,000 | 13,600 | <20 | <20 |
| 6 | 51,000 | 36,000 | 130 | 1,500 |
| 04-1360c | 27,000 | 36,000 | 95 | 140 |
| 04-3058c | 10,000 | 16,800 | 60 | 270 |
| 3 | 11,500 | 12,800 | 80 | 140 |
| 04-3062c | 9,500 | 8,700 | 90 | 150 |
| 17 | 12,700 | 9,100 | 140 | 1,480 |
| 04-1657F | 71,000 | 66,000 | 930 | 5,700 |
| 04-2132E | 75,000 | 93,000 | 1,300 | 3,000 |
| 21 | 10,500 | 29,000 | 180 | 1,100 |
| 04-0748D | 48,000 | 48,000 | 1,150 | 5,500 |
| 10 | 6,500 | 13,000 | 160 | 135 |
| 04-2213c | 6,700 | 12,000 | 180 | 1,000 |
| 22 | 10,000 | 14,000 | 400 | 1,000 |
| 04-0405J | 90,000 | 120,000 | 4,000 | 700 |
| 04-03552A | 12,800 | 97,000 | 1,400 | 5,300 |
| 18a | 7,500 | 29,000 | 1,100 | 2,900 |
| 04-2583b | 52,000 | 78,000 | 12,000 | 58,000 |
| NHSb | <20 | <20 | <20 | <20 |
ND50s correspond to dilutions required to reduce luc activity by 50%.
NHS, normal human sera (a pool of 9 normal sera tested as a control).
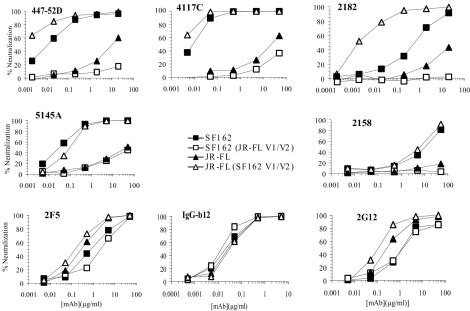
The preferential neutralization of the SF162 Env seen with most of the MAbs tested also correlated with the origin of the V1/V2 domain (Fig. 7 and Table 3). In contrast to the resistance of the parental JR-FL Env, all of the V3-specific MAbs neutralized the JR(SF V1/V2) chimera extremely efficiently and in several cases the chimera was neutralized considerably more efficiently than the SF162 Env parent. For example, the ND50 of 447-52D for the JR(SF V1/V2) chimera was more than 11,500-fold lower than for the JR-FL parent and almost 12 times lower than for SF162 and the ND50 for 2182 was 16,500-fold lower for this chimera than for JR-FL and 550-fold lower than for SF162. None of the V3 MAbs achieved 50% neutralization for the SF(JR V1/V2) chimera at the concentrations tested; when the comparison could be made, the SF(JR V1/V2) chimera was more resistant than the JR-FL parent.
FIG. 7.
Representative neutralization curves for MAbs against SF162 and JR-FL parental and JR(SF-V1/V2) and SF(JR-V1/V2) Env pseudotypes.
TABLE 3.
ND50s of MAbs for SF162 and JR-FL parental and chimeric viral pseudotypes
| Epitope | MAb ND50 | ND50 for pseudotypea:
|
|||
|---|---|---|---|---|---|
| SF162 | JR(SF V1/V2) | JR-FL | SF(JR V1/V2) | ||
| V3 | 447-52D | 0.015 | 0.0013 | 15 | >50 |
| 4117C | 0.015 | 0.005 | 35 | >50 | |
| 4148D | 0.042 | <0.01 | 100 | >100 | |
| 2191 | 0.004 | 0.002 | 18 | >5 | |
| 2219 | 0.003 | 0.002 | 24 | >5 | |
| 2182 | 1.1 | 0.002 | 33 | >50 | |
| V2 | 8.22.2 | 37 | 20 | >>100 | >>100 |
| 830A | 2 | 2 | >50 | >50 | |
| 2158 | 11 | 10 | >>50 | >>50 | |
| CD4-bs | IgG-b12 | 0.032 | 0.04 | 0.037 | 0.025 |
| 5145A | 0.04 | 0.18 | 50 | 50 | |
| 1125H | 1 | 0.8 | >>100 | >>100 | |
| sCD4 | 0.41 | 0.1 | 3.7 | >10 | |
| gp41 | 2F5 | 1.3 | 0.26 | 0.38 | 3.4 |
| CD4i | 17b | 16 | 6.5 | >>100 | >100 |
| 48d | 4.8 | 5.2 | >>100 | >>100 | |
| X5 | 0.54 | 0.25 | 47 | 19 | |
| Other | 2G12 | 2.4 | 0.20 | 0.38 | 2.5 |
Concentration of antibody (in micrograms per milliliter) required to reduce luciferase activity by 50%.
Similar effects of the V1/V2 domain on neutralizing potency were observed for MAbs to the V2 domain, for the CD4i epitopes, for two MAbs to the CD4-bs, and for sCD4 itself. However, the V1/V2 domain had a minimal effect on IgG-b12, 2G12, and 2F5, the three MAbs that efficiently neutralized the parental JR-FL pseudotype. IgG-b12 neutralized each of the parental and chimeric Env pseudotypes with similar potencies. 2G12 and 2F5 both neutralized JR-FL more efficiently than SF162, and relatively small effects were seen with the V1/V2-substituted chimeric Envs. Replacement of the JR-FL V1/V2 domain by the corresponding SF162 sequence resulted in less than 2-fold decreases in ND50s, and reciprocal replacement of the SF162 V1/V2 domain by the corresponding sequence of JR-FL resulted in a 2.6-fold increase in ND50 for MAb 2F5 and a 0.6-fold decrease in ND50 for 2G12. Thus, these three unusual MAbs with broad neutralizing activities differed from the more narrowly neutralizing antibodies in that they were not affected by V1/V2 sequence.
DISCUSSION
This study explored the factors responsible for the large difference in levels of neutralization sensitivity mediated by env genes derived from two related R5-tropic primary HIV-1 isolates. Despite the high degree of sequence homology of these genes, the ND50s of most of the human sera examined for SF162 Env were more than 100-fold lower than that for JR-FL Env and for many sera the differences were more than 1,000-fold. Differences in ND50s of more than 1,000-fold for the two Envs were also observed for MAbs directed against the V3 domain and the CD4-bs, with significant (although smaller) differences seen for MAbs to additional sites, including the V2 domain and several CD4-induced epitopes. Similar differences in neutralization efficiency were observed for selected human sera and MAbs when the neutralization assays were performed using human peripheral blood mononuclear cells (data not shown), indicating that these effects were not artifacts specific to engineered U87 cells. These differences were also seen for MAbs that possessed equivalent levels of affinity for soluble SF162 and JR-FL gp120s as measured by pseudoaffinity measurements (Fig. 3A), indicating that the selective neutralization sensitivity of those MAbs was not due to sequence polymorphisms at the epitopes themselves but to some other factor. Analyses of chimeric env genes in which the V1/V2 domains were exchanged demonstrated that the V1/V2 structure was the major determinant for this selective neutralization.
The Env protein possessing the JR-FL backbone with the SF162-derived V1/V2 sequence had sensitivity to neutralization by the large majority of sera similar to or greater than that seen with the SF162 parental Env, indicating that, once the effects of the V1/V2 domain were eliminated, the JR-FL backbone was not inherently more resistant to neutralizing antibodies in human sera than the SF162 backbone. In parallel, replacing the SF162 Env V1/V2 domain with the corresponding JR-FL-derived sequence significantly increased its resistance to neutralization, although this chimeric Env remained somewhat (2- to 12-fold) more sensitive to neutralization by half of the sera than the JR-FL parent. The effect of the V1/V2 domain on neutralization was particularly significant for the V3 MAbs. The very large ND50 ratios obtained for the V3-specific MAbs for SF162 and JR-FL Envs increased even more dramatically when V3 sequence variations present between these two strains were factored out. MAb 447-52D neutralized the Env chimera with the JR-FL backbone and the SF162 V1/V2 domain with >11,000-fold-higher potency than it did JR-FL, while MAb 2182 neutralized this chimera 16,500-fold more potently than it did JR-FL. These are examples in which a very potent neutralizing activity of an antibody possessing high affinity against its target epitope is almost completely negated by structural features outside the epitope itself.
Of interest were the exceptional human MAbs (IgG-b12, 2F5, and 2G12) and the occasional human sera that also possessed potent neutralizing activities against JR-FL and were not sensitive to the V1/V2 neutralization blocking effect. This correlation suggests that insensitivity to V1/V2 effects might be a determinant for such broad potency and further argues that the inability of other antibodies to broadly neutralize primary isolates might in many cases be due to V1/V2 blocking effects rather than to the absence or variation of the actual epitope. IgG-b12 and 2G12 have been reported to detect oligomeric JR-FL Env on the cell surface whereas nonneutralizing antibodies did not (15), suggesting that the neutralization activity of these MAbs may correlate with exposure of their epitopes on the oligomeric form of the envelope glycoprotein complex.
Only a few human sera possessed neutralizing activity that was relatively insensitive to the V1/V2 structure. The ND50 ratios for Envs with the SF162 backbone [SF162 versus SF(JR V1/V2)] were below 3 for three sera (04-03552A, 18a, and 04-2583b). Serum 04-2583b neutralized these two Envs equally well and also had an unusually high neutralizing titer for JR-FL (ND50 of 1:12,000). The nature of the antibodies mediating the potent neutralization of these sera is not known. They may be directed against epitopes related to the b12/2G12/2F5 sites or may be directed against other epitopes, possibly including some that have not yet been defined.
The inhibitory effect of the JR-FL V1/V2 domain on neutralization is consistent with a number of previous studies showing that mutations or deletions in the V1/V2 region of several cloned HIV-1 env genes can affect the sensitivity of those strains to neutralization by antibodies against multiple domains (5, 47, 54). Other studies indicated that for the SIVmac239 isolate, the V1/V2 region was a major factor inducing resistance to neutralization (23). While details regarding the effects of mutations at specific sites on neutralization by specific antibodies differ between these studies, their overall impact suggests that effects related to the V1/V2 modulation of neutralization sensitivity demonstrated in the present study might be widespread for both HIV-1 and simian immunodeficiency virus isolates. Several of these studies have suggested an important role for carbohydrates in conferring neutralization escape, either by altering the structure of gp120 or by shielding the major neutralization sites (5, 8, 24, 34). In this regard the difference in the number of N-linked glycans present in the V1/V2 domains of SF162 and JR-FL may be relevant: whereas JR-FL possesses seven glycans, SF162 has only four, an atypically low number for this domain (Fig. 5). JR-FL has three V1 glycans versus one for SF162; while both sequences share a highly conserved glycosylation site at position 156 and a glycan at position 188, JR-FL has two glycans (at positions 160 and 187) not present in the SF162 sequence whereas SF162 has a glycan at position 197 that is absent from the JR-FL sequence. It is possible that one or more of the glycans that are present in the JR-FL sequence but not in the SF162 sequence contribute to the effects described in this study.
These results do not preclude the possibility that for some primary Envs, changes at regions outside the V1/V2 domain can also induce neutralization resistance by related masking mechanisms. A recent analysis of HIV escape mutants evolving in response to autologous neutralizing antibodies showed that the mutations primarily involved changes in N-linked glycosylation at multiple sites, including the C terminus of V2, the C2 region, and the N-terminal base of V3 and V4 (53). These authors postulated that an evolving “glycan shield” consisting of N-linked glycans that sterically block access of neutralizing antibodies but not interactions with receptors necessary for infection controls sensitivity to neutralization.
Additional, more subtle aspects of V1/V2 structure may contribute to the observed effects of this region on neutralization. The C-terminal portion of the V1/V2 stem includes the β3 strand of the bridging sheet important for maintaining the global structure of gp120 (31), and a human MAb against gp120 that recognizes an epitope that is competed against by sCD4 and antibodies to epitopes in the CD4-bs, V2, and V3 domains has recently been described (55), suggesting that elements of these three domains exist in close proximity in gp120. Some of the amino acid variations between the two V1/V2 domains may affect these interactions. However, the inability of many of the MAbs studied to neutralize the resistant strains despite high-level affinity for soluble gp120 from that strain suggests that the neutralization-inhibitory effects of the V1/V2 domain are manifested in the context of the native oligomer rather than the monomer. Structural models of the Env trimer have been proposed that place the base of the V1/V2 loop of one subunit in proximity to the V3 loop of a neighboring subunit (30, 32). Thus, it can be envisioned that specific glycans or other structural features of the V1/V2 domain of one subunit might sterically interfere with access by antibodies to the V3 loop and other neutralization targets on an adjacent subunit on the functional spike.
Many of the current approaches towards developing improved HIV-1 vaccines are focused on designing modified gp120- or gp140-based immunogens that can induce high-level titers of antibodies against conserved neutralization epitopes. However, the demonstration that many immune human sera with high-level titers of antibodies against known neutralization epitopes fail to neutralize a virus expressing those epitopes suggests that the induction of similar antibodies (even when they possess high-level affinities and are present in high-level titers) might not be sufficient for broad protection. Vaccines capable of inducing antibodies against sites defined by the MAbs not sensitive to V1/V2 blocking may be more effective. This effectiveness could include specificities such as those of IgG-b12, 2G12, or 2F5. However, recent structural analyses of these MAbs indicate that they possess unusual structures that might not be easily induced (4, 55a, 57). Further characterization of human sera such as those examined in this study that possessed potent neutralizing activities not affected by V1/V2 blocking effects may allow the identification of additional neutralization sites in HIV-1 Env that might provide new targets amenable to vaccine development.
Acknowledgments
These studies were supported by U.S. Public Health Service grants AI46283 and AI50452 (to A.P.), AI51987 (to S.C.K.), and AI36085 and HL59725 (to S.Z.-P.). Additional support for these studies was provided by grants from the Department of Veterans Affairs and from the New York University Center for AIDS Research (funded by U.S. Public Health Service grant AI277742).
REFERENCES
- 1.Berman, P. W., A. M. Gray, T. Wrin, J. C. Vennari, D. J. Eastman, G. R. Nakamura, D. P. Francis, G. Gorse, and D. H. Schwartz. 1997. Genetic and immunologic characterization of viruses infecting MN-rgp120-vaccinated volunteers. J. Infect. Dis. 176:384-397. [DOI] [PubMed] [Google Scholar]
- 2.Bolognesi, D. P., and T. J. Matthews. 1998. Viral envelope fails to deliver? Nature 391:638-639. [DOI] [PubMed] [Google Scholar]
- 3.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. H. I. Parren, L. S. W. Sawyer, R. M. Hendry, N. Dunlop, and P. L. Nara. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 4.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 5.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cecilia, D., V. N. KewalRamani, J. O'Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, L. D. R., and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, B. K., K. Saksela, R. Andino, and D. Baltimore. 1994. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J. Virol. 68:654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng-Mayer, C., A. Brown, J. Harouse, P. A. Luciw, and A. J. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng-Mayer, C., M. Quiroga, J. W. Tung, D. Dina, and J. A. Levy. 1990. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J. Virol. 64:4390-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng-Mayer, C., C. Weiss, D. Seto, and J. A. Levy. 1989. Isolates of human immunodeficiency virus type 1 from the brain may constitute a special group of the AIDS virus. Proc. Natl. Acad. Sci. USA 86:8575-8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen, J. 2003. Public health: AIDS vaccine trial produces disappointment and confusion. Science 299:1290-1291. [DOI] [PubMed] [Google Scholar]
- 12.Conley, A. J., M. K. Gorny, J. A. Kessler II, L. J. Boots, M. Ossorio-Castro, S. Koenig, D. W. Lineberger, E. A. Emini, C. Williams, and S. Zolla-Pazner. 1994. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J. Virol. 68:6994-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conley, A. J., J. A. Kessler II, L. J. Boots, J.-S. Tung, B. A. Arnold, P. M. Keller, A. R. Shaw, and E. A. Emini. 1994. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proc. Natl. Acad. Sci. USA 91:3348-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connor, R. I., B. T. M. Korber, B. S. Graham, B. H. Hahn, D. D. Ho, B. D. Walker, S. H. Vermund, J. Mestecky, S. Jackson, E. Fenamore, Y. Cao, F. Gao, S. Kalams, K. J. Kunstman, D. McDonald, N. McWilliams, A. Trkola, J. P. Moore, and S. M. Wolinsky. 1998. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J. Virol. 72:1552-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilljam, G. 1993. Envelope glycoproteins of HIV-1, HIV-2, and SIV purified with Galanthus nivalis agglutinin induce strong immune responses. AIDS Res. Hum. Retrovir. 9:431. [DOI] [PubMed] [Google Scholar]
- 17.Gorny, M. K. 1994. Production of human monoclonal antibodies via fusion of Epstein-Barr virus-transformed lymphocytes with heteromyeloma, p. 276-281. In J. E. Celis (ed.), Cell biology: a laboratory handbook, vol. 2. Academic Press, San Diego, Calif. [Google Scholar]
- 18.Gorny, M. K., J. P. Moore, A. J. Conley, S. Karwowska, J. Sodroski, C. Williams, S. Burda, L. J. Boots, and S. Zolla-Pazner. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of HIV-1. J. Virol. 68:8312-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, S. Cohen, V. R. Polonis, W. J. Honnen, S. C. Kayman, C. Krachmarov, A. Pinter, and S. Zolla-Pazner. 2002. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J. Virol. 76:9035-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorny, M. K., J.-Y. Xu, V. Gianakakos, S. Karwowska, C. Williams, H. W. Sheppard, C. V. Hanson, and S. Zolla-Pazner. 1991. Production of site-selected neutralizing human monoclonal antibodies against the third variable domain of the HIV-1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 88:3238-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham, B. S., M. J. McElrath, R. I. Connor, D. H. Schwartz, G. J. Gorse, M. C. Keefer, M. J. Mulligan, T. J. Matthews, S. M. Wolinsky, D. C. Montefiori, S. H. Vermund, J. S. Lambert, L. Corey, R. B. Belshe, R. Dolin, P. F. Wright, B. T. Korber, M. C. Wolff, P. E. Fast, et al. 1998. Analysis of intercurrent human immunodeficiency virus type 1 infections in phase I and II trials of candidate AIDS vaccines. J. Infect. Dis. 177:310-319. [DOI] [PubMed] [Google Scholar]
- 22.He, Y., W. J. Honnen, C. P. Krachmarov, M. Burkhart, S. C. Kayman, J. Corvalan, and A. Pinter. 2002. Efficient isolation of novel human monoclonal antibodies with neutralizing activity against HIV-1 from transgenic mice expressing human IG loci. J. Immunol. 169:595-605. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, W. E., J. Morgan, J. Reitter, B. A. Puffer, S. Czajak, R. W. Doms, and R. C. Desrosiers. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 76:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, W. E., H. Sanford, L. Schwall, D. R. Burton, P. W. Parren, J. E. Robinson, and R. C. Desrosiers. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 77:9993-10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahn, J., K. S. Steimer, J. Baenziger, A. M. Duliege, M. Feinberg, T. Elbeik, M. Chesney, N. Murcar, D. Chernoff, and F. Sinangil. 1995. Clinical, immunologic, and virologic observations related to human immunodeficiency virus (HIV) type 1 infection in a volunteer in an HIV-1 vaccine clinical trial. J. Infect. Dis. 171:1343-1347. [DOI] [PubMed] [Google Scholar]
- 26.Kayman, S. C., Z. Wu, K. Revesz, H.-C. Chen, R. Kopelman, and A. Pinter. 1994. Presentation of native epitopes in the V1/V2 and V3 domains of HIV-1 gp120 by fusion glycoproteins containing fragments of gp120. J. Virol. 68:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kostrikis, L. G., Y. Cao, H. Ngai, J. P. Moore, and D. D. Ho. 1996. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J. Virol. 70:445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koyanagi, Y., S. Miles, R. T. Mitsuyasu, J. E. Merrill, H. V. Vinters, and I. S. Chen. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819-822. [DOI] [PubMed] [Google Scholar]
- 29.Krachmarov, C. P., S. C. Kayman, W. J. Honnen, O. Trochev, and A. Pinter. 2001. V3-specific polyclonal antibodies affinity purified from sera of infected humans effectively neutralize primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1737-1748. [DOI] [PubMed] [Google Scholar]
- 30.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 31.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, D. S. Burke, et al. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 36.McElrath, M., L. Corey, P. D. Greenberg, T. J. Matthews, D. C. Montefiori, L. Rowen, L. Hood, and J. I. Mullins. 1996. Human immunodeficiency virus type 1 infection despite prior immunization with a recombinant envelope vaccine regimen. Proc. Natl. Acad. Sci. USA 93:3972-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montefiori, D. C., R. G. Collman, T. R. Fouts, J. Y. Zhou, M. Bilska, J. A. Hoxie, J. P. Moore, and D. P. Bolognesi. 1998. Evidence that antibody-mediated neutralization of human immunodeficiency virus type 1 by sera from infected individuals is independent of coreceptor usage. J. Virol. 72:1886-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morikita, T., Y. Maeda, S.-I. Fujii, S. Matsushita, K. Obaru, and K. Takatsuki. 1997. The V1/V2 region of human immunodeficiency virus type 1 modulates the sensitivity to neutralization by soluble CD4 and cellular tropism. AIDS Res. Hum. Retrovir. 13:1291-1299. [DOI] [PubMed] [Google Scholar]
- 40.Moulard, M., S. K. Phogat, Y. Shu, A. F. Labrijn, X. Xiao, J. M. Binley, M. Y. Zhang, I. A. Sidorov, C. C. Broder, J. Robinson, P. W. Parren, D. R. Burton, and D. S. Dimitrov. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. USA 99:6913-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Brien, W. A., Y. Koyanagi, A. Namazie, J.-Q. Zhao, A. Diagne, K. Idler, J. A. Zack, and I. S. Y. Chen. 1990. HIV-1 tropism for mononuclear phagocytes can be determined by regions outside the CD4-binding domain. Nature 348:69-73. [DOI] [PubMed] [Google Scholar]
- 43.Pinter, A., W. J. Honnen, M. E. Racho, and S. A. Tilley. 1993. A potent, neutralizing human monoclonal antibody against a unique epitope overlapping the CD4-binding site of HIV-1 gp120 that is broadly conserved across North American and African virus isolates. AIDS Res. Hum. Retrovir. 9:985-996. [DOI] [PubMed] [Google Scholar]
- 44.Pinter, A., W. J. Honnen, and S. A. Tilley. 1993. Conformational changes affecting the V3 and CD4-binding domains of human immunodeficiency virus type 1 gp120 associated with env processing and with binding of ligands to these sites. J. Virol. 67:5692-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stamatatos, L., and C. Cheng-Mayer. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teng, N. N., K. S. Lam, F. C. Riera, and H. S. Kaplan. 1983. Construction and testing of mouse-human heteromyelomas for human monoclonal antibody production. Proc. Natl. Acad. Sci. USA 80:7308-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tilley, S. A., W. J. Honnen, M. E. Racho, M. Hilgartner, and A. Pinter. 1991. A human monoclonal antibody against the CD4 binding site of HIV-1 gp120 exhibits potent, broadly neutralizing activity. Res. Virol. 142:247-259. [DOI] [PubMed] [Google Scholar]
- 51.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas III, D. R. Burton, D. D. Ho, and J. P. Moore. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, C. B., L. Zhu, S. Holz-Smith, T. J. Matthews, and C. H. Chen. 2001. The role of the third beta strand in gp120 conformation and neutralization sensitivity of the HIV-1 primary isolate DH012. Proc. Natl. Acad. Sci. USA 98:15227-15232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zwick, M. B., R. Kelleher, R. Jensen, A. F. Labrijn, M. Wang, G. V. Quinnan, Jr., P. W. Parren, and D. R. Burton. 2003. A novel human antibody against human immunodeficiency virus type 1 gp120 is V1, V2, and V3 loop dependent and helps delimit the epitope of the broadly neutralizing antibody immunoglobulin G1 b12. J. Virol. 77:6965-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55a.Zwick, M. B., H. K. Komori, R. L. Stanfield, S. Church, M. Wang, P. W. Parren, R. Kunert, H. Katinger, I. A. Wilson, and D. R. Burton. 2004. The long third complementarity-determining region of the heavy chain is important in the activity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5. J. Virol. 78:3155-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zwick, M. B., P. W. Parren, E. O. Saphire, S. Church, M. Wang, J. K. Scott, P. E. Dawson, I. A. Wilson, and D. R. Burton. 2003. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J. Virol. 77:5863-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]