Abstract
Both the 5′ end of the Sindbis virus (SIN) genome and its complement in the 3′ end of the minus-strand RNA synthesized during virus replication serve as parts of the promoters recognized by the enzymes that comprise the replication complex (RdRp). In addition to the 5′ untranslated region (UTR), which was shown to be critical for the initiation of replication, another 5′ sequence element, the 51-nucleotide (nt) conserved sequence element (CSE), was postulated to be important for virus replication. It is located in the nsP1-encoding sequence and is highly conserved among all members of the Alphavirus genus. Studies with viruses containing clustered mutations in this sequence demonstrated that this RNA element is dispensable for SIN replication in cells of vertebrate origin, but its integrity can enhance the replication of SIN-specific RNAs. However, we showed that the same mutations had a deleterious effect on virus replication in mosquito cells. SIN with a mutated 51-nt CSE rapidly accumulated adaptive mutations in the nonstructural proteins nsP2 and nsP3 and the 5′ UTR. These mutations functioned synergistically in a cell-specific manner and had a stimulatory effect only on the replication of viruses with a mutated 51-nt CSE. Taken together, the results suggest the complex nature of interactions between nsP2, nsP3, the 5′ UTR, and host-specific protein factors binding to the 51-nt CSE and involved in RdRp formation. The data also demonstrate an outstanding potential of alphaviruses for adaptation. Within one passage, SIN can adapt to replication in cells of a vertebrate or invertebrate origin.
The Alphavirus genus of the Togaviridae family contains nearly 30 known members, including a number of human and animal pathogens (reviewed in reference 12). The majority of alphaviruses are transmitted by mosquitoes to higher vertebrates that serve as amplifying hosts. In insect vectors, alphaviruses cause persistent life-long infections and do not greatly affect the viability of their hosts (3). Accordingly, they establish persistent infections in cultured mosquito cells (2). In vertebrates, alphaviruses usually cause acute infections that often result in disease (14), and the infection of susceptible cultured vertebrate cells leads to a progressive cytopathic effect (CPE) and cell death (8, 20).
Sindbis virus (SIN) is a prototype member of the Alphavirus genus, and it has always been a highly valuable source of information about the mechanisms of alphavirus replication and virus-host cell interactions (reviewed in reference 44). This virus can productively replicate in a wide variety of cell lines of insect and vertebrate origins. Like other alphaviruses, SIN enters cells via receptor-mediated endocytosis (4). The fusion of viral and endosomal membranes leads to the release of nucleocapsids into the cytoplasm (10), and after their ribosome-mediated disassembly, viral genomes become capable of translation and replication (42, 49).
The SIN genome is a single-stranded RNA of positive polarity that is almost 12 kb long (43). It is capped at the 5′ end and contains a poly(A) tail at the 3′ end. The genome serves directly as a template for the synthesis of nonstructural proteins (nsPs) that form the enzyme complex (RdRp) required for viral genome replication and transcription of the subgenomic RNA. The 26S RNA corresponds to the 3′ one-third of the genome and encodes all of the viral structural proteins (36). Both replication of the viral genome and transcription of the subgenomic RNA are highly regulated. Viral nonstructural proteins are synthesized as two polyproteins, P123 and P1234. The latter is cleaved in cis by a papain-like protease, nsP2 (5, 13), to generate P123 and nsP4. In the early stages of infection, this partially processed protein complex, P123/nsP4, can efficiently synthesize full-length minus-strand genome intermediates, but it was shown to be inefficient for positive-strand RNA synthesis (17, 18). Further processing of P123 to nsP1, nsP2, and nsP3, also mediated by the nsP2 protease (40, 41), leads to transformation of the replicative enzyme complex into mature RdRp (containing nsP1, nsP2, nsP3, and nsP4) with a strongly increased ability to synthesize positive-sense genomes and subgenomic RNAs (19, 41). SIN replicative enzymes are highly selective and utilize only viral genomes, not cellular mRNAs, as templates, in spite of the presence of mRNAs in high concentrations. Moreover, the viral subgenomic RNA, whose sequence is identical to the 3′ end of the viral genome, is also excluded from replication, even though it accumulates to high levels in infected cells. This specificity of RNA replication is achieved via the recognition of cis-acting promoter elements that are present both in the viral genome and in the minus-strand genome intermediate.
Most of the cis-acting elements were previously identified as sequences that are conserved for all members of the Alphavirus genus, and the significance of their roles in RNA replication was later confirmed by reverse genetic experiments. First, the 24-nucleotide (nt) conserved sequence element (CSE) was identified upstream of the start of the subgenomic RNA (22, 28). The complement of this sequence in the negative-strand RNA was characterized as a transcriptional promoter. The second CSE, the 3′ 19-nt sequence followed by the poly(A) tail, was proposed as a core promoter for the synthesis of the genome-length negative-strand RNA (30). The activity of the 3′ CSE is also dependent on the 5′ untranslated region (UTR), and the interaction between the 3′ and the 5′ ends is critical for the initiation of minus-strand RNA synthesis (7). However, the complement of the 5′ UTR in the minus-strand RNA is also a promoter for the synthesis of positive-sense genomes (26, 29). Another cis-acting element that is essential for replication, the 51-nt CSE, is also found at the 5′ end of the SIN genomic RNA in the nsP1-encoding gene. Both the primary sequence of this CSE and its two-stem-loop secondary structure, in particular, are highly conserved among the alphaviruses (27). During SIN replication in mammalian cells, this element serves as a replication enhancer (7), but the exact mechanism of its functioning still remains obscure.
To further elucidate the role(s) of the 51-nt CSE in RNA replication and its interaction with the proteins involved in replication, we designed SIN viruses with clustered mutations in this sequence. These mutations destabilized the putative secondary structure of the RNA without changing the encoded amino acid sequence, and they affected virus replication in mosquito cells rather than in mammalian cells. Pseudorevertants of the virus with a mutated 51-nt CSE that were capable of growing in mosquito cells were selected and characterized. Adaptive mutations were identified in the genes encoding the viral nonstructural proteins and in the 5′ UTR. Our data further define the interaction of SIN-specific and host cell proteins with cis-acting elements of the viral genome.
MATERIALS AND METHODS
Cell cultures.
BHK-21 cells were obtained from Charles M. Rice (Rockefeller University, New York, N.Y.). These cells were propagated in alpha minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) and vitamins. Mosquito C710 cells were obtained from Henry Huang (Washington University, St. Louis, Mo). They were propagated in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated FBS and 10% tryptose phosphate broth.
Plasmid constructs.
The parental pToto1101 plasmid encoding the infectious wild-type SIN (SINwt) viral genome was described elsewhere (35). The pSIN43 plasmid, encoding the SIN43 viral genome with clustered silent mutations, was designed by standard recombinant DNA techniques. The introduced mutations are shown in Fig. 1. All other plasmids encoding SINwt and SIN43 genomes with adaptive mutations were prepared by standard PCR-based mutagenesis and cloning methods. All of the PCR fragments were initially cloned into the pRS2 plasmid and sequenced to exclude the possibility of spontaneous mutations. The details of the designed constructs are described in Results and the figure legends.
FIG. 1.
Mutations in 51-nt CSE of SIN43 virus genome (A) and computer-predicted (m-fold) secondary structures of 5′ ends of SINwt and SIN43 virus genomes (B). The proposed stem-loops in the SINwt structure are indicated as SL1 to -4. SL3 and SL4 represent the 51-nt CSE. Arrows indicate the positions of the initiating AUG codons.
The double subgenomic SIN replicons SINwt/GFP/Pac and SIN43/GFP/Pac had the same design as the previously described SINrep19/GFP replicon (1), but they did not have the P726→L mutation in nsP2 that makes replicons less cytopathic. The SIN43/GFP/Pac replicon also contained all of the clustered mutations present in the 5′ end of SIN43 (Fig. 1A).
RNA transcription.
Plasmids were purified by centrifugation in CsCl gradients. Prior to transcription, the viral genome-encoding plasmids were linearized with XhoI. RNAs were synthesized by SP6 RNA polymerase in the presence of a cap analog (35). The yield and integrity of the transcripts were monitored by gel electrophoresis under nondenaturing conditions. For electroporation, aliquots of reactions containing 1 μg of full-length transcripts were used without additional purification (23).
RNA transfections and infectious center assays.
In standard experiments, 1 μg of an in vitro-synthesized, full-length RNA transcript was used per electroporation. Tenfold dilutions of electroporated BHK-21 or C710 cells were seeded in six-well tissue culture plates containing 5 × 105 naïve BHK-21 or 106 C710 cells per well. After 2 h of incubation at 37°C (for BHK-21 cells) or 30°C (for C710 cells) in a 5% CO2 incubator, cells were overlaid with 2 ml of 0.6% Ultra-Pure agarose (Invitrogen) containing MEM supplemented with 2% FBS or with 2 ml of 0.6% tragacanth gum (ICN) containing MEM supplemented with 5% heat-inactivated FBS and 10% tryptose phosphate broth, for BHK-21 or C710 cells, respectively. Plaques were allowed to develop at 37°C (in BHK-21 cells) or 30°C (in C710 cells) and were stained with crystal violet after 1.5 days of incubation in BHK-21 cells or 2.5 days of incubation in C710 cells. The remaining electroporated cells were usually seeded into 35-mm-diameter dishes for radioactive labeling of RNAs and proteins, the generation of viral stocks, or virus growth analysis (see below).
Selection of SIN43 mutants capable of efficient growth in C710 cells.
The original stock of SIN43 virus was generated by the transfection of BHK-21 cells. Six plaques were isolated from the agarose overlay, viruses were eluted in 1.5 ml of alpha MEM supplemented with 1% FBS for several hours, and 0.5 ml was used to infect 5 × 105 BHK-21 cells in 35-mm-diameter dishes. After a comparison of growth rates in BHK-21 and C710 cells, one of the isolates was used for sequencing, and the same virus was also passaged three times in C710 cells at a multiplicity of infection (MOI) of ∼10 PFU/cell. Three plaques were randomly selected in C710 cells from the final stock (agarose was used for this experiment instead of tragacanth gum). These variants were additionally plaque purified in C710 cells, and then 5-ml stocks were generated in the same cells and used for further characterization and sequencing.
Sequencing of viral genomes.
For sequencing, viruses were purified by ultracentrifugation through 20% sucrose at 55,000 rpm for 2 h in an SW-60 rotor at 4°C. Pellets were suspended in 100 μl of phosphate-buffered saline (PBS) containing 1% FBS, and RNAs were isolated by use of TRIzol according to the procedure recommended by the manufacturer (Invitrogen). Ten overlapping (approximately 1-kb long) fragments representing the entire viral genome were synthesized by a reverse transcription-PCR procedure, purified by agarose gel electrophoresis, and sequenced by using the same primers as those used for PCR (the sequences of the primers will be provided upon request). DNA fragments representing the 5′- and 3′-terminal sequences were synthesized by use of a commercially available FirstChoice RLM-RACE kit according to the procedure recommended by the manufacturer (Ambion). Fragments were purified by agarose gel electrophoresis and cloned into the plasmid pRS2. Multiple independent clones were sequenced to determine possible variations in the 5′ and 3′ ends of the genomes.
Virus growth analysis.
BHK-21 and C710 cells were seeded at concentrations of 5 × 105 and 106 cells/35-mm-diameter dish, respectively. After 4 h of incubation at an appropriate temperature, monolayers were infected at the MOIs indicated in the figure legends for 1 h, washed three times with PBS supplemented with 1% FBS, and overlaid with 1 ml of complete medium. At the indicated times postinfection, the medium was replaced with fresh medium, and virus titers in the harvested samples were determined by plaque assays on BHK-21 or C710 cells, as indicated in the figure legends.
In many experiments, to exclude the effect of accumulation of additional adaptive mutations, we analyzed virus growth directly after the transfection of 1 μg of RNA into C710 or BHK-21 cells. One-fifth of the electroporated cells were seeded into a 35-mm-diameter dish and incubated at 37°C (BHK-21 cells) or 30°C (C710 cells) for 1 h. The medium was then replaced with 1 ml of fresh medium and continued to be replaced at the indicated times postelectroporation. Virus titers in the harvested samples were determined by plaque assays with BHK-21 cells or C710 cells, as indicated in the figure legends. It should be mentioned that the SINwt virus has a lower infectivity for C710 cells, and titers of the same viral stocks determined in BHK-21 cells were 20- to 50-fold higher than those in C710 cells.
RNA analysis.
One-fifth of the C710 cells that were electroporated with 1-μg samples of different RNAs were seeded into 35-mm-diameter dishes and incubated at 30°C. At 18.5 h posttransfection (when the virus was growing exponentially), SIN virus-specific RNAs were labeled with [3H]uridine as described in the legend to Fig. 7. RNAs were isolated from the cells by use of the TRIzol reagent, as recommended by the manufacturer (Invitrogen), denatured with glyoxal in dimethyl sulfoxide, and analyzed by agarose gel electrophoresis using previously described conditions (7).
FIG. 7.
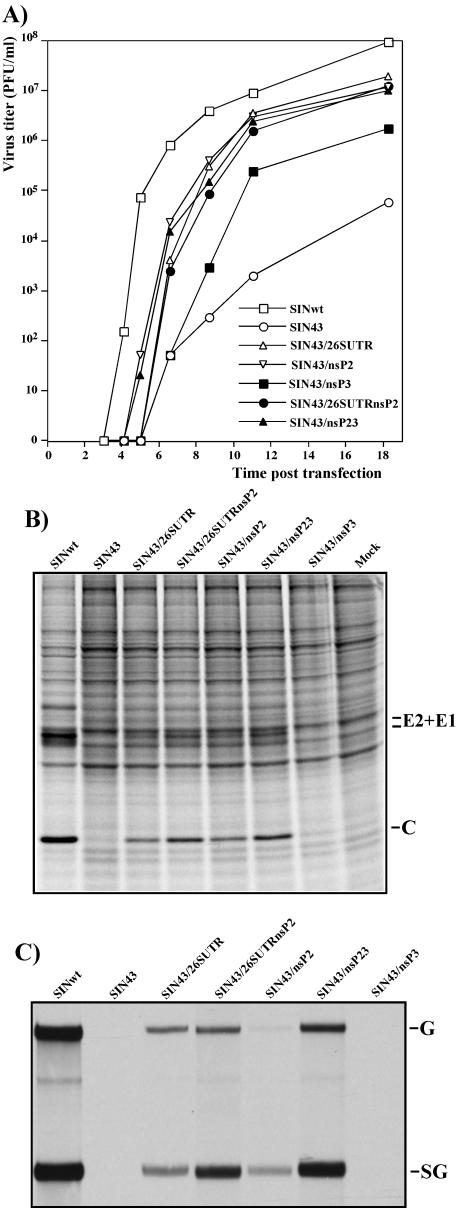
Phenotypes of SIN43-derived viruses containing single or double adaptive mutations in genomes. (A) Single-step viral growth curves after electroporation of 1 μg of in vitro-synthesized RNAs into C710 cells. At the indicated times, the medium was replaced and virus titers were determined in C710 cells, as described in Materials and Methods. Protein synthesis (B) and viral RNA replication (C) are shown for C710 cells transfected with 1 μg of the RNAs. At 18.5 h posttransfection, the cells were labeled with [35S]methionine and analyzed in a sodium dodecyl sulfate-10% polyacrylamide gel as described in Materials and Methods. For RNA analysis, at 18.5 h posttransfection, the medium in other wells was replaced with 1 ml of alpha MEM supplemented with 10% FBS, dactinomycin (1 μg/ml), and [3H]uridine (20 μCi/ml). After 3 h of incubation at 30°C, RNAs were isolated from the cells and analyzed by agarose gel electrophoresis as described in Materials and Methods.
Analysis of protein synthesis.
One-fifth of the C710 cells that were electroporated with 1-μg samples of different RNAs were seeded into 35-mm-diameter dishes and incubated at 30°C. At 18.5 h posttransfection (when the virus was still growing exponentially), the cells were washed three times with PBS and then incubated for 1 h at 37°C in 1 ml of RPMI medium that lacked methionine and was supplemented with 0.1% FBS and 20 μCi of [35S]methionine. After the incubation, the cells were scraped from the dish into PBS, pelleted by centrifugation, and dissolved in 150 μl of standard loading buffer. Aliquots of the samples (10 μl) were analyzed in sodium dodecyl sulfate-10% polyacrylamide gels. After electrophoresis, the gels were dried and analyzed by autoradiography.
RESULTS
The 51-nt CSE is critical for SIN replication in mosquito cells.
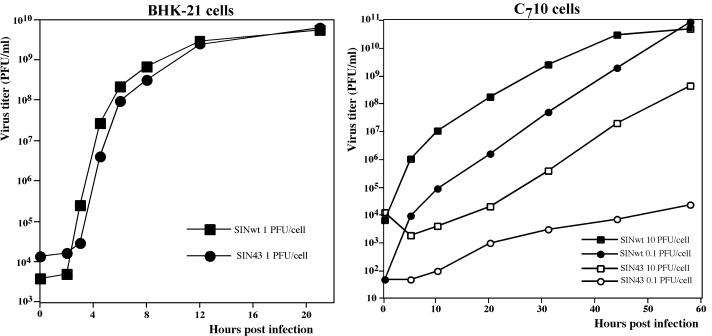
As a part of our previous studies (7, 9), we performed extensive mutagenesis of the fragment located downstream of the 5′ UTR (which includes the 51-nt CSE) in the genome of SINwt (Toto1101). The clustered silent mutations (Fig. 1A) in the SIN43 virus strongly modified the sequence of the CSE and the corresponding computer-predicted secondary structure (Fig. 1B) but did not change the encoded protein sequence. In the infectious center assay with BHK-21 cells, the in vitro-synthesized SIN43 and SINwt RNAs had the same infectivities (1 × 106 to 2 × 106 PFU/μg), and both rescued viruses were capable of replicating in BHK-21 cells at comparable rates and to similar final titers (Fig. 2A). However, the SIN43 virus demonstrated a very different ability to propagate in mosquito C710 cells compared to SINwt (Fig. 2B). The increase in viral titers was detected only if the infection was performed at an MOI of >0.1 PFU/cell and after a 24-h or longer delay. The growth rate of the SIN43 virus in C710 cells infected at an MOI of 10 PFU/cell was very similar to that of a SINwt infection performed at a 3-to-4-order of magnitude lower MOI (0.01 to 0.001 PFU/cell). This fact suggested that either the SIN43 virus stock harvested directly after RNA transfection of BHK-21 cells contained a small fraction of viruses (most probably pseudorevertants) capable of replicating in mosquito cells or those pseudorevertants were generated with a low efficiency during SIN43 replication.
FIG. 2.
Growth curves of SINwt and SIN43 viruses in BHK-21 and C710 cells. BHK-21 and C710 cells were infected at the indicated MOIs as described in Materials and Methods. The medium was harvested and replaced at the indicated time points. Released virus titers were measured by a plaque assay on BHK-21 cells. These data represent one of four reproducible experiments.
SIN replicons with clustered mutations in the 51-nt CSE replicate inefficiently in mosquito cells.
Our previous data suggested that the 51-nt CSE served as a replication enhancer during the replication of virus-specific RNAs in vertebrate cells (7). In order to test whether the clustered mutations in the CSE completely abolished the replication of viral RNA in cells of a mosquito origin (and the detected virus release was only due to pseudorevertants generated during the in vitro transcription) or downregulated it to a low level, we designed SIN replicons encoding two subgenomic promoters driving the expression of the green fluorescent protein (GFP) and Pac genes (Fig. 3). The SINrep43/GFP/Pac genome contained the entire cluster of the above-described mutations in the 51-nt CSE, and the control SINrepwt/GFP/Pac replicon had the same design, but with the authentic sequence of the 51-nt CSE. Upon transfection into the cells, these replicons were expected to express both GFP and Pac. Pac expression was employed to detect low levels of RNA replication, because in our previous studies, we noticed that even inefficient expression of this gene makes cells resistant to puromycin (1, 6). GFP expression was used to evaluate the level of viral RNA replication. The in vitro-synthesized RNAs were electroporated into C710 cells, and puromycin selection was applied at 12 h posttransfection. The transfection of 1 μg of RNA made approximately 30% of the cells in both samples resistant to puromycin (the naïve C710 cells died within 24 h of puromycin selection) (Fig. 3A). However, Purr cells transfected with SINrep43/GFP/Pac expressed a very low level of GFP that was barely detectable by both flow cytometry and fluorescence microscopy compared to the very efficient expression observed for the cells transfected with SINrepwt/GFP/Pac (Fig. 3B). Similarly, BHK-21 cells transfected with 1 μg of SINrep43/GFP/PacRNA demonstrated a lower level of GFP expression than SINrepwt/GFP/Pac-transfected cells (Fig. 3C) at an early time posttransfection. However, by 12 h the difference was no longer obvious (data not shown).
FIG. 3.
Schematic representation of double subgenomic SINrepwt/GFP/Pac and SINrep43/GFP/Pac replicons and their replication in C710 cells. (A) C710 cells were transfected with 1 μg of in vitro-synthesized RNAs. At 12 h posttransfection, puromycin selection (10 μg/ml) was applied, and at 16 h postelectroporation (before the Pur selection was completed), GFP expression was analyzed by flow cytometry (B) and fluorescence microscopy. +, transfection of 1 μg of SINrepwt/GFP/Pac or SINrep43/GFP/Pac replicon RNA produced about 30% C710 cells that were resistant to puromycin. The SINrep43/GFP/Pac-transfected cells expressed very low levels of GFP (+/−) compared to the GFP levels detected in the SINrepwt/GFP/Pac-transfected cells (+++). (C) BHK-21 cells were transfected with 1 μg of in vitro-synthesized RNAs. At 6 h posttransfection, GFP expression was evaluated by flow cytometry.
Together, these data indicate that the 51-nt CSE is likely to function as an enhancer of the replication of SIN-specific RNAs not only in mammalian cells but in mosquito cells as well. However, in contrast to the case for BHK-21 cells, its integrity is critical for SIN virus replication and release from C710 cells.
SIN genomes with mutated 51-nt CSE accumulate adaptive mutations during replication in C710 cells.
To study the adaptation of the SIN43 mutant for replication in C710 cells and to identify the mutations that accumulated in the viral genome during virus adaptation, we intended to select efficiently replicating variants. To exclude from the analysis mutations that were randomly generated during in vitro transcription by SP6 RNA polymerase (6), we selected the variants from the homogeneous isolates of the SIN43 virus. For this purpose, six plaques formed by the SIN43 virus in BHK-21 cells were randomly selected. All of them contained viruses that replicated very inefficiently in C710 cells, and at 36 h postinfection with an MOI of 10 PFU/cell, virus titers reached only 106 PFU/ml, compared to >1010 PFU/ml observed for SINwt (data not shown). In contrast to SINwt, they were also incapable of causing CPE and forming plaques in this cell line (data not shown). This was an indication that these variants were most likely the same and represented the major fraction in the population. One of the plaque-purified isolates, SIN43/1, was used further for selection of the variants that were adapted for growth in C710 cells. The stock of SIN43/1 virus, generated in BHK-21 cells, was passaged three times in C710 cells (see Materials and Methods for details), and variants that attained the ability to cause CPE and form plaques in this cell line were isolated. Three randomly selected plaques (SIN43/1/1, SIN43/1/2, and SIN43/1/3) were used for further characterization and sequencing of the viral genomes.
All three variants demonstrated efficient replication in C710 cells, and their final titers reached values that were similar to those observed for SINwt and were 4 orders of magnitude higher than the titer of the parental SIN43/1 variant (Fig. 4A). In contrast to the original SIN43 or SIN43/1 viruses, which were incapable of causing CPE in C710 cells, the selected variants efficiently formed plaques in this cell line (Fig. 4B). Importantly, the accumulated mutations that adapted SIN for replication in C710 cells strongly affected the ability of viruses to propagate and to cause CPE in BHK-21 cells (Fig. 4B). SIN43/1/1 generated easily detected, but heterogeneous, plaques, indicating a significant level of genetic instability. Plaques of SIN43/1/2 were very small, and the pinpoint plaques of SIN43/1/3 developed only after a >3-day incubation. Thus, all three viral isolates adapted for growth in mosquito cells were distinguishably different, and two of them demonstrated strongly reduced replication in mammalian cells.
FIG. 4.
Single-step growth curves of plaque-purified SIN43/1 isolates adapted for growth in C710 cells and analysis of plaque morphology of different viruses in C710 and BHK-21 cells. (A) C710 cells were infected with SINwt, SIN43/1, and the selected SIN43/1/1, SIN43/1/2 and SIN43/1/3 viruses at an MOI of 1 PFU/cell. At the indicated times, the medium was replaced and the virus titers were determined on BHK-21 cells. (B) Plaque morphology of indicated viruses compared by titration of the same viral stocks on BHK-21 and C710 cells. The stocks of SINwt and SIN43/1 viruses were prepared in BHK-21 cells, and the stocks of the SIN43/1/1, SIN43/1/2, and SIN43/1/3 variants were generated in C710 cells.
Mutations adapting SIN43 for replication in a mosquito-derived cell line.
To identify the adaptive mutations, we determined the consensus sequences of the entire genome of SIN43/1, the original virus used for selection of C710-adapted mutants, and the genomes of the SIN43/1/1, SIN43/1/2, and SIN43/1/3 derivatives. The SIN43/1 sequence corresponded to the original SIN43 construct designed for this study. The mutations discovered in the C710-adapted plaque isolates are shown in Fig. 5. First, no additional changes were found in the 51-nt CSE of the selected variants compared to the original SIN43/1 (and SIN43) genome. Second, all of the variants had a common single-amino-acid substitution in the N-terminal region of the SIN nonstructural protein nsP2 (Fig. 5A). They contained a Lys substitution for Glu at nsP2 residue 118. In addition, the SIN43/1/1 virus also had a mutation, a substitution of Thr for Ile at residue 286, in another nonstructural protein, nsP3 (Fig. 5B), and both SIN43/1/2 and SIN43/1/3 had mutated 5′ UTRs in the viral genomic RNA, in which the very 5′-terminal 9 nt were replaced with nt 1 to 52 of the SIN subgenomic RNA (Fig. 5C). Surprisingly, this insertion also included the initiating AUG codon of SIN 26S RNA and created an alternative, 23-codon open reading frame that was not in frame with the SIN nsP1-coding sequence (Fig. 5C). However, the initiation of translation from this additional AUG apparently did not profoundly affect virus replication in mosquito cells. Compared to SIN43/1/2, the SIN43/1/3 virus contained an additional mutation in the subgenomic RNA. The insertion of three adenosines into the A7 sequence present in the E2 glycoprotein coding sequence led to the appearance of an additional Lys at position 147 of this protein. This amino acid most likely also altered the ability of the virus to form plaques in BHK-21 cells, and SIN43/1/3 developed pinpoint plaques only after a prolonged incubation. However, there was a very small chance that the mutation in the SIN E2 glycoprotein had any compensatory effect on the mutations in the 51-nt CSE, and its role was not further investigated.
FIG. 5.
Sequence alignments and mutations found in SIN43/1/1, SIN43/1/2, and SIN43/1/3 variants adapted for growth in C710 cells. (A) Alignment of nsP2 residues 100 to 139 (SIN numbering) for several alphaviruses. The adaptive mutation found in all three variants is indicated. (B) Alignment of nsP3 residues 273 to 292 (SIN numbering). The mutation found in the SIN43/1/1 variant is indicated. Aura, Aura virus (39); ONN, O'nyong nyong virus (21); SFV, Semliki Forest virus (45); VEE, Venezuelan equine encephalitis virus (15); EEE, Eastern equine encephalitis virus (48). Residues identical to those in the SIN sequence are indicated by dashes. (C) Sequence alignment of the SIN43/1/2 and SIN43/1/3 5′ UTRs with the SINwt 5′ UTR and SIN subgenomic 26S 5′ UTR. The sequence representing an alternative open reading frame in the genomes of adapted variants is underlined.
Adaptive mutations function synergistically.
To evaluate the effects of the observed mutations on the ability of the SIN43 mutant to replicate in C710 cells, we incorporated them into an infectious SIN43 cDNA (Fig. 6). Constructs contained either the single site mutation in nsP2, nsP3, or the 5′ UTR (SIN43/nsP2, SIN43/nsP3, and SIN43/26SUTR, respectively) or both mutations found in the selected adapted pseudorevertants (SIN43/nsP23 and SIN43/26SUTRnsP2). In order to determine the mutations that were critical for the adaptation of SIN43 for replication in C710 cells, we compared the infectivities of all of the in vitro-synthesized RNAs in infectious center assays performed with C710 cells. Because of the very high rate of SIN evolution during replication, the levels of RNA replication of all mutants, their growth rates, and their ability to cause CPE (plaque formation) were tested directly after RNA transfection. The results of these experiments, shown in Fig. 6A, demonstrate that the point mutation in nsP2 and the replacement of the 5′ UTR increased the infectivities of the RNAs to a nearly SINwt level, but the effect of the mutation in nsP3 alone was less obvious (the infectivity of SIN43/nsP3 RNA became only 10-fold higher than that of SIN43). However, two mutations were required to make viruses cytopathic for C710 cells (Fig. 6B). Only SIN43/nsP23 and SIN43/26SUTRnsP2 caused rapid CPE development in C710 cells, as did the original SIN43/1/1 and SIN43/1/2 variants. As seen in the original plaque isolates, the SIN43/nsP23 mutant retained the ability to form plaques in BHK-21 cells and the plaques of SIN43/26SUTRnsP2 were hardly detectable. This was most likely a result of the insertion of the 26S UTR into the genomic 5′ end, because the single mutant SIN43/26SUTR also developed pinpoint plaques in BHK-21 cells and both 5′ end mutants rapidly generated large-plaque-forming pseudorevertants (data not shown), which were not further studied.
FIG. 6.
Replication of SIN43 virus variants, containing different adaptive mutations in the genomes, in C710 and BHK-21 cells. (A) Schematic representation of viral genomes, infectivities of in vitro-synthesized RNAs in C710 and BHK-21 cells in the infectious center assay, and sizes of plaques formed on both cell lines in the same test. Filled boxes indicate the positions of the introduced adaptive mutations and the presence of clustered mutations in the 51-nt CSE. NA, nonapplicable, because of a lower infectivity of the RNAs and heterogeneous plaque sizes, indicating a pseudorevertant appearance. The data represent one of three reproducible experiments. (B) Presentation of different levels of CPE development at 48 h postelectroporation of 1 μg of in vitro-synthesized RNAs into C710 cells.
An analysis of virus growth rates in C710 cells did not reveal strong differences between the designed mutants (Fig. 7A). However, viruses containing only the nsP2 mutation or both the nsP2 and nsP3 mutations initiated virus release from the cells earlier than other mutants. Both the SIN43 and SIN43/nsP3 variants generated pseudorevertants, and in agreement with results of the infectious center assay, SIN43/nsP3 made them more efficiently and hence demonstrated faster growth.
The level of RNA replication and the synthesis of viral proteins correlated better with the observed differences in CPE development (Fig. 7B and C). Variants containing any single mutation in the genome were less efficient in RNA replication, transcription of the subgenomic RNA, and synthesis of virus-specific proteins than viruses with two adaptive mutations. The single-site mutants, SIN43/26SUTR and SIN43/nsP2, grew as efficiently as SIN43/26SUTRnsP2 and SIN43/nsP23, respectively. However, they caused slower CPE development and formed smaller plaques in C710 cells (Fig. 6A and B). This fact indicated that mutations found in the genomes of the SIN43 variants adapted for growth in C710 cells functioned synergistically and were likely more important in terms of an increase in cytopathogenicity of viruses with the mutated 51-nt CSE than just for their replication in mosquito cells.
Adaptive mutations have no enhancing effect on SINwt replication.
To further elucidate the role(s) of the identified adaptive mutations on SIN RNA replication, we tested them in the context of the SINwt genome backbone (Fig. 8). All of the in vitro-synthesized viral RNAs had very similar infectivities in C710 cells, indicating that neither the adaptive amino acid changes in nsP2 and nsP3 nor the insertion of the 26S RNA sequence into the 5′ UTR of the SIN genome was lethal for SIN replication in this cell line. However, the single-site mutants SINwt/nsP3 and SINwt/26SUTR and the double mutants SINwt/26SUTRnsP2 and SINwt/nsP23 had significantly slower growth rates than SINwt. In particular, SINwt/nsP23 produced pinpoint plaques in C710 cells and replicated 2 orders of magnitude slower than SINwt.
FIG. 8.
Replication of SINwt variants containing different adaptive mutations in genomes in C710 and BHK-21 cells. (A) Schematic representation of viral genomes, infectivities of in vitro-synthesized RNAs in C710 and BHK-21 cells in the infectious center assay, and sizes of plaques formed on both cell lines. Filled boxes indicate the positions of the introduced mutations found in SIN43 variants adapted for growth in C710 cells. (B) Single-step viral growth curves after electroporation of 1 μg of in vitro-synthesized RNAs into C710 cells. At the indicated times, the medium was replaced and virus titers were determined on BHK-21 cells, as described in Materials and Methods.
All of the single and double mutants made in the context of the SINwt genome were also tested for the ability to propagate in BHK-21 cells. It was not surprising to find that the positioning of the 26S 5′ UTR to the 5′ end of the SINwt genome strongly diminished virus replication. Both SINwt/26SUTR and SIN43/26SUTRnsP2 were slower growing (data not shown), formed very small plaques, and rapidly reverted to a large-plaque-forming phenotype.
DISCUSSION
In spite of a great deal of progress that has been made in our understanding of the regulation of alphavirus RNA replication, critical details about the interaction of viral nonstructural proteins with cis-acting elements of their genomes and with host factors involved in the replication process remain obscure. Particularly poorly understood is the functioning of the 51-nt CSE. This highly conserved RNA sequence is located between nt 154 and 205 in the SIN genome and is predicted to form two stem-loop structures with very similar conformations among all studied members of the Alphavirus genus. The conservation of this sequence in distantly related alphaviruses (27) and in the naturally occurring SIN and Semliki Forest virus defective interfering RNAs (16, 24, 25) suggested its importance for the replication of the virus-specific RNAs, and the most attractive hypothesis to explain this importance was that this element determines the selective replication of the alphavirus genomes, but not the subgenomic RNAs.
In our earlier work, we demonstrated that the SIN 51-nt CSE is not a part of the core promoter for either plus- or minus-strand SIN genome synthesis in mammalian cells (7). It served as a replication enhancer, and multiple mutations in this fragment that destabilized the secondary structure did not abolish the replication of SIN genome RNA, but downregulated RNA synthesis between 5- and 10-fold. Accordingly, some decreases in replication rates and final titers of mutant viruses were detected both after infection of mice and in some cell lines (9). However, the detected differences were within the 10-fold range. Similarly, the clustered silent mutations in the 51-nt CSE of the Venezuelan equine encephalitis virus genome also do not significantly attenuate the virus (Scott Weaver, personal communication).
Previously, it was shown that point mutations that destabilize the predicted secondary structure of the 51-nt CSE downregulate SIN replication more strongly in C636 cells than in mammalian cells (27). In agreement with these data, we demonstrated here that clustered mutations in the SIN 51-nt CSE had a deleterious effect on virus replication in mosquito C710 cells. The defect was not on the level of viral particle formation but was in the replication of SIN-specific RNAs, because the SINrep43/GFP/Pac replicon with a mutated 51-nt CSE was capable of making cells resistant to puromycin, but produced a very low level of GFP. In the case of viral SIN43 RNA, this replication level was not sufficient to develop a productive infection, and the release of the SIN43 virus from C710 cells was below 1 PFU/cell (compared to the 1 × 104 to 5 × 104 PFU/cell production observed for SINwt in this cell line). Accordingly, the mutant was also incapable of causing CPE or plaque formation. These data strongly suggested that the integrity of the sequence located downstream of the SIN 5′ UTR (which includes the 51-nt CSE) was critical for SIN RNA replication in C710 cells.
The presence of multiple mutations in the 51-nt CSE of SIN43 essentially eliminated the occurrence of true revertants. However, variants capable of efficient growth in C710 cells were generated. These viruses accumulated adaptive mutations in the amino-terminal part of nsP2 (E118→K), in nsP3 (T286→I), and in the 5′ UTR. In other independent selection experiments, all of the randomly picked pseudorevertants of the SIN43 virus with increased replication efficiencies in C710 cells also contained mutations in the amino-terminal part of the nsP2 (E212→G, E155→A, or E131→K) (data not shown). The exact role of these mutations was not sufficiently characterized. However, we can hypothesize that changes in the amino-terminal fragment of nsP2, which is not conserved among alphaviruses and has not been shown before to have any particular functions, were essential for the replication of SIN43 RNAs in mosquito cells.
During the last few years, the number of known nsP2 activities has been continuously growing. This protein acts as an RNA helicase and has ATPase, GTPase, and RNA 5′-triphosphatase activity (11, 37, 47). Moreover, the carboxy-terminal domain of nsP2 possesses the proteinase activity (5, 13) that orchestrates the sequential processing of the nonstructural polyprotein, which is required for the switch of the activity of the replicative complex from the minus to the plus strand and subgenomic RNA synthesis (19, 41). nsP2 appears to be also directly involved in the initiation of subgenomic RNA synthesis (31). In addition, alphavirus nsP2 is produced in a 10- to 20-fold excess compared to nsP4, the catalytic subunit of the RdRp, and is equally distributed all over the cell, including the nucleus (38). The data presented in this paper suggest that nsP2 is also involved in the binding of the replicative complex to SIN RNA, and the mutations in the fragment that includes the 51-nt CSE require adaptive mutations in the nsP2 coding sequence in order to promote virus replication in mosquito cells.
The E118→K mutation in nsP2 alone was not sufficient to make the SIN43 mutant cytopathic for C710 cells. The additional change in nsP3 (T286→I) had a strong synergistic effect on RNA replication, the synthesis of viral structural proteins, and the development of CPE (and plaque formation), despite its effect on SIN43 replication in the absence of the nsP2 E118→K substitution being nearly undetectable.
The replacement of nt 1 to 9 in the 5′ UTR of some pseudorevertants with nt 1 to 52 of the subgenomic RNA was a second way to strongly enhance the effect of the nsP2 E118→K mutation. The addition of nt 1 to 52 to the 5′ end of SIN43 (SIN43/26SUTR) made the virus replicate more efficiently. However, together the mutations in nsP2 and the 5′ UTR increased the replication of SIN43 (SIN43/26SUTRnsP2 variant) in C710 cells to a level comparable to that of SINwt, suggesting that nsP2 may play a role in the proper positioning of the RdRp during replication of the genome with a new 5′ UTR. Previously, the presence of the 26S UTR was detected in one of the naturally occurring SIN DI RNAs (46), but the overall sequence and the computer-predicted secondary structure of the 5′ end of that DI RNA were very different from those found for SIN43/1/2 and SIN43/1/3. In addition, that DI RNA was generated in mammalian cells, and the complete data about the sequence of the nonstructural proteins of helper SIN are unavailable.
The results of the present work correlate with the previously published hypothesis that the activity of the promoter elements located in the 5′ end of the SIN genome and, particularly, in the 3′ end of the minus-strand intermediate depends on binding of the protein factors of both cellular and viral origin (32, 33). It was shown that the 5′-terminal 425 nt of the plus-strand RNA bind proteins that are essential for minus-strand RNA synthesis (7), and the works of Pardigon et al. unambiguously demonstrated that the 3′-terminal 250 nt of the SIN negative-strand RNA contain at least four binding sites for the mosquito homolog of the La autoantigen (32, 34). The clustered mutations present in the original SIN43 genome strongly changed the sequences of binding domains 2 and 3. Binding domain 1 in the SIN/43/1/2 variant also contained a replacement of the 3′-terminal nt 1 to 9, the most critical for La autoantigen binding. In spite of these extensive changes, SIN could adapt for replication in C710 cells by changing two amino acids (nsP2 E118→K and nsP3 T286→I) or by mutating the same amino acid in nsP2 when nt 1 to 52 of the SIN 26S RNA were inserted into the 5′ UTR. Those mutations functioned in cell- and virus-specific modes. They increased the replication of SIN43, but not SINwt, and the profound stimulatory effect was observed only in mosquito cells, not in mammalian cells. Thus, the adaptive mutations were critical only if the 51-nt CSE was significantly altered and, most likely, incapable of binding one or more copies of the mosquito cell-specific protein involved in RdRp formation. We can speculate that nsP2 is a protein factor that binds to a host protein(s) that is essential for the assembly of the replicative complexes on SIN RNA. The elimination of a number of protein-binding sites in the 3′ end of the minus-strand RNA and/or in the 5′ end of the viral genome might lead to selection of the mutated form of nsP2 with an increased affinity for the residual C710-specific protein factors (interacting, for instance, with domain 4 located between nt 190 and 250 in the 3′ end of the genome intermediate). Alternatively, it is also possible that mutations in the 51-nt CSE require a change in the positioning of the nsP2 in RdRp that is achieved through its conformational change and the replacement of the 5′ UTR or the nsP3 mutation. Viral genomes (or their minus-strand counterparts) with the wt sequence of the 51-nt CSE bind a complete set of protein factors, and as a result, the introduced adaptive mutations had no stimulatory effect on the replication of SINwt. In the case of SINwt/nsP23, the mutations even downregulated virus replication in C710 cells. This was not surprising, because any advantageous mutation in the nsPs could undoubtedly be selected during previous SIN evolution under natural conditions, and our modifications altered the structure of wt RdRp. The discovered mutations also did not promote SINwt replication in mammalian BHK-21 cells, and the replication of SIN43 viruses with a mutated 5′ UTR (SIN43/26SUTR and SIN43/26SUTRnsP2) in this cell line was strongly affected. Taken together, the data suggest that (i) the 26S 5′UTR is highly beneficial for virus replication only in mosquito cells, in the presence of mosquito cell-specific protein factors that likely bind to this sequence, and (ii) mutated forms of nsP2 and nsP3 promote the replication of viral genomes only with a mutated 51-nt CSE in mosquito cells. Our understanding of the structure of the SIN replicative complex remains incomplete, and based on the available data, it is difficult to interpret the nsP2-nsP3 interaction, or particularly, the costimulatory effects of the nsP2 mutation and the replacement of the 5′ terminus of the SIN43 genome. However, the new 51-nt CSE mutants and selected pseudorevertants open up the possibility for us to analyze the spectrum of cellular proteins binding to the positive and negative strands of the SIN 5′ end, but not to the SIN43 derivative, and to identify the host factor(s) that is critical for RNA replication. The study of multiple protein-protein and RNA-protein interactions, another element of our research, is now in progress.
In conclusion, we have shown that a SIN virus with a mutated sequence located downstream of the 5′ UTR and including the 51-nt CSE replicates efficiently in mammalian cells and is incapable of productive replication in cells of a mosquito origin. This virus accumulates adaptive mutations in the genome fragment encoding the nonstructural proteins. The mutations in the amino-terminal fragment of the nsP2 gene appear to be most important for promoting SIN43 genome replication in C710 cells. However, a mutation in nsP3 or the replacement of the 5′ terminus of the genomic 5′ UTR with the 5′ UTR of the subgenomic 26S RNA has additional stimulatory effects. The accumulated mutations function in cell- and virus-specific manners. They are critical only for SIN43 replication in mosquito cells and can have a deleterious effect on the replication of viruses with a mutated 51-nt CSE in mammalian cells. Besides enhancing our understanding of the 51-nt CSE, the results have possible practical applications. They suggest the possibility of developing attenuated vaccine strains of alphaviruses with a restricted host range.
Acknowledgments
We thank Scott Weaver for his critical reading of the manuscript.
This work was supported by Public Health Service grant AI50537.
REFERENCES
- 1.Agapov, E. V., I. Frolov, B. D. Lindenbach, B. M. Prägai, S. Schlesinger, and C. M. Rice. 1998. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc. Natl. Acad. Sci. USA 95:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, D. T., and L. D. Condreay. 1986. Replication of alphaviruses in mosquito cells, p. 171-207. In S. Schlesinger and M. J. Schlesinger (ed.), The Togaviridae and Flaviviridae. Plenum Press, New York, N.Y.
- 3.Chamberlain, R. W., and W. D. Sudia. 1961. Mechanisms of transmission of viruses by mosquitoes. Annu. Rev. Entomol. 61:371-390. [DOI] [PubMed] [Google Scholar]
- 4.DeTulleo, L., and T. Kirchhausen. 1998. The clathrin endocytic pathway in viral infection. EMBO J. 17:4585-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding, M., and M. J. Schlesinger. 1989. Evidence that Sindbis virus nsP2 is an autoprotease which processes the virus nonstructural polyprotein. Virology 171:280-284. [DOI] [PubMed] [Google Scholar]
- 6.Frolov, I., E. Agapov, T. A. Hoffman, Jr., B. M. Prágai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frolov, I., R. Hardy, and C. M. Rice. 2001. cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frolov, I., and S. Schlesinger. 1994. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J. Virol. 68:1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frolova, E. I., R. Z. Fayzulin, S. H. Cook, D. E. Griffin, C. M. Rice, and I. Frolov. 2002. Roles of nonstructural protein nsP2 and alpha/beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76:11254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glomb-Reinmund, S., and M. Kielian. 1998. The role of low pH and disulfide shuffling in the entry and fusion of Semliki Forest virus and Sindbis virus. Virology 248:372-381. [DOI] [PubMed] [Google Scholar]
- 11.Gomez de Cedron, M., N. Ehsani, M. L. Mikkola, J. A. Garcia, and L. Kaariainen. 1999. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 448:19-22. [DOI] [PubMed] [Google Scholar]
- 12.Griffin, D. E. 2001. Alphaviruses, p. 917-962. In D. M. Knipe et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 13.Hardy, W. R., and J. H. Strauss. 1989. Processing the nonstructural proteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and trans. J. Virol. 63:4653-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston, R. E., and C. J. Peters. 1996. Alphaviruses, p. 843-898. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 15.Kinney, R. M., B. J. B. Johnson, J. B. Welch, K. R. Tsuchiya, and D. W. Trent. 1989. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology 170:19-30. [DOI] [PubMed] [Google Scholar]
- 16.Lehtovaara, P., H. Söderlund, S. Keränen, R. F. Pettersson, and L. Kääriäinen. 1982. Extreme ends of the genome are conserved and rearranged in the defective interfering RNAs of Semliki Forest virus. J. Mol. Biol. 156:731-748. [DOI] [PubMed] [Google Scholar]
- 17.Lemm, J. A., and C. M. Rice. 1993. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J. Virol. 67:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemm, J. A., and C. M. Rice. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemm, J. A., T. Rümenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine, B., Q. Huang, J. T. Isaacs, J. C. Reed, D. E. Griffin, and J. M. Hardwick. 1993. Conversion of lytic to persistent alphavirus infection by the bcl-2 cellular oncogene. Nature 361:739-742. [DOI] [PubMed] [Google Scholar]
- 21.Levinson, R. S., J. H. Strauss, and E. G. Strauss. 1990. Complete sequence of the genomic RNA of O'nyong-nyong virus and its use in the construction of alphavirus phylogenetic trees. Virology 175:110-123. [DOI] [PubMed] [Google Scholar]
- 22.Levis, R., S. Schlesinger, and H. V. Huang. 1990. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J. Virol. 64:1726-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monroe, S. S., J.-H. Ou, C. M. Rice, S. Schlesinger, E. G. Strauss, and J. H. Strauss. 1982. Sequence analysis of cDNAs derived from Sindbis virions and of defective interfering particles. J. Virol. 41:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monroe, S. S., and S. Schlesinger. 1984. Common and distinct regions of defective-interfering RNAs of Sindbis virus. J. Virol. 49:865-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niesters, H. G. M., and J. H. Strauss. 1990. Defined mutations in the 5′ nontranslated sequence of Sindbis virus RNA. J. Virol. 64:4162-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niesters, H. G. M., and J. H. Strauss. 1990. Mutagenesis of the conserved 51-nucleotide region of Sindbis virus. J. Virol. 64:1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou, J. H., C. M. Rice, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1982. Sequence studies of several alphavirus genomic RNAs in the region containing the start of the subgenomic RNA. Proc. Natl. Acad. Sci. USA 79:5235-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou, J. H., E. G. Strauss, and J. H. Strauss. 1983. The 5′-terminal sequences of the genomic RNAs of several alphaviruses. J. Mol. Biol. 168:1-15. [DOI] [PubMed] [Google Scholar]
- 30.Ou, J. H., E. G. Strauss, and J. H. Strauss. 1981. Comparative studies of the 3′-terminal sequences of several alpha virus RNAs. Virology 109:281-289. [DOI] [PubMed] [Google Scholar]
- 31.Paessler, S., R. Z. Fayzulin, M. Anishchenko, I. P. Greene, S. C. Weaver, and I. Frolov. 2003. Recombinant Sindbis/Venezuelan equine encephalitis virus is highly attenuated and immunogenic. J. Virol. 77:9278-9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pardigon, N., E. Lenches, and J. H. Strauss. 1993. Multiple binding sites for cellular proteins in the 3′ end of Sindbis alphavirus minus-sense RNA. J. Virol. 67:5003-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pardigon, N., and J. H. Strauss. 1992. Cellular proteins bind to the 3′ end of Sindbis virus minus-strand RNA. J. Virol. 66:1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pardigon, N., and J. H. Strauss. 1996. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J. Virol. 70:1173-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice, C. M., and J. H. Strauss. 1981. Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc. Natl. Acad. Sci. USA 78:2062-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rikkonen, M., J. Peranen, and L. Kaariainen. 1994. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2. J. Virol. 68:5804-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rikkonen, M., J. Peranen, and L. Kaariainen. 1994. Nuclear targeting of Semliki Forest virus nsP2. Arch. Virol. 9(Suppl.):369-377. [DOI] [PubMed] [Google Scholar]
- 39.Rumenapf, T., E. G. Strauss, and J. H. Strauss. 1995. Aura virus is a New World representative of Sindbis-like viruses. Virology 208:621-633. [DOI] [PubMed] [Google Scholar]
- 40.Shirako, Y., and J. H. Strauss. 1990. Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123. Virology 177:54-64. [DOI] [PubMed] [Google Scholar]
- 41.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 185:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh, I., and A. Helenius. 1992. Role of ribosomes in Semliki Forest virus nucleocapsid uncoating. J. Virol. 66:7049-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92-110. [DOI] [PubMed] [Google Scholar]
- 44.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takkinen, K. 1986. Complete nucleotide sequence of the non-structural protein genes of Semliki Forest virus. Nucleic Acids Res. 14:5667-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsiang, M., S. S. Monroe, and S. Schlesinger. 1985. Studies of defective interfering RNAs of Sindbis virus with and without tRNAAsp sequences at their 5′ termini. J. Virol. 54:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasiljeva, L., A. Merits, P. Auvinen, and L. Kaariainen. 2000. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J. Biol. Chem. 275:17281-17287. [DOI] [PubMed] [Google Scholar]
- 48.Weaver, S. C., A. Hagenbaugh, L. A. Bellew, S. V. Netesov, V. E. Volchkov, G. J. Chang, D. K. Clarke, L. Gousset, T. W. Scott, D. W. Trent, et al. 1993. A comparison of the nucleotide sequences of Eastern and Western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses. Virology 197:375-390. [DOI] [PubMed] [Google Scholar]
- 49.Wengler, G., and C. Gros. 1996. Analyses of the role of structural changes in the regulation of uncoating and assembly of alphavirus cores. Virology 222:123-132. [DOI] [PubMed] [Google Scholar]