Abstract
The latency-associated transcript (LAT) is required for efficient reactivation of herpes simplex virus type 1 from latent infection in the rabbit eye model, but LAT's mechanism of action is unknown. In addition to reactivation, the LAT region seems to correspond to multiple functions, with some LAT deletion mutants exhibiting increased virulence, increased neuronal death, and restricted establishment of latency. While a LAT promoter deletion mutant (17ΔPst) seems to be primarily restricted in reactivation in the rabbit, subtle effects on virulence or the establishment of latency cannot be precluded at the normal high levels of virus inoculum used in the rabbit model. Since such additional LAT phenotypes may be more evident with lower doses of virus, we evaluated the influence of initial viral inoculum and LAT expression on the progression of acute infection and the establishment of latency. We have assayed both virus recovery rates and viral genome loads in rabbit corneas and trigeminal ganglia. Our results show that (i) in the corneas and trigeminal ganglia, the maximum amount of virus present during acute infection is independent of the LAT genotype and inoculum dose, although greater viral yields are obtained earlier with higher inoculum doses, and (ii) the range in numbers of latent genomes detected in the ganglia is independent of the inoculum dose and the LAT genotype and therefore no difference in establishment of latency is observed.
Herpes simplex virus type 1 (HSV-1) establishes latency in neurons of sensory ganglia innervating the site of initial infection. The virus can reactivate spontaneously or under conditions of stress to cause a recurrent infection. During latency, the genome forms an episome in neuronal nuclei from which no viral replication occurs (19, 26). Approximately one-third of the latently infected neurons express high levels of a single transcript, termed the latency-associated transcript (LAT) (12, 18). This transcript is important for reactivation, even though LAT does not seem to encode a protein (14, 17).
While LAT is required for efficient reactivation in animal models, its mechanism is not well understood. One factor that complicates these analyses is that observations vary depending on the animal model (24) and HSV strain (20, 28) used. The two most common models employed are the rabbit and mouse. In the rabbit eye model, latency is established in trigeminal ganglia (TG) following corneal inoculation. Reactivation, either spontaneous or induced by iontophoresis of epinephrine, is scored by recovery of infectious virus in the tear film (1, 13, 21). In the mouse model, latency is established in the trigeminal or dorsal root ganglia following inoculation of corneas or rear footpads, respectively. Viral reactivation from ganglia can be induced by thermal stress, as demonstrated by the presence of infectious virus in the ganglia, or by explant cocultivation of dissected ganglia on cultured cells (30, 32).
Mutants with large LAT deletions have been reported to have reduced numbers of latent viral genomes in neurons of both mice and rabbits (23, 25, 29, 33). This suggests that functions corresponding to the LAT region are involved in the establishment of latency. In contrast, mutants with smaller LAT deletions, such as 17ΔPst (a LAT promoter mutant) and 17Δ348 (a 5′ exon deletion mutant), do not demonstrate significant differences in total numbers of latent HSV-1 genomes (2, 3, 8). This suggests that either the establishment function in the LAT region maps to a region independent of the LAT promoter (LAP1) or that a defect in establishment exhibited by the mutants with smaller deletions was below the limit of detection in the previous studies.
The possibility existed that the dose of virus used in rabbit infections, which involve a relatively large inoculum (1× 105 to 5 × 105 PFU/eye), may mask subtle replication or establishment deficits inherent in these LAT mutants. Therefore, the course of the acute infection in the rabbit eye model was examined using 1,000-fold-lower inoculation doses of 17ΔPst and the corresponding rescue strain. Differences in acute infection kinetics and levels of establishment of latency were not detected by this method. The observation that peak establishment occurs with even low-dose inocula suggests that saturation of latent sites occurs relatively early. To determine the contribution of the initial inoculum to establishment, rabbits were infected with a nonreplicating HSV-1 recombinant, KD6 (ICP4−). While this recombinant is capable of establishing latency in the rabbit TG following ocular infection, the total number of latent genomes is much lower than that seen after infection with wild-type virus, indicating that peripheral replication contributes to maximal establishment of latency.
MATERIALS AND METHODS
Cells and viruses.
Virus was propagated on cultured rabbit skin (RS) cells. Titers of viral stocks were determined on RS cells grown in minimal essential medium supplemented with 5% fetal bovine serum and antibiotics (34). Acute infection titers in eye swabs, corneas, and TG were determined on primary rabbit kidney cells grown in minimal essential medium supplemented with 7% fetal bovine serum and antibiotics (15). The following HSV-1 genotypes previously described were used in these experiments: wild-type strain 17syn+; 17ΔPst, a recombinant with a 202-bp portion of the LAT promoter (nucleotides 118,664 to 118,866) deleted, and the corresponding rescue strain, 17ΔPstR (8); 17Δ348, a LAT recombinant with bases 119,007 to 119,355 deleted, and the corresponding rescue strain, 17Δ348R (3); RHA-6, a recombinant expressing the 5′ portion of LAT by virtue of having nucleotides 120,290 to 120,467 removed and replaced with a 442-bp fragment of simian virus 40 encoding the cleavage-polyadenylation signal site (3); and KD6, a recombinant in which both copies of the ICP4 coding sequence have been deleted to yield a nonreplicating virus (9). The KD6 stocks were propagated on complementing E5 cells (a generous gift from N. DeLuca) (6), and the number of ICP4+ revertants was determined by passage and titration on RS cells (nonpermissive for ICP4− mutants). All stocks used in this study had less than one revertant per 106 PFU of ICP4− plaques.
Infections.
Lightly scarified rabbit eyes were inoculated with the indicated number of PFU in 25-μl aliquots. Rabbits were sacrificed between 1 and 7 days postinfection (dpi) for acute studies, and their corneas and TG were harvested. Latently infected TG were recovered from rabbits 40 dpi. All data presented in individual Tables 2 to 4 and Fig. 1 are results from separate and independent experiments, each performed on groups of rabbits that were infected and analyzed at the same time.
TABLE 2.
Relative amounts of viral DNA (expressed as the ratios of VP5 DNA to actin DNA) at a high dose of inoculation (500,000 PFU)a
| dpi | Mean value ± SEM in corneas for:
|
Mean value ± SEM in ganglia for:
|
||
|---|---|---|---|---|
| 17ΔPst | 17ΔPstR | 17ΔPst | 17ΔPstR | |
| 1 | 1.51 ± 0.54 | 1.19 ± 0.99 | 0.12 ± 0.12 | 0.23 ± 0.19 |
| 2 | 2.29 ± 0.76 | 1.40 ± 0.94 | 0.65 ± 0.26 | 0.37 ± 0.28 |
| 3 | 2.11 ± 0.32 | 2.38 ± 0.59 | 1.10 ± 0.26 | 1.86 ± 0.66 |
| 5 | 2.31 ± 0.64 | 1.59 ± 0.18 | 1.80 ± 0.36 | 1.74 ± 0.39 |
| 7 | 2.16 ± 1.30 | 2.01 ± 0.27 | 0.80 ± 0.20 | 0.54 ± 0.40 |
| 14 | 0.44 ± 0.14 | 0.36 ± 0.34 | 0.43 ± 0.30 | 0.21 ± 0.23 |
Rabbits' eyes were inoculated with 500,000 PFU of 17ΔPst or 17ΔPstR (rescue strain). At the indicated times postinfection, the rabbits (two rabbits per virus per time point) were sacrificed and corneas (four per virus per time point) and TG (four per virus per time point) were dissected. Total DNA was isolated from the tissue and amplified with VP5 and actin gene primer sets in combination. The relative amounts of viral DNA (ratios of VP5 DNA to actin DNA) were determined by densitometry.
TABLE 4.
Relative amounts of viral DNA present in TG during latency in rabbits infected with different doses of virusa
| Virus, dose | Rabbit tattoo no. (left or right TG)b | HSV-1 DNA (mean no. of genome equivalents) | Amt of viral DNA (mean ± SEM)c |
|---|---|---|---|
| 17ΔPst, 500 PFU | A3 (L) | 30,000 | 18,300 ± 7,888 |
| A3 (R) | 2,000 | ||
| A5 (L) | 40,000 | ||
| A5 (R) | 1,200 | ||
| 17ΔPstR (rescue strain), 500 PFU | A9 (L) | 800 | 12,200 ± 7,888 |
| A9 (R) | 8,000 | ||
| A10 (L) | 30,000 | ||
| A10 (R) | 10,000 | ||
| 17ΔPst, 50,000 PFU | A26 (L) | 1,200 | 10,750 ± 7,888 |
| A26 (R) | 1,800 | ||
| A30 (L) | 3,000 | ||
| A30 (R) | 11,000 | ||
| 17ΔPstR (rescue strain), 50,000 PFU | A31 (L) | 8,000 | 16,500 ± 7,888 |
| A31 (R) | 3,000 | ||
| A32 (L) | 15,000 | ||
| A32 (R) | 40,000 |
Rabbits were inoculated with the indicated doses of 17ΔPstR or 17ΔPst in both eyes. Total DNA was isolated from latently infected ganglia (40 dpi) and analyzed by PCR amplification with actin and VP5 gene primer sets. Data are from four TG per dose per virus per time point.
L, left; R, right.
Relative amounts of viral DNA are expressed as the number of genome equivalents of HSV determined following semiquantitative PCR for the HSV DNA polymerase gene and are standardized to the amount of cellular actin present in each sample. Standard curves were generated using known amounts of HSV polymerase target DNA in order to calculate the number of genomes present in each sample (see Materials and Methods). Means and standard errors of the mean (SEM) were calculated as described in Materials and Methods.
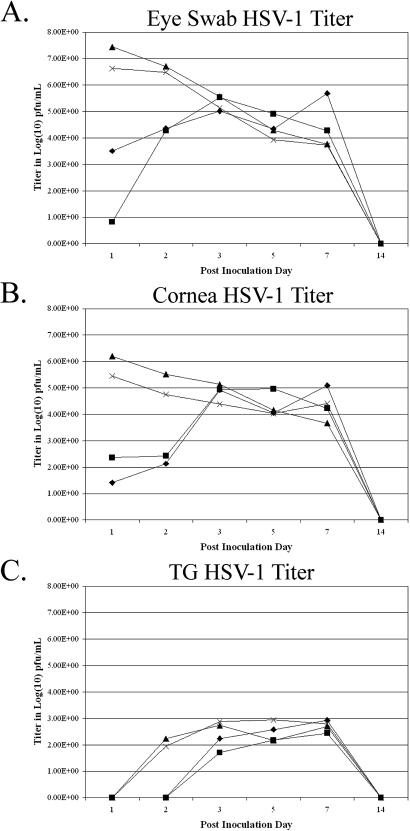
FIG. 1.
Titers of infectious virus detected in eye swabs, corneas, and TG during acute infections following inoculation with high and low doses of LAT+ and LAT− viruses. Rabbits were inoculated with 500,000 or 500 PFU of either 17ΔPst (LAT−) or 17ΔPstR (LAT+). At the indicated times, eye swabs were taken, the rabbits were sacrificed, and corneas and TG were dissected. Virus titers were determined by standard plaque assays and are expressed as the log titer of infectious virus present in the eye swabs (A), corneas (B), and TG (C). Diamonds, 17ΔPst (500 PFU); squares, 17ΔPstR (500 PFU); triangles, 17ΔPst (50,000 PFU); X's, 17ΔPstR (50,000 PFU).
DNA extraction.
Dissected corneas or ganglia were incubated with 0.6 ml of extraction buffer (25 mM EDTA, 100 mM NaCl, 1% sodium dodecyl sulfate, 10 mM Tris [pH 7.5]) and 50 μl of proteinase K solution (15 mg/ml) overnight at 48°C. DNA was extracted three times with phenol-chloroform (1:1) and once with chloroform. DNA was precipitated with ethanol overnight and pelleted by centrifugation. The pellet was washed once with 70% ethanol, air dried, and dissolved in 200 μl of water.
Analysis of the relative amounts of viral DNA by PCR.
Semiquantitative PCR analysis incorporating [α-32P]dCTP is able to detect 1 pg of purified HSV-1 DNA by comparison to a control plasmid containing a subcloned fragment of the VP5 gene. When purified viral DNA was mixed with uninfected ganglia, fewer than 1,000 viral genomes could be detected. This PCR method was also able to detect the viral DNA from a single infected cell. Actin gene primer sets were used to amplify DNA corresponding to cellular genomes to normalize product intensities. The signals were determined by densitometry, and the ratios were calculated (2).
Amplification by PCR was carried out as previously described (2) by using the primer sets illustrated in Table 1 for the actin and HSV-1 VP5 genes. The products were radiolabeled for autoradiography and image quantitation by addition of 0.2 μCi of [α-32P]dCTP. The reactions were carried out in an M. J. Research thermal cycler as follows: denaturation, 94°C for 30 s; annealing, 55°C for 30 s; and extension, 72°C for 60 s. The final cycle was terminated with a 10-min extension step. For each reaction, we used 20 μl (10%) of the DNA sample and the final volume of the reaction mixture was 100 μl. One-fifth of the amplified product (corresponding to 2% of the original material) was fractionated on 6% polyacrylamide gels in Tris-borate-EDTA buffer. The PCR signals were visualized by scanning an appropriately exposed autoradiogram by using a Deskcan II scanner (Hewlett-Packard). The signals were quantified by densitometry by using IP Lab Gel software (Signal Analysis Corporation) in accordance with operational instructions.
TABLE 1.
PCR primers
| Gene target | Primer pair | Product size (bp) |
|---|---|---|
| HSV-1 VP5 gene | 5′-TGAACCCCAGCCCCAGAAACC-3′ | 149 |
| 5′-CGAGTAAACCATGTTAAGGACC-3′ | ||
| HSV-1 ICP4 gene | 5′-CTGATCACGCGGCTGCTGTACACC-3′ | 144 |
| 5′-GGTGATGAAGGAGCTGCTGTTGCG-3′ | ||
| HSV-1 DNA poly- merase gene | 5′-CATCACCGACCCGGAGAGC-3′ | 92 |
| 5′-GGGCCAGGCGCTTGTTGGTGTA-3′ | ||
| Rabbit actin gene | 5′-AAGATCTGGCACCACACCTT-3′ | 110 |
| 5′-CGAACATGATCTGGGTCATC-3′ |
PCR analysis to determine relative levels of latent viral DNA and wild-type revertants.
For these experiments, PCR primers specific for the HSV-1 DNA polymerase gene were used to quantitate latent HSV-1 genomes, and the cellular actin gene served as an internal standard for normalizing levels of latent viral DNA among samples. PCR primers specific for the HSV-1 ICP4 gene (Table 1) were also used for analysis of the KD6 viral recombinant to confirm that the HSV-1 genomes detected were not due to wild-type revertants. PCRs were performed in a 50-μl final volume consisting of 40.5 μl of sterile H2O, 1 μl each of both forward and reverse primers (600 ng/μl), 1 μl of deoxynucleoside triphosphates (1.25 mM each), 5 μl of 10× AS buffer [Tris-Cl, KCl, (NH4)2SO4, 15 mM MgCl2 (pH 8.7); Qiagen], 1 μl of respective DNA sample, and 0.5 μl of HotStar Taq DNA polymerase (5 U/μl; Qiagen). The amplification profile consisted of a step at 95°C for 15 min to activate the Taq, followed by one cycle of 94, 55, and 72°C for 3 min, followed by 30 identical cycles of 1 min each (Ericomp Twinblock System, Easy Cycler). PCR products were resolved on 5% polyacrylamide gels, stained with SYBR Green (Molecular Probes), and scanned with a Storm PhosphorImager (Molecular Dynamics) using a 450-nm-wavelength laser. Relative numbers of latent genomes were determined by establishing the ratio of HSV-1 polymerase product to cellular actin within each sample. Viral polymerase-specific PCR products were compared to a plasmid titration mixture containing the subcloned target sequence spiked into processed, uninfected rabbit TG tissue. The signal intensity of each sample was compared to that of this titration mixture to determine the relative number of latent HSV-1 molecules in each sample. Dilutions (twofold) of all samples were performed to determine the appropriate amount of sample yielding a linear response and falling within the linear range of the standard curve.
Statistical analyses.
Results in Tables 2, 3, and 4 were analyzed using factorial analyses of variance with within-subject (nesting of tissue and virus strain combinations within an animal) arrangement of treatments. Post hoc evaluation of means following a significant overall model fit and significant interactions was conducted using protected t tests and a simulation method to correct alpha levels for the number of comparisons carried out (10).
TABLE 3.
Relative amounts of viral DNA in corneas and TG during acute infections following low-dose inoculation with viruses of different LAT genotypesa
| Virus | dpi | Mean value ± SEMb in:
|
|
|---|---|---|---|
| Corneas | Ganglia | ||
| 17syn+ | 1 | 0.21 ± 0.12 | 0.03 ± 0.01 |
| 2 | 0.82 ± 0.55 | 0.03 ± 0.02 | |
| 3 | 0.88 ± 0.46 | 0.07 ± 0.04 | |
| 5 | 0.79 ± 0.87 | 0.49 ± 0.48 | |
| 7 | 1.42 ± 0.49 | 0.50 ± 0.31 | |
| 21 | 0.22 ± 0.09 | 0.22 ± 0.13 | |
| 17ΔPst | 1 | 0.27 ± 0.16 | 0.03 ± 0.30 |
| 2 | 0.37 ± 0.24 | 0.08 ± 0.30 | |
| 3 | 0.51 ± 0.36 | 0.05 ± 0.30 | |
| 5 | 1.44 ± 0.56 | 0.39 ± 0.30 | |
| 7 | 0.83 ± 0.79 | 0.16 ± 0.30 | |
| 21 | 0.30 ± 0.23 | 0.25 ± 0.31 | |
| 17Δ348 | 1 | 0.40 ± 0.28 | 0.04 ± 0.04 |
| 2 | 0.23 ± 0.22 | 0.03 ± 0.04 | |
| 3 | 0.36 ± 0.27 | 0.03 ± 0.04 | |
| 5 | 0.80 ± 0.55 | 0.31 ± 0.04 | |
| 7 | 0.83 ± 0.70 | 0.21 ± 0.21 | |
| 21 | 0.23 ± 0.12 | 0.28 ± 0.21 | |
| 17Δ348R | 1 | 0.30 ± 0.33 | 0.03 ± 0.01 |
| 2 | 0.59 ± 0.47 | 0.03 ± 0.01 | |
| 3 | 0.92 ± 0.67 | 0.17 ± 0.35 | |
| 5 | 1.83 ± 0.69 | 0.61 ± 0.42 | |
| 7 | 1.83 ± 1.45 | 0.74 ± 0.70 | |
| 21 | 0.22 ± 0.10 | 0.22 ± 0.01 | |
| RHA-6 | 1 | 0.10 ± 0.13 | 0.04 ± 0.03 |
| 2 | 0.07 ± 0.09 | 0.03 ± 0.02 | |
| 3 | 0.67 ± 0.35 | 0.07 ± 0.04 | |
| 5 | 1.20 ± 0.35 | 0.46 ± 0.34 | |
| 7 | 0.79 ± 0.65 | 0.57 ± 0.34 | |
| 21 | 0.15 ± 0.11 | 0.33 ± 0.15 | |
Rabbit eyes were inoculated with 500 PFU of 17syn+, 17ΔPst, 17Δ348, 17Δ348R, and RHA-6. At the indicated dpi, corneas and TG (four each per virus per time point) were dissected and the relative amounts of viral DNA were determined.
Relative amounts of viral DNA are presented as the ratios of the HSV VP5 gene to the cellular actin gene as determined by PCR (see Materials and Methods). Means and standard errors of the mean (SEM) are presented as least-squares mean values and were calculated as described in Materials and Methods.
RESULTS
Acute replication in rabbit corneas and TG in high- versus low-dose infections.
The contributions of both LAT expression and inoculation dose were analyzed over the course of acute ocular infection of rabbits with either 500 or 500,000 PFU of 17ΔPst or 17ΔPstR (rescue strain)/eye. Infectious virus yields during the acute infection were measured in tear swabs, corneas, and TG (Fig. 1). At high viral doses (5 × 105 PFU), titers were highest in the tears and corneas on the first dpi. These levels tended to reach a lower plateau by days 3 through 7, and the virus was undetectable by day 14. Virus titers in TG increased during the first 3 days of infection, followed by 3 days (days 3 to 7 postinfection) of sustained virus titers, with the peak occurring during this period. As in the case of the corneas, virus was not detectable by day 14. Infection of rabbits with an inoculum of 500 PFU resulted in the detection of less infectious virus in the eye swabs and corneas at 1 and 2 dpi; however, by day 3, the amounts of infectious virus present in these samples were indistinguishable from those in the samples from rabbits infected with 5 × 105 PFU (Fig. 1A and B). A similar lag was evident in the ability to detect infectious virus in TG of rabbits receiving the 500-PFU inoculum (Fig. 1C), and it was not until days 5 to 7 that TG from rabbits infected with 500 PFU of each virus contained amounts of infectious virus similar to those contained in the TG from rabbits infected with 5 × 105 PFU. When the replication curves of the two different viruses, 17ΔPst and 17ΔPstR, were compared, they were roughly colinear and not significantly different for either the eye swabs, corneas, or TG. So, while the infecting dose clearly affected the initial infection kinetics, it did not significantly alter maximal virus yields. In addition, the ability to express LAT had no identifiable effects on acute replication in the eyes or TG.
Analysis of viral DNA levels in corneas and TG during acute infection.
While the use of 1,000-fold-lower inoculum doses of 17ΔPst and its rescue strain did not identify any differences in viral yields during the acute infection, the possibility remained that there might be detectable differences in genome loads. PCR analysis to determine the relative amounts of viral DNA present in corneas and TG following both high-dose (5 × 105 PFU per eye) and low-dose (500 PFU per eye) infection was performed. The relative amounts of viral DNA present in corneas and TG following high-dose infection did not show significant differences based on LAT genotypes at any time points (Table 2). The course of infection was then examined following a much lower dose infection (500 PFU per eye). In general, the amounts of HSV-1 DNA detected in the corneas versus those detected in the TG paralleled the findings from infectious virus assays. As with the high-titer infections, relative amounts of HSV-1 DNA in corneas were greater than those in TG during the entire acute infection course (Table 3). Comparison of the data in Tables 2 and 3 revealed a delay in the increases in viral DNA in the lower-dose infections, and the peak values for viral DNA occurred at the same time points as in the infectious virus assays (Fig. 1). Since the assay results for viral DNA seemed to parallel the data obtained for infectious virus and also permitted the detection of viral genomes as the virus entered latency, we evaluated several different LAT mutations in a low-dose infection by using this method of analysis. In addition to the LAT promoter deletion recombinant, 17ΔPst, the recombinants 17Δ348, its rescue strain, and RHA-6 were included in this analysis. These other two recombinants differ in LAT expression and/or reactivation phenotypes; 17Δ348 expresses LAT but exhibits significant reactivation impairment following epinephrine induction, whereas RHA-6, which contains a simian virus 40 cleavage-polyadenylation sequence in the middle of the 2.0-kb LAT intron, expresses LAT and reactivates normally (2).
Rabbits inoculated with 500 PFU of reactivation-impaired viral recombinants (17Δ348 and 17ΔPst) demonstrated significantly decreased amounts of viral DNA in TG during the acute phase of infection compared to rabbits inoculated with the wild type, 17syn+, and RHA-6 (Table 3). At day 5 postinfection, the mean value for the reactivation-impaired mutants (0.35 ± 0.19 [ratio of VP5 DNA to actin DNA]) was marginally significantly different (P = 0.068) from that for the normal reactivators (0.56 ± 0.38). Mean values for HSV DNA at day 7 (0.29 ± 0.18 for reactivation-impaired viruses and 0.63 ± 0.31 for normally reactivating viruses) were again significantly different (P = 0.006), but by the time the active acute infection had cleared (21 days), all TG values were statistically indistinguishable for all of the viruses tested. Therefore, during the initial phase of the low-dose infection there was a transient period (days 3 to 7) during which somewhat less viral DNA was detected in the TG following infection with the LAT recombinants containing deletions in the LAT region. As the infection progressed and then resolved (day 21), this difference was no longer seen.
The relative amounts of latent viral DNA in TG of rabbits infected with the wild type or LAT mutants were similar regardless of infecting dose.
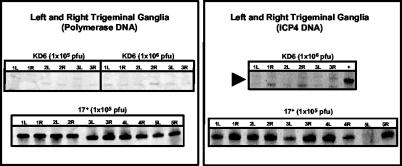
The amount of viral DNA in ganglia following clearance of the acute infection suggested that viral genome loads in the ganglia were independent of LAT genotype and infecting dose. This observation was extended to a strict latency time point by using semiquantitative PCR to carefully compare relative amounts of latent viral DNA over a range of infecting doses (Fig. 2 and Table 4). Rabbit corneas inoculated with 500 to 50,000 PFU/eye were sacrificed 40 dpi to determine the amount of latent HSV-1. Comparison of 17ΔPst with its rescue strain at an inoculum of 500 PFU resulted in mean numbers of genome equivalents that overlapped when standard error and statistical analyses were applied (P = 0.94; least-squares means analysis; see Materials and Methods). A similar comparison of the mean numbers of HSV-1 genome equivalents of these two recombinants following a 50,000-PFU infection indicated that that there was no statistical significance assignable to differences in the latent infections established by 17ΔPst and 17ΔPstR (P = 0.95). Next, an analysis of differences in numbers of latent genomes present as a function of infecting inoculum was performed. Comparisons of 17ΔPst at 500 versus 50,000 PFU and 17ΔPstR at 500 versus 50,000 PFU resulted in P values of 0.94 and 0.97, respectively. In summary, no statistical difference in numbers of viral genomes was detected as a function of either LAT genotype or initial virus dose. As with the high-titer infections examined in Table 2, neither dose nor LAT genotype affected DNA levels in latently infected TG.
FIG. 2.
HSV-1 DNA detected in the TG of rabbits 30 days after infection with high and low doses of LAT+ and LAT− viruses. Total TG DNA was isolated from rabbits infected with 50,000 or 500 PFU of either 17ΔPst or 17ΔPstR, and HSV-1 DNA was detected by PCR analysis as described in Materials and Methods. HSV-1 DNA was detected using primers specific for the HSV-1 DNA polymerase gene, and primers specific for the rabbit β-actin gene were used as an internal control. A titration mixture of dilutions of a cloned target plasmid containing the HSV-1 DNA polymerase target sequences was spiked into DNA extracted from an uninfected rabbit TG to generate a standard curve.
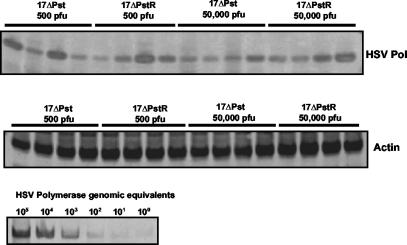
A nonreplicating HSV-1 recombinant established a latent infection in the TG but at lower levels than wild-type virus.
The analysis of the course of the acute infection as a function of dose seemed to indicate that, in the rabbit eye model, the ultimate amount of DNA that established latency in the TG was only a small fraction of the amount that reached the ganglia during the entire course of the acute infection. This result was not surprising; however, comparison of the relative levels of DNA accumulation observed in the high-dose and low-dose infections suggested that a “saturating threshold” of HSV DNA in the ganglia, or the ultimate amount of latent DNA, might actually be reached relatively early during the acute infection. This raised the question as to the relative role that the input inoculum might have on the establishment of a latent infection, particularly the normal high-dose inocula used in the rabbit model. To further assess the contribution of input inoculum versus the need for ocular replication for efficient establishment of latency, a nonreplicating (ICP4−) HSV-1 recombinant (KD6) was used. The amount of HSV-1 DNA was determined by PCR using TG from rabbits inoculated with 105 or 106 PFU of this virus at 14 dpi (Fig. 3). While TG of rabbits inoculated with KD6 contained detectable HSV genomes, overall numbers were lower than those observed using replication-competent HSV-1 strain 17syn+. PCR analysis of these ganglia (using primers specific for the ICP4 gene) indicated that the DNA present was not due to ICP4 revertants (see Materials and Methods). These results demonstrated that while nonreplicating HSV-1 recombinants could seed the TG and establish a latent infection, replication was required to achieve wild-type levels of establishment. These data also suggested that while a high-dose inoculum can result in a significant amount of HSV-1 DNA in the TG at 1 dpi, much of this DNA (and the DNA that ultimately establishes a latent infection in the rabbit TG) is the product of replication.
FIG. 3.
HSV-1 DNA detected in the TG of rabbits 14 days after infection with a nonreplicating HSV-1 recombinant. Total TG DNA was isolated from rabbits infected with 500,000 PFU of either KD6, a nonreplicating (ICP4−) recombinant, or wild-type 17syn+. The left panels show HSV-1 DNA samples obtained using primers specific for the HSV-1 DNA polymerase gene and primers specific for the rabbit β-actin gene as an internal control. The right panels show PCR analysis of the same samples using primers specific for the ICP4 gene and β-actin as the internal control. The dash indicates the location of the ICP4-specific product. L, left TG; R, right TG.
DISCUSSION
LAT has been suggested to play a role in protecting neurons from death or apoptosis during the initial stages of establishment (23, 33; R. L. Thompson and N. M. Sawtell, Letter, Science 289:1651, 2000). These observations have been made with mutants that carry deletions extending from the entire LAT promoter into the 2.0-kb intron and that often display altered virulence. While we have never observed such effects with the 202-bp LAT promoter mutant (17ΔPst), the statistical power required for discerning threefold (or less) establishment or virulence defects is difficult to achieve in the rabbit model (2). The goal of this study was to determine whether subtle deficits in replication or establishment were detectable using inocula of 500 and 50,000 PFU, doses that are 10- and 1,000-fold lower than normal 17ΔPst inocula in the rabbit eye model. The hope was that additional multiple rounds of replication permitted by the lower inoculum doses might amplify subtle replicative or establishment defects.
No significant differences in the amounts of infectious virus produced during the acute infection in corneas and ganglia or in the numbers of latent genomes in rabbit TG were observed. We did observe a slight, but statistically significant, decrease in DNA accumulation at days 3 to 7 of the acute infection in the case of several of the LAT mutations that are correlated with reactivation defects. The fact that DNA levels in the TG were comparable to those for the normally reactivating viruses at day 21 (and during latency) suggests that this DNA accumulation defect was transient and that 17ΔPst's defect in reactivation in the rabbit eye model was not simply the result of less DNA being present in the ganglia during latency. While our statistical analyses cannot rule out the possibility that 17ΔPst may have a very subtle reduction in overall establishment of latency, we would argue that it is unlikely that a decreased amount of DNA alone is the primary basis of the dramatic restriction in reactivation displayed by LAT mutants.
One possible explanation for our not seeing the effect on establishment reported for other LAT deletion mutants is that the other studies have employed recombinants with relatively large deletions (25, 33). The fact that these other deletions encompass not only the LAT promoter but also the 5′ exon and part of the intron suggests that the primary effect on establishment observed in these systems may be mediated by a distinct genetic element that lies outside of the 202-bp LAT promoter deletion in 17ΔPst. Previous studies from the Fraser and Glorioso labs have shown that a promoter element (LAP2) exists downstream of the primary latent LAT promoter and that this promoter is active in acutely infected ganglia (5, 11, 22). It should be pointed out that while the LAP1 deletion in 17ΔPst eliminates almost all latent LAT expression, transcription from the LAP2 promoter can still be detected in acute ganglia. Therefore, our study does not eliminate the contribution of this element to the course of the acute and/or establishment phases of infection and may therefore suggest a role for this downstream region in these processes.
Another observation made in this study is that lower (and probably more physiologically relevant) doses of viruses are sufficient to efficiently establish latency in the rabbit TG. It is interesting that increasing inoculum does not decrease the scatter in total levels of establishment observed in the rabbit TG over a range of doses. This scatter is likely due to variability in the numbers of nerve termini that are physically accessible to the initial inoculum and local replication of the virus in the cornea. The fact that 17ΔPst and 17PstR show similar wide and overlapping ranges of establishment in the rabbit TG but that 17ΔPst exhibits a 5- to 10-fold reduction in the number of rabbits or eyes that can be adrenergically induced to reactivate (3, 16) highlights long-standing observation that, at least in the rabbit, the absolute genome load seems to be secondary to the genotype of the HSV strain in determining the potential for reactivation.
While this suggests that the level of establishment, as measured by the amount of HSV-1 DNA present in the TG during latency, is not the primary defect in 17ΔPst's ability to reactivate, it does not rule out the idea that LAT plays some role in establishment. In fact, it is very possible that 17ΔPst may be altered in a function that substantially impacts the quality of HSV-1 establishment, such as the efficient regulation of transcription or accessibility of the HSV latent genome, a possibility first suggested by Chen et al. (4). It is also possible that 17ΔPst alters the establishment program, perhaps resulting in pushing of the HSV latent infection to populations of neurons that are less permissive for induced reactivation. It should be pointed out that the numbers of latently infected neurons, phenotypic distribution, and the numbers of genome copies per neuron have not been analyzed with these mutants in the rabbit. These have been shown to be critical parameters defining the potential to reactivate in the mouse (27, 28). Future studies will be required to investigate how these parameters are altered in the case of 17ΔPst.
Another interesting finding was that the amount of HSV-1 DNA detected in the corneas remained high at 21 dpi. While we did not examine latent-stage (28 dpi or later) corneas from rabbits infected at low doses (such as the day-21 corneas for which results are shown in Table 3), a previous study that examined reactivation of LAT+ viruses versus that of LAT− viruses in the rabbit model revealed that (i) there were relatively high amounts of HSV DNA detected in the corneas of rabbits infected with 17syn+ and the 17ΔPst rescue strain and, interestingly, (ii) there was approximately 10-fold less HSV DNA in the corneas of rabbits infected with the LAT promoter deletion recombinant 17ΔPst. In contrast, no significant differences in amounts of HSV DNA present in the TG from rabbits infected with these three viruses were detected (7). One interpretation of these data is that the presence of HSV-1 DNA in the corneas is actually the result of persistent seeding that is the result of reactivation from the TG and the fact that less 17ΔPst was detected in the corneas at latent-stage times suggests that this virus's decreased ability to reactivate results in substantially less seeding of the corneas. Our findings in the present study that there were relatively high (and comparable) amounts of HSV DNA in the corneas of rabbits infected with both LAT+ and LAT− viruses at days 14 and 21 suggests that by day 21 the DNA resulting from the acute infection-establishment phase of the latent infection had not yet cleared from the corneas. Indeed, this supports the rationale of waiting until at least 28 dpi for analysis of latency.
This study provided the additional opportunity to monitor the course of an HSV-1 ocular infection in the rabbit as a function of dose. Not surprisingly, peak acute titers in the tears, corneas, and TG were delayed by several days when lower inocula were used. Interestingly, peak levels of viral DNA in the TG were reached slightly earlier, suggesting that maximum establishment of the latent DNA pool occurs fairly early and at relatively low inoculation doses. This in turn suggests that corneas provide a limited number of entry sites into the nervous system (or number of available neuronal termini), which become saturated relatively quickly. To address this question more directly, a nonreplicating virus, KD6, was used (9, 31). Since this virus cannot undergo any replication in the cornea, it allows assessment of the amount of viral DNA delivered to the TG as a direct function of input. Results indicate that while significant establishment of latency is achieved, even doses of 106 PFU yield approximately a 10-fold lower amount of DNA than that seen with a lower inoculum of 17syn+. This indicates that while a nonreplicating virus can establish latency in TG, replication is required to establish maximal latent infections. This requirement is likely due to mechanical barriers that must be overcome to efficiently gain access to the nerve termini projecting to the TG. While infecting the corneal surface (even with scarification) provides access to many nerve termini, replication and cell-to-cell spread are much more important factors.
Acknowledgments
This work was supported by PHS grants AI06246 and AI48633 (D.C.B.), CA11861 (E.K.W.), and EY06311 (J.M.H.) from the National Institutes of Health. J.M.H. is a Research to Prevent Blindness Scientific Investigator, and J.M.L. was supported by NIH postdoctoral fellowship EY06996.
We gratefully acknowledge H. Thompson for statistical expertise, M. Banaszak, M. Rice, and M. Simpson-Evans for technical expertise, and A. Amelio, N. Kubat, L. Gary, N. Giordani, A. Gussow, P. McAnany, and Z. Zeier for help with the preparation of the manuscript and figures.
REFERENCES
- 1.Berman, E. J., and J. M. Hill. 1985. Spontaneous ocular shedding of HSV-1 in latently infected rabbits. Investig. Opthalmol. Vis. Sci. 26:587-590. [PubMed] [Google Scholar]
- 2.Bloom, D. C., G. B. Devi-Rao, J. M. Hill, J. G. Stevens, and E. K. Wagner. 1994. Molecular analysis of herpes simplex virus type 1 during epinephrine induced reactivation of latently infected rabbits in vivo. J. Virol. 68:1283-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom, D. C., J. T. Hill, E. K. Wagner, L. F. Feldman, and J. G. Stevens. 1996. A 348-bp region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J. Virol. 70:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, S. H., M. F. Kramer, P. A. Schaffer, and D. M. Coen. 1997. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 71:5878-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, X., M. C. Schmidt, W. F. Goins, and J. C. Glorioso. 1995. Two herpes simplex virus type 1 latency-active promoters differ in their contributions to latency-associated transcript expression during lytic and latent infections. J. Virol. 69:7899-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devi-Rao, G. B., J. S. Aguilar, M. K. Rice, H. Garza, D. C. Bloom, J. M. Hill, and E. K. Wagner. 1997. Herpes simplex virus genome replication and transcription during induced reactivation in the rabbit eye. J. Virol. 71:7039-7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devi-Rao, G. B., D. C. Bloom, J. G. Stevens, and E. K. Wagner. 1994. Herpes simplex virus type 1 DNA replication and gene expression during explant induced reactivation of latently infected murine sensory ganglia. J. Virol. 68:1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobson, A. T., T. P. Margolis, F. Sedarati, J. G. Stevens, and L. T. Feldman. 1990. A latent, nonpathogenic HSV-1-derived vector stably expresses beta-galactosidase in mouse neurons. Neuron 5:353-360. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, D., and J. J. Berry. 1987. The efficiency of simulation-based multiple comparisons. Biometrics 43:913-928. [PubMed] [Google Scholar]
- 11.Goins, W. F., L. R. Sternberg, K. D. Croen, P. R. Krause, R. L. Hendricks, D. J. Fink, S. E. Straus, M. Levine, and J. C. Glorioso. 1994. A novel latency-active promoter is contained within the herpes simplex virus type 1 UL flanking repeats. J. Virol. 68:2239-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gressens, P., and J. R. Martin. 1994. In situ polymerase chain reaction: localization of HSV-2 DNA sequences in infections of the nervous system. J. Virol. Methods 46:61-83. [DOI] [PubMed] [Google Scholar]
- 13.Hill, J. M., J. B. Dudley, Y. Shimomura, and H. E. Kaufman. 1986. Quantitation and kinetics of induced HSV-1 ocular shedding. Curr. Eye Res. 5:241-246. [DOI] [PubMed] [Google Scholar]
- 14.Hill, J. M., F. Sedarati, R. T. Javier, E. K. Wagner, and J. G. Stevens. 1990. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174:117-125. [DOI] [PubMed] [Google Scholar]
- 15.Hill, J. M., R. Wen, and W. P. Halford. 1998. Pathogenesis and molecular biology of HSV latency and ocular reactivation in the rabbit, p. 291-316. In S. M. Brown and A. R. MacLean (ed.), Herpes simplex virus protocols, vol. 10. Humana Press, Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 16.Jarman, R. G., J. M. Loutsch, G. B. Devi-Rao, M. E. Marquart, M. P. Banaszak, X. Zheng, J. M. Hill, E. K. Wagner, and D. C. Bloom. 2002. The region of the HSV-1 latency-associated transcript required for epinephrine-induced reactivation in the rabbit does not include the 2.0 kb intron. Virology 292:59-69. [DOI] [PubMed] [Google Scholar]
- 17.Leib, D. A., C. L. Bogard, V. M. Kosz, K. A. Hicks, D. M. Coen, D. M. Knipe, and P. A. Schaffer. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J. Virol. 63:2893-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehta, A., J. Maggioncalda, O. Bagasra, S. Thikkavarapu, P. Saikumari, T. Valyi-Nagy, N. W. Fraser, and T. M. Block. 1995. In situ DNA PCR and RNA hybridization of herpes simplex virus sequences in trigeminal ganglia of latently infected mice. Virology 206:633-640. [DOI] [PubMed] [Google Scholar]
- 19.Mellerick, D. M., and N. W. Fraser. 1987. Physical state of the latent herpes simplex virus genome in a mouse model system: evidence suggesting an episomal state. Virology 158:265-275. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell, B. M., D. C. Bloom, R. J. Cohrs, D. H. Gilden, and P. G. Kennedy. 2003. Herpes simplex virus-1 and varicella-zoster virus latency in ganglia. J. Neurovirol. 9:194-204. [DOI] [PubMed] [Google Scholar]
- 21.Nesburn, A. B., J. H. Elliot, and H. M. Leibowitz. 1967. Spontaneous reactivation of experimental herpes simplex keratitis in rabbits. Arch. Ophthalmol. 78:523-529. [DOI] [PubMed] [Google Scholar]
- 22.Nicosia, M., S. L. Deshmane, J. M. Zabolotny, T. Valyi-Nagy, and N. W. Fraser. 1993. Herpes simplex virus type 1 latency-associated transcript (LAT) promoter deletion mutants can express a 2-kilobase transcript mapping to the LAT region. J. Virol. 67:7276-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perng, G. C., C. Jones, J. Ciacci-Zanella, M. Stone, G. Henderson, A. Yukht, S. M. Slanina, F. M. Hofman, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287:1500-1503. [DOI] [PubMed] [Google Scholar]
- 24.Perng, G. C., S. M. Slanina, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2001. The effect of latency-associated transcript on the herpes simplex virus type 1 latency-reactivation phenotype is mouse strain-dependent. J. Gen. Virol. 82:1117-1122. [DOI] [PubMed] [Google Scholar]
- 25.Perng, G. C., S. M. Slanina, A. Yukht, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2000. The latency-associated transcript gene enhances establishment of herpes simplex virus type 1 latency in rabbits. J. Virol. 74:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rock, D. L., and N. W. Fraser. 1983. Detection of HSV-1 genome in central nervous system of latently infected mice. Nature 302:523-525. [DOI] [PubMed] [Google Scholar]
- 27.Sawtell, N. M. 1998. The probability of in vivo reactivation of herpes simplex virus type 1 increases with the number of latently infected neurons in the ganglia. J. Virol. 72:6888-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawtell, N. M., D. K. Poon, C. S. Tansky, and R. L. Thompson. 1998. The latent herpes simplex virus type 1 genome copy number in individual neurons is virus strain specific and correlates with reactivation. J. Virol. 72:5343-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawtell, N. M., and R. L. Thompson. 1992. Herpes simplex virus type 1 latency associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J. Virol. 66:2157-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawtell, N. M., and R. L. Thompson. 1992. Rapid in vivo reactivation of herpes simplex virus in latently infected murine ganglionic neurons after transient hyperthermia. J. Virol. 66:2150-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sedarati, F., T. P. Margolis, and J. G. Stevens. 1993. Latent infection can be established with drastically restricted transcription and replication of the HSV-1 genome. Virology 192:687-691. [DOI] [PubMed] [Google Scholar]
- 32.Stevens, J. G., and M. L. Cook. 1971. Latent herpes simplex virus in spinal ganglia of mice. Science 173:843-845. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, R. L., and N. M. Sawtell. 2001. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J. Virol. 75:6660-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran, R. K., P. T. Lieu, S. Aguilar, E. K. Wagner, and D. C. Bloom. 2002. Altering the expression kinetics of VP5 results in altered virulence and pathogenesis of herpes simplex virus type 1 in mice. J. Virol. 76:2199-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]