Abstract
All gammaherpesviruses encode a virion glycoprotein positionally homologous to Epstein-Barr virus gp350. These glycoproteins are thought to be involved in cell binding, but little is known of the roles they might play in the whole viral replication cycle. We have analyzed the contribution of murine gammaherpesvirus 68 (MHV-68) gp150 to viral propagation in vitro and host colonization in vivo. MHV-68 lacking gp150 was viable and showed normal binding to fibroblasts and normal single-cycle lytic replication. Its capacity to infect glycosaminoglycan (GAG)-deficient CHO-K1 cells and NS0 and RAW264.7 cells, which express only low levels of GAGs, was paradoxically increased. However, gp150-deficient MHV-68 spread poorly through fibroblast monolayers, with reduced cell-free infectivity, consistent with a deficit in virus release. Electron microscopy showed gp150-deficient virions clustered on infected-cell plasma membranes. MHV-68-infected cells showed reduced surface GAG expression, suggesting that gp150 prevented virions from rebinding to infected cells after release by making MHV-68 infection GAG dependent. Surprisingly, gp150-deficient viruses showed only a transient lag in lytic replication in vivo and established normal levels of latency. Cell-to-cell virus spread and the proliferation of latently infected cells, for which gp150 was dispensable, therefore appeared to be the major route of virus propagation in an infected host.
Herpesviruses are large, complex pathogens that use a range of different glycoproteins to spread between cells and between hosts (34). A common first step in infection is virion adsorption to cell surface glycosaminoglycans (GAGs), such as heparan sulfate, which are widespread on epithelial surfaces (5). Herpesvirus-GAG interactions are thought to concentrate virions on cell surfaces and so promote specific protein receptor binding, which is then followed by membrane fusion (32). Cell binding by purified virions and recombinant viral glycoproteins has been studied extensively in vitro, but relatively little is known about how individual herpesvirus glycoproteins contribute to infection of the natural host. Here simple fluid-filled spaces are rare, extracellular matrix is abundant, and viral spread is probably limited to defined anatomical pathways. A significant role for direct cell-to-cell spread in herpesvirus infections is implied by pseudorabies virus that lacks glycoprotein D and infects cells poorly as free virus propagating efficiently in mice (28) and by herpes simplex virus that lacks glycoprotein E (required only for cell-to-cell spread of virus) disseminates inefficiently from the rat cornea (14).
Our understanding of in vivo glycoprotein function is especially limited with gammaherpesviruses such as Epstein-Barr virus (EBV) and the Kaposi's sarcoma-associated herpesvirus (KSHV), whose narrow host ranges have largely precluded studies of pathogenesis. In vitro lytic replication systems have also proved difficult to establish. Nonetheless, it is important for practical purposes, such as vaccination, to understand how viral glycoproteins function in vivo and hence what might be achieved by glycoprotein-specific antibodies. EBV gp350 is a target for neutralizing antibody (43) and is the basis for a vaccine currently in clinical trials (31). Recombinant gp350 can inhibit EBV infection of some cell types (24), but EBV lacking gp350 is still infectious to both B cells and epithelial cells (19). A gH/gL/gp42 complex that binds to HLA class II molecules (35) is probably the major route of entry into B cells (17, 21, 22). Interestingly, virus produced by B cells tends to lack gp42 and to preferentially infect epithelial cells (9). Here, gp350 may have a more important role. Integrin binding by the BMRF2 gene product (44) may also contribute to infection in some circumstances. The positional homolog of gp350 in KSHV, gp35/37, binds GAGs (6, 45) and is therefore likely to be involved in the initial steps of cell binding. However, gB, gL, and gH of KSHV are sufficient for cell fusion in a transfection-based assay (29), suggesting that gp35/37 does not play an essential role in virus attachment. KSHV gB binds cellular integrins (4) and may be a key receptor binding protein. EBV lacking gB or gH is nonviable (18, 26). Thus, as with herpes simplex virus (13), gB and gH/L are probably key core members of the gammaherpesvirus cell binding-membrane fusion machinery. However, this leaves unanswered the question of what role gp350 binding to CR2 (41) or gp35/37 binding to GAGs might play in infection.
The murine gammaherpesvirus-68 (MHV-68) provides a valuable tool with which to investigate how gammaherpesvirus genes function in pathogenesis. MHV-68 naturally infects free-living mice (7; D. Blaskovic, M. Stancekova, J. Svobodova, and J. Mistrikova, Letter, Acta Virol. 24:468, 1980), and its colonization of conventional laboratory strains conforms to the pattern expected of a natural gammaherpesvirus infection, namely, a mononucleosis-like acute infection syndrome followed by lifelong asymptomatic latency (39). After intranasal infection, MHV-68 replicates in epithelial cells in the lung and spreads to lymphoid tissues to infect mainly B lymphocytes and macrophages (40). Establishment of latency in splenic B cells is accompanied by a self-limiting splenomegaly, and the virus persists predominantly in latently infected memory B cells (15, 23, 47).
The MHV-68 gp150 is a virion glycoprotein encoded by the M7 gene (37). As with gp350, DNA sequence analysis indicates that gp150 is a type I transmembrane protein with an N-terminal globular domain and a long, heavily O-glycosylated, proline-rich stalk. MHV-68 gp150 is a target for neutralizing antibody (37), but vaccination with gp150, despite inhibiting lytic viral replication, achieved only an eightfold reduction in peak latency titers (38). Thus, either inadequate levels of immunity were achieved by vaccination or there is an intrinsic limit on the protection possible with immunity to gp150, particularly with regard to B-cell infection. In order to understand the possible benefits and limitations of gp150-specific immunity, we generated and characterized MHV-68 with a disrupted M7 gene. This has provided a first opportunity to identify the role of one of the gammaherpesvirus gp350 positional homologs in the complete replication cycle, including binding, penetration, release, and the establishment of latency. The results suggested first, that MHV-68 infection of epithelial cells is a complex, multistage process that requires cell surface GAGs; second, that the requirement for GAGs involves gp150 and incorporates a possible mechanism of virion release; and third, that in vivo host colonization is relatively independent of gp150 and, by implication, independent of virion release.
MATERIALS AND METHODS
Mice.
Female BALB/c and C57BL/6 mice were purchased from Harlan U.K., Ltd. (Bicester, United Kingdom), and housed at the Biological Services Unit of the Cambridge University Department of Pathology. All procedures were carried out in accordance with Home Office Project Licence 80/1579. The female mice were infected at 6 to 12 weeks of age, either intranasally with 2 × 104 PFU or intraperitoneally with 105 PFU of MHV-68.
Cell lines.
BHK-21 cells (American Type Culture Collection [ATCC] CCL-10), NIH 3T3-CRE cells (36), L929 cells (ATCC CCL-1), CHO-K1 cells (ATCC CCL-61), the GAG-deficient derivative CHO-pgsA-745 (ATCC CRL-2242), NS0 cells (ATCC CRL-11177), RAW264.7 cells (ATCC TIB-71), and murine embryonic fibroblasts (MEFs) harvested at 13 to 14 days of gestation were grown in Dulbecco's modified Eagle medium (Invitrogen, Paisley, United Kingdom) supplemented with 2 mM glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 10% fetal calf serum (PAA Laboratories, Linz, Austria) (complete medium). The medium for MEFs was further supplemented with 50 μM β-mercaptoethanol.
Viral mutagenesis.
The M7 gene was disrupted in an MHV-68 genomic bacterial artificial chromosome (BAC) in accordance with published protocols (1). Briefly, a kanamycin resistance gene flanked by Flp recombinase target (FRT) sites was amplified from BsaI-linearized pCP15 by PCR (Expand Hi Fidelity; Roche, Lewes, United Kingdom), using primers with 50-bp 5′ extensions corresponding to nucleotides 69603 to 69651 and 69852 to 69803, respectively, of the MHV-68 genome (Genbank accession numbers em_vrl: u97553 and em_vrl: af105037). The purified PCR product was electroporated into Escherichia coli JC8679 containing the wild-type (WT) MHV-68 BAC to allow RecE/T-mediated recombination of the kanamycin resistance gene into the gp150 coding sequence. Recombinant BACs were isolated from chloramphenicol- and kanamycin-resistant colonies and transformed into E. coli DH10B for easier manipulation. The chloramphenicol- and kanamycin-resistant E. coli DH10B cells were then transformed with a temperature-sensitive Flp recombinase expression plasmid, pCP20, to excise the kanamycin resistance gene. This left 167 bp of foreign sequence in the BAC, comprising a single FRT site, in-frame stop codons, and flanking plasmid sequence, in place of 150 bp of the M7 gene (M7−FRT). The E. coli cells were cured of pCP20 by growth at 43°C. Successful Flp recombination was checked by replicate plating of bacteria onto chloramphenicol and chloramphenicol plus kanamycin and confirmed by restriction enzyme mapping of BAC DNA. To make a revertant virus (M7REV), an unmutated MHV-68 BglII genomic clone (genomic coordinates, 67744 to 73044) was ligated into the RecA+ SacB+ temperature-sensitive shuttle vector pST76K-SR and transformed into E. coli DH10B containing the M7−FRT BAC. Cointegrants were selected by growth in chloramphenicol plus kanamycin at 43°C. Cointegrant resolution was selected by growth in chloramphenicol and 5% sucrose at 30°C. Colonies were then screened by restriction enzyme digestion for resolution to a wild-type genomic pattern. An independent M7 mutant was also made by the pST76K-SR shuttle methodology. The BglII genomic fragment was ligated into pSP73 (Promega UK, Southampton, United Kingdom), cut at an internal AfeI site (genomic coordinate, 69743) and phosphatased with shrimp alkaline phosphatase (Roche Diagnostics); i.e., 5′ phosphate groups were removed. An oligonucleotide containing multiple stop codons in all reading frames (CTAGCTAGCTAGCTGGATCCGAATTCGGATCCAGCTAGCTAGCTAG) was self-annealed, phosphorylated with polynucleotide kinase (New England Biolabs, Hitchen, United Kingdom), and ligated into the AfeI site. The mutant BglII genomic fragment was then subcloned into pST76K-SR and recombined into the MHV-68 BAC as described above. After cointegrant resolution, colonies were screened for the insertion of an additional BamHI site (underlined in the oligonucleotide sequence above) into the M7 gene. All BACs were reconstituted into infectious virus by transfecting BAC DNA into BHK-21 cells using Fugene-6 (Roche Diagnostics). We grew green fluorescent protein-positive (GFP+) virus stocks directly. To grow GFP− virus stocks, we first removed the loxP-flanked BAC/GFP cassette by passaging the virus through NIH 3T3-CRE cells until GFP+ cells were no longer visible. The BAC cassette compromises MHV-68 replication in vivo but not in vitro (2). All virus stocks were prepared in BHK-21 cells. Infected cells were sonicated after being harvested. Cell debris was then pelleted by low-speed centrifugation (1,000 × g; 3 min) and discarded. Virions were pelleted by high-speed centrifugation (38,000 × g; 90 min) and stored in aliquots at −70°C.
Southern blotting.
Viral DNA was isolated from infected BHK-21 cells as previously described (10). Briefly, DNA was extracted from pelleted virions by alkaline lysis, digested with either BamHI or SacI, electrophoresed on a 0.8% agarose gel, and transferred to positively charged nylon membranes (Roche Diagnostics). A [32P]dCTP-labeled probe (APBiotech, Little Chalfont, United Kingdom) was generated by random primer extension (Nonaprimer kit; Qbiogene, Bingham, United Kingdom) from a 7,380-bp SacI genomic fragment (coordinates, 68,650 to 76,030) according to the manufacturer's instructions. Membranes were hybridized with probe (65°C; 18 h), washed to a stringency of 0.2% SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate, and exposed to X-ray film.
Immunoblotting.
MHV-68-specific monoclonal antibodies (MAbs) were generated from spleen cells of mice previously infected with ORF73-deficient MHV-68 (16), using polyethylene glycol-mediated fusion with NS0 cells and selection in medium with hypoxanthine and azaserine according to established protocols (20). The characterization of these and other MHV-68 glycoprotein-specific hybridomas will be described in detail elsewhere (unpublished data). Virus samples were lysed in 1% Triton X-100-50 mM Tris-Cl (pH 7.4)-150 mM NaCl supplemented with complete medium and protease inhibitors (Roche Diagnotics). A volume equivalent to 105 infectious units was then electrophoresed on a 10% denaturing polyacrylamide gel, transferred to a polyvinyl difluoride membrane (Perbio Science, Tattenhall, United Kingdom), and immunoblotted with either rabbit anti-MHV-68 polyclonal antibody (PAb) (39) or mouse MAb against gp150, ORF4, or gB. Detection was with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) or horseradish peroxidase-conjugated goat anti-rabbit IgG PAb serum (Perbio Science) and development with enhanced chemiluminescence reagents (APBiotech).
Viral titers.
Titers of infectious virus were determined by plaque assay (10). Briefly, 10-fold virus dilutions were incubated on BHK-21 monolayers for 2 to 3 h. The inoculum was removed, and the monolayers were overlaid with Dulbecco's modified Eagle medium containing 0.3% carboxymethylcellulose. After 4 days, the monolayers were fixed in 10% formaldehyde and stained with 0.1% toluidine blue. Plaques were counted using a plate microscope. To assay viral replication in vitro, MEF or BHK-21 monolayers were infected at 5 PFU per cell or 0.01 PFU per cell. After 2 h the inoculum was removed and the cell monolayers were acid washed (40 mM citric acid, pH 3, 135 mM NaCl, 10 mM KCl) to remove extracellular virus and incubated in complete medium at 37°C with 5% CO2 before being harvested and frozen for subsequent plaque assay. To specifically assay cell-free virus, supernatants were cleared by low-speed centrifugation (1,000 × g; 3 min) and frozen separately. Lungs from infected mice were frozen, thawed, homogenized in 1 ml of complete medium, frozen, thawed, and sonicated. Tissue debris was pelleted (1,000 × g; 3 min), and the infectious virus in homogenate supernatants was measured by plaque assay. Latent virus in spleens was measured by infectious-center assay (11). Spleens were disrupted into single-cell suspensions and cultured on MEF monolayers, which were fixed and stained after 6 days.
Electron microscopy.
BHK-21 cells were infected with WT, M7−FRT, and M7REV viruses (5 PFU per cell; 16 h); washed with PBS; and fixed in 4% glutaraldehyde-100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 7.4)-2 mM CaCl2 for 30 min on ice. Subsequent processing followed published methods (33). The cells were washed three times in 50 mM Tris-Cl (pH 7.4)-2 mM CaCl2, treated with 1% osmium tetroxide for 30 min, rinsed in double-distilled H2O, stained in 2% uranyl acetate for 30 min, rinsed in double-distilled H2O, dehydrated in methanol, infiltrated with Spurr's epoxy resin over 5 days, and cured by incubation at 60°C. Sections (50 nm thick) were cut using a Leica Ultracut S and mounted on uncoated copper grids, counterstained with uranyl acetate and lead citrate, and viewed with a Philips CM100 transmission electron microscope. Images were recorded on Kodak 4489 cut film.
Assays of infection kinetics.
BHK-21 cells (8 × 105) were infected with WT or M7−FRT virus (250 PFU) at 37°C. At different times after infection, the cells were washed either with PBS to remove unbound virions or with citrate buffer (pH 3) to remove both bound and unbound extracellular virus. Infectious virus in the cultures was then quantitated by plaque assay. MHV-68 replicates poorly in CHO-K1 cells and does not form plaques on RAW264.7 or NS0 cells, so in some experiments we used viruses that retained the BAC/GFP cassette at the left end of the genome and used GFP expression, quantitated by flow cytometry, to count infected cells. The cells were exposed to virus for 2 to 3 h, the inoculum was removed, and the cells were cultured in complete medium for 18 h before GFP+ cells were counted. To investigate the effect of soluble heparin on virus infection, GFP+ viruses were incubated with heparin for 1 h at 37°C and mixed with cells for 2 h with the heparin still present. The cells were then washed with PBS to remove unbound virus and heparin and incubated for 18 h in complete medium before flow cytometric analysis.
Flow cytometry.
Cells infected with GFP+ viruses were washed (0.1% BSA, 0.01% azide in PBS) and analyzed directly for green-channel fluorescence. For specific staining, cells were incubated for 30 min on ice with anti-CD16/32 MAb (BD-Pharmingen, San Diego, Calif.) plus 5% nonimmune serum matching the species of any conjugated antibody. Primary antibody staining was for 1 h on ice, using biotinylated rat anti-mouse syndecan-1 (BD-Pharmingen), rabbit anti-MHV-68 serum (39), fluorescein isothiocyanate (FITC)-conjugated mouse anti-heparan sulfate clone F58-10E4 (Seikagaku Corp., East Falmouth, Mass.), or rat anti-mouse transferrin receptor MAb (Serotec, Oxford, United Kingdom). Unconjugated primary antibodies were detected with FITC-conjugated swine anti-rabbit Ig PAb or FITC-conjugated rabbit anti-rat Ig PAb (DAKO A/S, Ely, United Kingdom). After being washed, the cells were analyzed on a FACSCalibur using Cellquest software (Becton Dickinson, Oxford, United Kingdom). Graphs were plotted with FCSPress version 1.3 (http://www.fcspress.com).
RESULTS
Generation of gp150-deficient MHV-68 mutants.
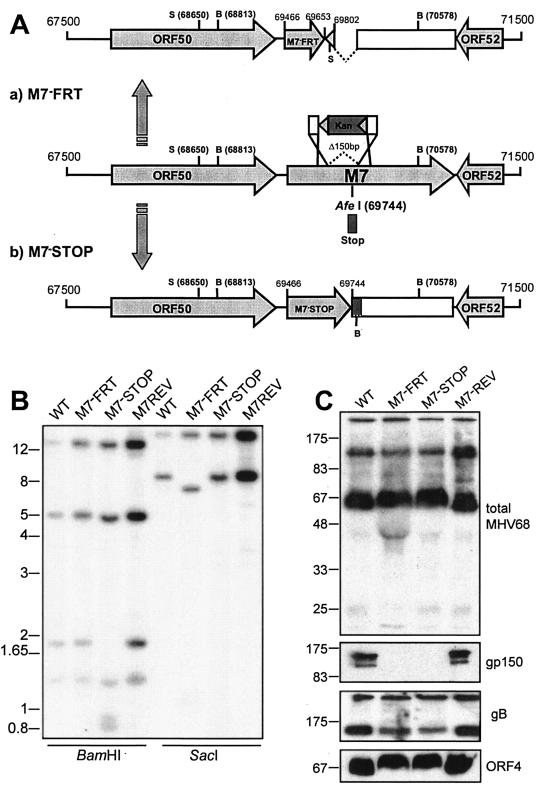
We mutated the MHV-68 M7 gene by inserting an FRT site plus short flanking plasmid sequences (167 bp total) in place of 150 bp of viral sequence (M7−FRT) (Fig. 1A). This introduced a premature stop codon into M7, thereby terminating gp150 translation after 62 amino acids. gp150 has a predicted 18-amino-acid signal sequence, so a 44-amino-acid protein fragment could still have been secreted from infected cells. However, such a small polypeptide would presumably have little structure. More importantly, because gp150 has a C-terminal membrane anchor, the truncated protein would lack any attachment to the viral envelope. The M7−FRT mutant was reverted (M7REV) by homologous recombination with an unmutated genomic clone spanning the M7 gene. An independent mutant (M7−STOP) was made by inserting an oligonucleotide with multiple stop codons in all reading frames at genomic coordinate 69744, thereby terminating gp150 after 93 amino acids. Again a 75-amino-acid fragment could still have been secreted but would not have been incorporated into virions. Southern blots confirmed the expected genomic structure for all viruses (Fig. 1B), and immunoblotting with a gp150-specific MAb confirmed a lack of gp150 on M7−FRT and M7−STOP virions (Fig. 1C).
FIG. 1.
Mutagenesis of the MHV-68 M7 gene encoding gp150. (A) The M7−FRT mutant was made using RecE/T to insert an FRT-flanked kanamycin resistance (Kan) gene in place of genomic coordinates 69653 to 69802. The resistance gene was then removed with Flp recombinase, leaving a single FRT site plus short flanking plasmid sequence, including a SacI restriction site. This mutant was reverted by RecA-mediated replacement of the M7 locus with an unmutated BglII genomic clone (67744 to 73044). The M7−STOP mutant was made by inserting an oligonucleotide containing multiple stop codons in all three frames into an AfeI site (genomic coordinate 69744) of the BglII clone. The oligonucleotide also included a BamHI restriction site. The mutated BglII clone was then recombined into the WT MHV-68 BAC using RecA. SacI (S) and BamHI (B) restriction sites are indicated. (B) Southern blots of viral DNA, digested with BamHI or SacI and probed with a SacI genomic fragment (68650 to 76030). BamHI digestion of M7−STOP cut the WT 1,765-bp band into 931- and 834-bp fragments; SacI digestion of M7−FRT cut the WT 7,380-bp band into 6,370- and 1,010-bp fragments. (C) Immunoblot analysis of gp150-deficient viruses. Titers of GFP+ virus stocks were determined by infecting BHK-21 cells with serial fivefold virus dilutions. The proportion of BHK-21 cells expressing GFP after 16 h was determined by flow cytometry. There was no significant difference between the GFP titer and the plaque titer for either M7− or M7+ viruses. Unreduced lysates equivalent to 105 GFP+ infectious units were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted for virion components, using either a rabbit serum raised against whole virus (total MHV-68) or MAbs specific for gp150, gB, and ORF4, as indicated.
Immunoblotting with a polyclonal serum raised against whole virus showed little difference between M7− and M7+ viruses. (Distortion of the gel at ∼50 kDa for M7− samples, also seen on the ORF4 immunoblots, corresponded to bovine albumin, since undiluted M7− virus was required for a titer equivalent to that of diluted M7+ virus.) Thus, in the absence of gp150, there was no indication of an imbalance between virus stock infectivity and virus stock protein content, as would be expected if there were more defective particles. As expected, viruses with a disruption of M7 lacked gp150. There was also no evidence of an increase in gB or ORF4 (complement control protein) content in stocks of gp150-deficient MHV-68: if anything, gB levels were somewhat reduced.
Impaired propagation of cell-free virus in the absence of gp150.
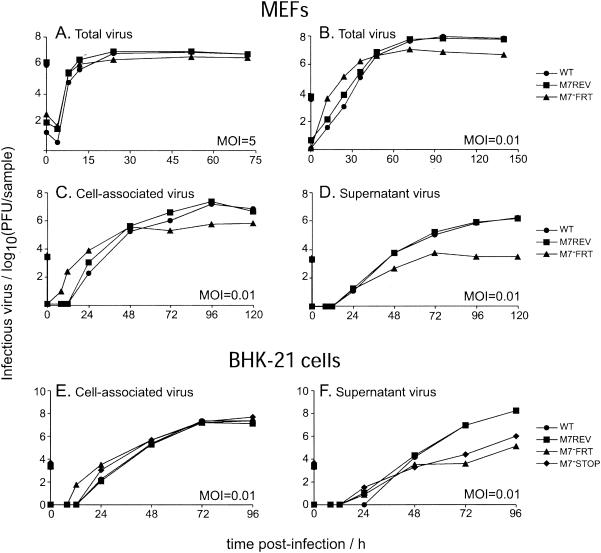
We were able to recover both the M7−FRT and M7−STOP viruses from transfected BACs without difficulty, so gp150 was not essential for MHV-68 lytic replication in vitro. However, the M7 mutants grew to lower titers than WT or M7REV viruses. There was no significant single-cycle replication deficit (Fig. 2A), suggesting that a lack of gp150 led to a defect in either entry into or release from infected cells. The plaque size under semisolid medium was normal (data not shown), implying that cell-to-cell virus spread was intact. Low-multiplicity growth (Fig. 2B) consistently showed higher titers of M7−FRT virus at early time points (up to 24 h) than WT or M7REV. However, M7−FRT growth reached a plateau at 48 h, while WT and M7REV growth continued up to 96 h and so reached higher titers. In order to understand this phenomenon better, we determined the titers of cell-associated and supernatant virus separately during low-multiplicity growth (Fig. 2C and D). This established that the M7−FRT virus detected at early times was cell associated and that M7−FRT supernatant virus titers were more severely reduced than cell-associated titers. By 120 h, WT and M7REV viruses were approximately equally distributed between supernatant and cells, while the M7−FRT virus was 90% cell associated. A similar phenotype was observed with the independent mutant (M7−STOP) and with growth in BHK-21 cells, as well as MEFs (Fig. 2E and F).
FIG. 2.
In vitro growth of gp150-deficient viruses. Subconfluent MEF (A to D) or BHK-21 cell (E to F) monolayers were infected for 2 h at a multiplicity of infection of 5 or 0.01, acid washed, and incubated in complete medium. At the indicated times, titers of cells and supernatants were determined by plaque assay on BHK-21 cells, either as a pooled sample (A and B) or separately (C to F). (A to D) WT (•), M7−FRT (▴), and M7REV (▪) viruses. (E and F) We also assayed the independent mutant M7−STOP (⧫).
The predominant reduction in cell-free virus with the M7 mutants suggested that a lack of gp150 impaired virion release. The higher titers of gp150-deficient viruses at early time points of low-multiplicity growth were also consistent with a release deficit: newly formed gp150-deficient virus that remained associated with infected cells would have remained infectious, while released WT and M7REV viruses would have been absorbed by the large excess of uninfected cells. Rapid entry into eclipse phase would thus have conferred a short half-life on WT and M7REV virions until the pool of uninfected cells was diminished.
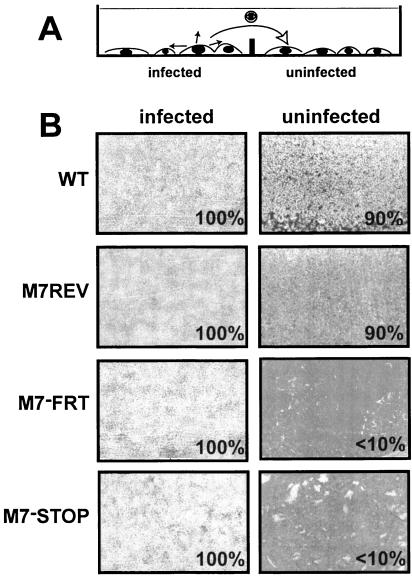
We tested more specifically the capacities of the M7 mutants to escape from infected cells by assaying the transfer of infection between cell populations connected only by medium (Fig. 3). BHK-21 cells were seeded on either side of a separating barrier, and only those on one side of the barrier were infected. Extracellular input virus was removed after 2 h, and medium was added to cover both the fibroblast monolayers and the divide between them. Thus, virus could spread from cell to cell within the infected population but could reach the uninfected population only by diffusion or on convection currents across the dividing barrier. Although detached infected cells might also be carried across the divide by convection and obscure differences in virus release, we reasoned that this would occur much less readily than with cell-free virions. A clear difference in spread was apparent between the M7− and M7+ viruses. By the time the infected populations were entirely destroyed, WT and M7REV viruses had destroyed almost all of both populations, whereas the cytopathic effects with M7−FRT and M7−STOP viruses were largely confined to the initially infected populations. Only discrete plaques were visible on the initially uninfected monolayers.
FIG. 3.
Fluid phase virus spread. (A) A total of 4 × 105 infected (multiplicity of infection, 0.01) and 4 × 105 uninfected BHK-21 cells were seeded on opposite sides of a 6-cm-diameter dish, separated by a physical barrier but connected by medium so that virus could not spread directly from the infected to the uninfected population but only via their common supernatant. (B) After 4 days, the cells were fixed and stained. Representative areas are shown; the numbers indicate the percentage of each monolayer showing cytopathic effects.
Kinetics of gp150-deficient virion uptake.
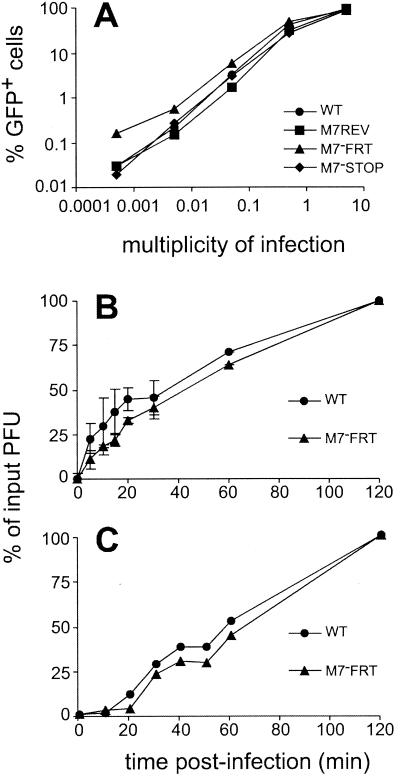
Because the best-characterized positional homologue of gp150—EBV gp350—binds to cells and enhances the efficiency of infection, the main alternative possibility to a release deficit was that M7− virions were released but poorly infectious. We therefore assayed the kinetics of infection by M7−FRT virions (Fig. 4). First, we compared the capacities of M7− and M7+ viruses to infect fibroblasts at different multiplicities (Fig. 4A). No differences were observed. We then kept a fixed low multiplicity of infection and limited the time available for binding and entry. Cells were infected at 37°C and, at various times afterward, were washed either with PBS to remove unbound extracellular virus (Fig. 4B) or with acid to remove both bound and unbound extracellular virus (Fig. 4C). There was little or no difference between the kinetics of infection in M7−FRT and WT viruses. An attachment or penetration deficit therefore seemed unlikely to account for the reduced growth of the M7−FRT virus.
FIG. 4.
Virus binding and penetration and effect of virion multiplicity on infection. (A) BHK-21 cells were infected for 2 h with dilutions of GFP+ viruses and analyzed for GFP expression by flow cytometry 16 h later. (B and C) Subconfluent BHK-21 cell monolayers were infected at room temperature with 250 PFU of virus. After the times indicated, the monolayers were washed either with PBS (B) or with citrate buffer (pH 3) (C). The cells were then overlaid with semisolid medium, and after 4 days they were fixed and stained. PBS washes should remove only unattached virions, whereas acid washes should denature proteins and so remove all virus that has not fused with the plasma membrane or been endocytosed. The 20-min time lag between panels B and C, therefore, corresponds to the time required for virions to enter cells. The data shown are representative of at least two independent experiments. Means and standard deviations of triplicate samples are shown in panel B.
Association of gp150-deficient virions with infected-cell surfaces.
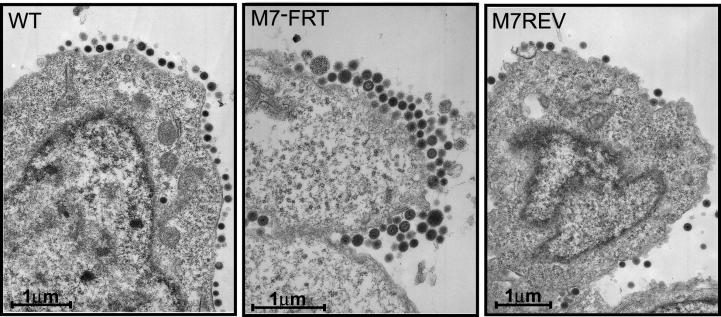
Further support for a gp150-associated virion release deficit came from electron microscopy of infected fibroblasts. BHK-21 cells were infected overnight and then examined for virions and viral nucleocapsids. Since we looked only at infected cells, the excess WT and M7REV virus released into the supernatant was discarded. Nevertheless, a difference was still apparent between M7− and M7+ viruses. gp150-deficient virions were piled up on the extracellular surfaces of infected-cell plasma membranes to form multiple layers of tightly packed virions. In contrast, WT and M7REV virions never formed more than a single layer on infected-cell surfaces (Fig. 5). We saw no increased accumulation of intracellular M7− virions or nucleocapsids, suggesting that egress was normal in the absence of gp150, and input gp150-deficient virus was not stranded at the cell surface, since neither M7− nor M7+ virions were seen on cell surfaces 8 h after infection (data not shown). The most likely explanation was therefore that gp150-deficient virions either failed to escape from the cell surface or rapidly rebound to it after release.
FIG. 5.
Electron microscopy of virus-infected cells. BHK-21 cells were infected (16 h; multiplicity of infection, 5) with WT, M7−FRT, or M7REV viruses as indicated. The pictures shown are representative of at least 10 sections per sample. The heterogeneity in the cross-sectional area of the M7−FRT virions presumably reflects some tangential cuts, as would be expected with a densely packed population. We saw no evidence for abnormal virion morphologies in the absence of gp150.
Enhanced infection of GAG-deficient cells by gp150-deficient MHV-68.
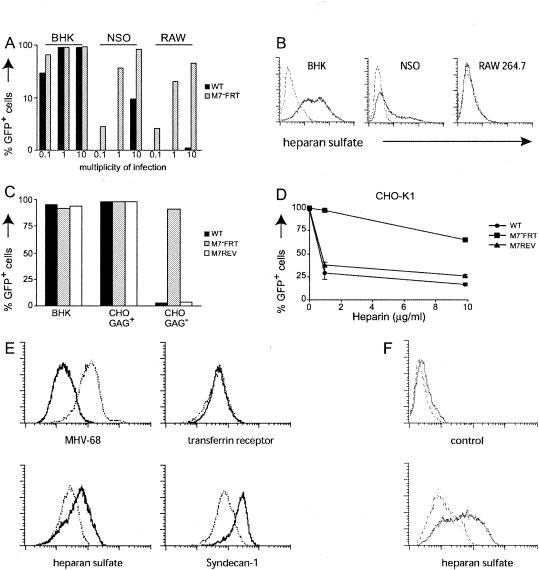
Herpesviruses typically infect several different cell types, and EBV uses different glycoproteins to infect different cell types (44, 46). MHV-68 is found in B cells and macrophages, in addition to epithelial cells. We therefore sought further information about how gp150 might function by comparing the capacities of WT and M7−FRT viruses to infect NS0 myeloma cells and RAW264.7 macrophages (Fig. 6A). We used virus-driven GFP expression as an indicator of infection, since neither cell type supports MHV-68 plaque formation. Surprisingly, the M7−FRT virus showed considerably enhanced infection of NS0 and RAW264.7 cells. Similar enhancement of infection was observed with the M7−STOP virus (data not shown).
FIG. 6.
Interactions between MHV-68 and GAGs. (A) BHK-21 (fibroblast), NS0 (myeloma), and RAW264.7 (monocyte-macrophage) cells were infected at 0.1, 1, or 10 PFU per cell as indicated and analyzed for GFP expression 16 h later. (B) BHK-21, NS0, and RAW264.7 cells were detached from tissue culture flasks without trypsin and stained for cell surface heparan sulfate (solid lines). The dashed lines represent unstained cells. (C) BHK-21 cells, CHO-K1 cells (CHO GAG+), and the GAG-deficient derivative CHO-pgsA-745 (CHO GAG−) were infected at 1 PFU per cell with GFP+ viruses and analyzed for GFP expression 16 h later. (D) GFP+ viruses (106 PFU) were incubated with 0, 1, or 10 μg of soluble heparin/ml for 1 h at 37°C. The virus-heparin mixtures were then incubated on CHO-K1 cell monolayers for 2 h. Unbound virus was removed by washing the cells with PBS, and the cells were analyzed for GFP expression by flow cytometry 16 h later. (E) L929 cells were left uninfected (solid lines) or infected (10 PFU per cell; 16 h) with WT MHV-68 (dotted lines). Each population was then analyzed by flow cytometry for cell surface expression of viral antigens (MHV-68) to assess infection, for transferrin receptor to check that there was no global surface protein down-regulation, and for heparan sulfate and syndecan-1 to assess GAG expression. (F) Similar down-regulation of GAGs was observed on BHK-21 cells infected (1 PFU per cell; 16 h) with either WT (dotted lines) or M7−FRT (dashed lines) virus. The solid lines represent uninfected cells.
By analogy with KSHV gp35/37, one possible function of gp150 is GAG binding, and one obvious difference between BHK-21 cells and NS0 or RAW264.7 cells was their levels of cell surface GAGs (Fig. 6B). We therefore tested whether GAGs differentially influenced the capacity of M7− and M7+ viruses to infect CHO-K1 fibroblasts (Fig. 6C and D). WT MHV-68 was found to be highly dependent on GAGs for the infection of CHO-K1 cells, as it infected GAG-deficient CHO-pgsA-745 cells to only a very limited extent (Fig. 6C). In contrast, the M7−FRT virus infected CHO-pgsA-745 and CHO-K1 cells with similar efficiencies. The M7−FRT virus was also relatively resistant to inhibition by soluble heparin (Fig. 6D), consistent with an interaction between gp150 and GAGs. The residual inhibition of M7−FRT infection by heparin suggested that MHV-68 has an additional GAG-binding protein—the KSHV gB binds heparin (3), and the MHV-68 gB has a heparin binding motif—but that this interaction makes relatively little contribution to the efficiency of infection.
The independence of the M7−FRT virus of cell surface GAGs suggested a possible explanation for its impaired release. Flow cytometric analysis of L929 and BHK-21 fibroblasts established that at 16 h post-MHV-68 infection, the time of maximal virus release (25), GAG levels were reduced (Fig. 6E and F). There was also a marked loss of syndecan-1 (Fig. 6E), the major protein to which the GAGs furthest away from the cell surface are attached (5); these more distal GAGs are those most likely to be involved in virus attachment (8). Infection with gp150-deficient virus also reduced cell surface GAGs (Fig. 6F). Since WT virus infected GAG-deficient cells poorly, GAG shedding would have prevented its rebinding to infected cells after release. No such limitation applied to gp150-deficient virus, which infected GAG− and GAG+ cells equally well.
Replication of gp150-deficient MHV-68 in vivo.
We next tested the capacity of gp150-deficient MHV-68 viruses to replicate in vivo (Fig. 7). Because host colonization is a stringent test of viral fitness, in vitro growth deficits normally translate into substantial pathogenesis deficits. The natural route of MHV-68 transmission is unknown. Insofar as natural infection is likely first to reach mucosal epithelial cells and then spread to lymphoid tissue, intranasal virus inoculation provides a reasonably realistic model. This leads to lytic replication in alveolar epithelial cells, followed by latency amplification in lymphoid sites, such as the spleen (39). These two processes are relatively independent: a virus with fairly normal lytic replication can fail in latency amplification (25), and a virus grossly deficient in lytic replication, although somewhat delayed in reaching lymphoid tissue, still establishes normal latency (11).
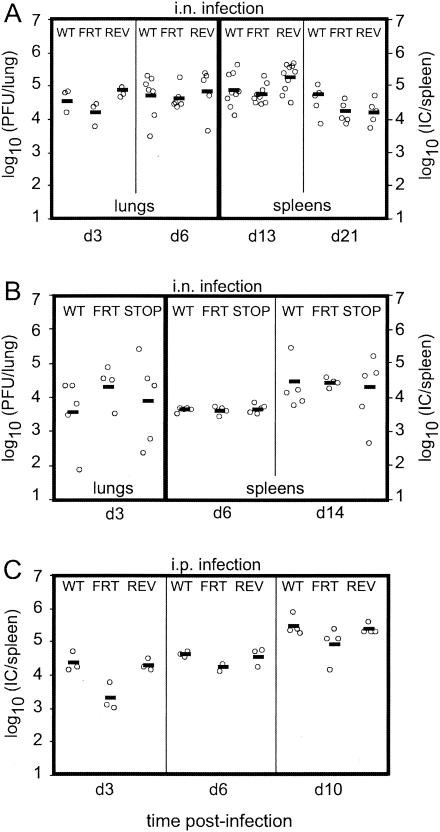
FIG. 7.
In vivo growth of gp150-deficient MHV-68. Mice were infected either intranasally (2 × 104 PFU) (A and B) or intraperitoneally (105 PFU) (C) with WT, M7−FRT (FRT), M7REV (REV), or M7−STOP (STOP) virus. At the times indicated, amounts of lytic virus in the lungs were determined by plaque assay, and amounts of latent virus in spleens were determined by infectious-center assay (IC). Each data point represents an individual mouse. In panel C, data from one of two equivalent experiments are shown. The horizontal bars indicate values.
We first compared the capacities of wild-type virus, the M7−FRT mutant, and its revertant to replicate lytically and to establish latency following intranasal inoculation. Surprisingly, the M7−FRT virus showed no significant deficit in either lytic or latent replication. We then tested the M7−STOP mutant in parallel with wild-type virus and the M7−FRT mutant. Again, no deficit was observed. Finally, we tested the replicative fitness of the M7−FRT mutant after intraperitoneal virus inoculation. This is probably a less stringent test of viral fitness than intranasal inoculation, since the normal mucosal barrier to infection is bypassed. Nevertheless, it provides a slightly different test, in that virus reaches the spleen before strong immunity to lytic cycle antigens has been established, allowing lytic, as well as latent, spread in this site. Apart from a possible early lag in replication, no gp150-related deficit was observed and normal levels of latency were achieved. gp150 was therefore entirely dispensable for entry into a naive host and for subsequent lytic and latent viral propagation. By implication, host colonization was relatively independent of the need for efficient virion release from infected cells.
DISCUSSION
We disrupted the MHV-68 M7 gene, which encodes the gp150 positional homolog of EBV gp350, in order to define the role of gp150 in viral replication and pathogenesis. gp150-deficient MHV-68 was viable, and we found no evidence for a deficit in cell binding or entry. Indeed, a lack of gp150 removed the normal dependence of cell-free MHV-68 on GAGs for efficient infection. However, gp150-deficient mutants were poorly released from infected cells. Because GAG expression was decreased on MHV-68-infected cells, we surmise that the impaired release reflected enhanced rebinding to (or a failure ever to be released from) infected cells in the absence of gp150. Thus, gp150 provided a “trapdoor” mechanism of cell binding by covering critical receptor binding proteins on the virion until it interacted with GAGs. gp150 thus regulated the attachment of virions to cell surfaces. Despite their in vitro growth deficits, gp150-deficient viruses showed little or no in vivo deficit, suggesting that here the predominant mode of virus propagation is direct spread from cell to cell, for which gp150 was not required.
The generally high affinity of viruses for their receptors inevitably creates a problem of virion release from infected cells. The MHV-68 gp150 provided an example of how this might be solved by a herpesvirus. Our model is that the long, extended stalk of gp150 places its N-terminal domain some distance from the virion surface so as to be a point of initial cell contact, probably with GAGs. gp150 binding then makes available a cell binding epitope, perhaps on gB or gH. gp150 inhibits binding in the absence of GAG interaction, so by ensuring low GAG levels on infected cells, MHV-68 allows gp150+ virions to be efficiently released. In the absence of gp150, high-affinity cell binding is constitutively available, leading to a release deficit. This model predicts that gp150 will also limit the access of virus-specific antibody to cell binding epitopes on the virion and thus, that some antibodies should selectively neutralize gp150-deficient virions.
A masking of virion receptor binding proteins is clearly not the only possible explanation for the gp150-deficient phenotype. One alternative was that higher levels of a receptor binding protein were incorporated into virions as a consequence of gp150 being lost. Glycoprotein B is a receptor binding protein of KSHV (4), and high gB expression enhances EBV infection of various cell types (27). However, we found no evidence for an absence of gp150 increasing the representation of gB or that of the ORF4 gene product, another viral glycoprotein. It remains possible that the abundance of another, untested viral glycoprotein was increased, but we saw no evidence of this by immunoblotting with rabbit anti-MHV-68 PAb. Also, it is not clear how higher levels of a receptor binding protein would selectively enhance the infection of GAG− cells by gp150-deficient MHV-68 (Fig. 6). Another possibility is that gp150 has a signaling function, akin to that of EBV gp350 (12, 42). But while some such function cannot be excluded, it seemed unlikely to account for the major features of the M7− MHV-68 phenotype: a lack of GAG dependence and impaired virion release late in infection.
The dependence of WT MHV-68 on GAGs for infection raised the question of how it normally manages to infect relatively GAG-deficient cells, such as B lymphocytes. Direct cell-to-cell spread is one possibility, since this does not necessarily require the same receptor binding interactions as infection by cell-free virions. Also, B-cell infection may not be a common event, since infected B cells can proliferate to generate a large latent pool. Thus, there could be a very specific route of B-cell infection, for example, from mucosal surfaces via M cells (48). Another possibility is that MHV-68 produces gp150-deficient particles from at least some cell types.
The fact that gp150 was entirely dispensable for the colonization of a single host obviously does not imply that it has no role in vivo. The efficient production of cell-free virus is probably most important in transmission between hosts, and gp150 may have an important role here, ensuring that virions are efficiently shed from mucosal surfaces. The gammaherpesviruses transmit on the basis of chronic low-level infectivity (49), a process intrinsically difficult to measure in a reproducible way. The MHV-72 isolate of MHV-68 appears to be released into milk during acute infection of pregnant mice, with viral DNA detectable by nested PCR of suckling mouse stomach contents (30). However, this was observed only in the rather unusual circumstance of high-dose acute infection just before delivery, and there was no evidence for infection being established in breast-fed pups. Thus, it remains uncertain whether infectious virions are shed in milk and whether breast feeding is a physiological route of MHV-68 transmission. Our attempts to reproducibly transmit MHV-68 from persistently infected mice have so far been unsuccessful.
The fairly normal host colonization by gp150-deficient MHV-68 did imply that propagation within an individual host is relatively independent of the need for virion release. In this regard, the infection of cultured fibroblast lines is perhaps not a good model of in vivo lytic spread, something that has been observed with the replication of thymidine kinase-deficient MHV-68 mutants (11). In order to develop strategies of virus control, it is important to identify key mechanisms of in vivo dissemination and to model them as far as possible in vitro. To that extent, the limited capacity of gp150-deficient virions to be released may make gp150-deficient viruses a useful tool for assaying the control of direct cell-to-cell virus spread.
Acknowledgments
We thank Helena Browne for helpful advice, Susanna Colaco for generating the MHV-68-specific hybridomas, and Jeremy Skepper for assistance with electron microscopy.
This work was supported by a project grant (059601) from the Wellcome Trust and a cooperative group (G9800903) component grant (G9901295) from the Medical Research Council United Kingdom. B.D.D.L. is supported by the Portuguese Foundation for Science and Technology. P.G.S. is an MRC/Academy of Medical Sciences Clinician Scientist (G108/462).
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler, H., M. Messerle, and U. H. Koszinowski. 2001. Virus reconstituted from infectious bacterial artificial chromosome (BAC)-cloned murine gammaherpesvirus 68 acquires wild-type properties in vivo only after excision of BAC vector sequences. J. Virol. 75:5692-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2001. Human herpesvirus 8 envelope-associated glycoprotein B interacts with heparan sulfate-like moieties. Virology 284:235-249. [DOI] [PubMed] [Google Scholar]
- 4.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2002. Integrin α3β1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108:407-419. [DOI] [PubMed] [Google Scholar]
- 5.Bernfield, M., M. Gotte, P. W. Park, O. Reizes, M. L. Fitzgerald, J. Lincecum, and M. Zako. 1999. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68:729-777. [DOI] [PubMed] [Google Scholar]
- 6.Birkmann, A., K. Mahr, A. Ensser, S. Yaguboglu, F. Titgemeyer, B. Fleckenstein, and F. Neipel. 2001. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 75:11583-11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blasdell, K., C. McCracken, A. Morris, A. A. Nash, M. Begon, M. Bennett, and J. P. Stewart. 2003. The wood mouse is a natural host for Murid herpesvirus 4. J. Gen. Virol. 84:111-113. [DOI] [PubMed] [Google Scholar]
- 8.Bobardt, M. D., A. C. Saphire, H. C. Hung, X. Yu, B. Van der Schueren, Z. Zhang, G. David, and P. A. Gallay. 2003. Syndecan captures, protects, and transmits HIV to T lymphocytes. Immunity 18:27-39. [DOI] [PubMed] [Google Scholar]
- 9.Borza, C. M., and L. M. Hutt-Fletcher. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. [DOI] [PubMed] [Google Scholar]
- 10.Bridgeman, A., P. G. Stevenson, J. P. Simas, and S. Efstathiou. 2001. A secreted chemokine binding protein encoded by murine gammaherpesvirus-68 is necessary for the establishment of a normal latent load. J. Exp. Med. 194:301-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman, H. M., B. de Lima, V. Morton, and P. G. Stevenson. 2003. Murine gammaherpesvirus 68 lacking thymidine kinase shows severe attenuation of lytic cycle replication in vivo but still establishes latency. J. Virol. 77:2410-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Addario, M., T. A. Libermann, J. Xu, A. Ahmad, and J. Menezes. 2001. Epstein-Barr virus and its glycoprotein-350 upregulate IL-6 in human B-lymphocytes via CD21, involving activation of NF-κB and different signaling pathways. J. Mol. Biol. 308:501-514. [DOI] [PubMed] [Google Scholar]
- 13.Davis-Poynter, N., S. Bell, T. Minson, and H. Browne. 1994. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J. Virol. 68:7586-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingwell, K. S., C. R. Brunetti, R. L. Hendricks, Q. Tang, M. Tang, A. J. Rainbow, and D. C. Johnson. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flano, E., I. J. Kim, D. L. Woodland, and M. A. Blackman. 2002. Gamma-herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J. Exp. Med. 196:1363-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fowler, P., S. Marques, J. P. Simas, and S. Efstathiou. 2003. ORF73 of murine herpesvirus-68 is critical for the establishment and maintenance of latency. J. Gen. Virol. 84:3405-3416. [DOI] [PubMed] [Google Scholar]
- 17.Haan, K. M., W. W. Kwok, R. Longnecker, and P. Speck. 2000. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J. Virol. 74:2451-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrold, R. E., A. Marchini, S. Fruehling, and R. Longnecker. 1996. Glycoprotein 110, the Epstein-Barr virus homolog of herpes simplex virus glycoprotein B, is essential for Epstein-Barr virus replication in vivo. J. Virol. 70:2049-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janz, A., M. Oezel, C. Kurzeder, J. Mautner, D. Pich, M. Kost, W. Hammerschmidt, and H. J. Delecluse. 2000. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 74:10142-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Köhler, G., and C. Milstein. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256:495-497. [DOI] [PubMed] [Google Scholar]
- 21.Li, Q., S. M. Turk, and L. M. Hutt-Fletcher. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Q., M. K. Spriggs, S. Kovats, S. M. Turk, M. R. Comeau, B. Nepom, and L. M. Hutt-Fletcher. 1997. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 71:4657-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marques, S., S. Efstathiou, K. G. Smith, M. Haury, and J. P. Simas. 2003. Selective gene expression of latent murine gammaherpesvirus 68 in B lymphocytes. J. Virol. 77:7308-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maruo, S., L. Yang, and K. Takada. 2001. Roles of Epstein-Barr virus glycoproteins gp350 and gp25 in the infection of human epithelial cells. J. Gen. Virol. 82:2373-2383. [DOI] [PubMed] [Google Scholar]
- 25.May, J. S., H. M. Coleman, B. Smillie, S. Efstathiou, and P. G. Stevenson. 2004. Forced lytic replication impairs host colonization by a latency-deficient mutant of murine gammaherpesvirus-68. J. Gen. Virol. 85:137-146. [DOI] [PubMed] [Google Scholar]
- 26.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuhierl, B., R. Feederle, W. Hammerschmidt, and H. J. Delecluse. 2002. Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc. Natl. Acad. Sci. USA 99:15036-15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peeters, B., J. Pol, A. Gielkens, and R. Moormann. 1993. Envelope glycoprotein gp50 of pseudorabies virus is essential for virus entry but is not required for viral spread in mice. J. Virol. 67:170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pertel, P. E. 2002. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol. 76:4390-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raslova, H., M. Berebbi, J. Rajcani, A. Sarasin, J. Matis, and M. Kudelova. 2001. Susceptibility of mouse mammary glands to murine gammaherpesvirus 72 (MHV-72) infection: evidence of MHV-72 transmission via breast milk. Microb. Pathog. 31:47-58. [DOI] [PubMed] [Google Scholar]
- 31.Rickinson, A. B. 1995. Immune intervention against virus-associated human cancers. Ann. Oncol. 6(Suppl. 1):69-71. [DOI] [PubMed] [Google Scholar]
- 32.Shukla, D., and P. G. Spear. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Investig. 108:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skepper, J. N. 2000. Immunocytochemical strategies for electron microscopy: choice or compromise. J. Microsc. 199:1-36. [DOI] [PubMed] [Google Scholar]
- 34.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spriggs, M. K., R. J. Armitage, M. R. Comeau, L. Strockbine, T. Farrah, B. Macduff, D. Ulrich, M. R. Alderson, J. Mullberg, and J. I. Cohen. 1996. The extracellular domain of the Epstein-Barr virus BZLF2 protein binds the HLA-DR beta chain and inhibits antigen presentation. J. Virol. 70:5557-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevenson, P. G., J. S. May, X. G. Smith, S. Marques, H. Adler, U. H. Koszinowski, J. P. Simas, and S. Efstathiou. 2002. K3-mediated evasion of CD8+ T cells aids amplification of a latent gamma-herpesvirus. Nat. Immunol. 3:733-740. [DOI] [PubMed] [Google Scholar]
- 37.Stewart, J. P., N. J. Janjua, S. D. Pepper, G. Bennion, M. Mackett, T. Allen, A. A. Nash, and J. R. Arrand. 1996. Identification and characterization of murine gammaherpesvirus 68 gp150: a virion membrane glycoprotein. J. Virol. 70:3528-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart, J. P., N. Micali, E. J. Usherwood, L. Bonina, and A. A. Nash. 1999. Murine gamma-herpesvirus 68 glycoprotein 150 protects against virus-induced mononucleosis: a model system for gamma-herpesvirus vaccination. Vaccine 17:152-157. [DOI] [PubMed] [Google Scholar]
- 39.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 73:2347-2356. [DOI] [PubMed] [Google Scholar]
- 40.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 41.Tanner, J., J. Weis, D. Fearon, Y. Whang, and E. Kieff. 1987. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 50:203-213. [DOI] [PubMed] [Google Scholar]
- 42.Tanner, J. E., C. Alfieri, T. A. Chatila, and F. Diaz-Mitoma. 1996. Induction of interleukin-6 after stimulation of human B-cell CD21 by Epstein-Barr virus glycoproteins gp350 and gp220. J. Virol. 70:570-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorley-Lawson, D. A., and C. A. Poodry. 1982. Identification and isolation of the main component (gp350-gp220) of Epstein-Barr virus responsible for generating neutralizing antibodies in vivo. J. Virol. 43:730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tugizov, S. M., J. W. Berline, and J. M. Palefsky. 2003. Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat. Med. 9:307-314. [DOI] [PubMed] [Google Scholar]
- 45.Wang, F. Z., S. M. Akula, N. P. Pramod, L. Zeng, and B. Chandran. 2001. Human herpesvirus 8 envelope glycoprotein K8.1A interaction with the target cells involves heparan sulfate. J. Virol. 75:7517-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, X., W. J. Kenyon, Q. Li, J. Mullberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willer, D. O., and S. H. Speck. 2003. Long-term latent murine gammaherpesvirus 68 infection is preferentially found within the surface immunoglobulin D-negative subset of splenic B cells in vivo. J. Virol. 77:8310-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf, J. L., D. H. Rubin, R. Finberg, R. S. Kauffman, A. H. Sharpe, J. S. Trier, and B. N. Fields. 1981. Intestinal M cells: a pathway for entry of reovirus into the host. Science 212:471-472. [DOI] [PubMed] [Google Scholar]
- 49.Yao, Q. Y., A. B. Rickinson, and M. A. Epstein. 1985. A re-examination of the Epstein-Barr virus carrier state in healthy seropositive individuals. Int. J. Cancer 35:35-42. [DOI] [PubMed] [Google Scholar]