Abstract
The atypical Nef protein (NefF12) from human immunodeficiency virus type 1 strain F12 (HIV-1F12) interferes with virion production and infectivity via a mysterious mechanism. The correlation of these effects with the unusual perinuclear subcellular localization of NefF12 suggested that the wild-type Nef protein could bind to assembly intermediates in late stages of viral replication. To test this hypothesis, Nef from HIV-1NL4-3 was fused to an endoplasmic reticulum (ER) retention signal (NefKKXX). This mutant NefKKXX protein recapitulated fully the effects of NefF12 on Gag processing and virion production, either alone or as a CD8 fusion protein. Importantly, the mutant NefKKXX protein also localized to the intermediate compartment, between the ER and the trans-Golgi network. Furthermore, Nef bound the GagPol polyprotein in vitro and in vivo. This binding mapped to the C-terminal flexible loop in Nef and the transframe p6* protein in GagPol. The significance of this interaction was demonstrated by a genetic assay in which the release of a mutant HIV-1 provirus lacking the PTAP motif in the late domain that no longer binds Tsg101 was rescued by a Nef.Tsg101 chimera. Importantly, this rescue as well as incorporation of Nef into HIV-1 virions correlated with the ability of Nef to interact with GagPol. Our data demonstrate that the retention of Nef in the intermediate compartment interferes with viral replication and suggest a new role for Nef in the production of HIV-1.
Human immunodeficiency virus type 1 (HIV-1), the causative agent of AIDS, encodes 16 distinct proteins that are expressed differentially during the viral replicative cycle. Initially, the early regulatory proteins Tat and Rev and the so-called negative effector (Nef) are translated from multiply spliced mRNA species. During late phases, singly spliced or unspliced transcripts direct the expression of viral accessory proteins (viral proteins R and U [Vpr and Vpu] and viral infectivity factor [Vif]), as well as structural group-specific antigen (Gag), Gag and polymerase (GagPol), and envelope (Env) polyproteins (see reference 20 and references therein).
The expression of Gag and GagPol is regulated tightly by ribosomal frameshifting that enables the precursor polyproteins to be expressed from the same unspliced genomic mRNA (25). Frameshifting is promoted by a “slippery” sequence (U UUU UUA), which occurs at the junction between nucleocapsid (NC) and the spacer peptide (p1) of Gag and ensures that only 5% of the transcripts give rise to GagPol (1, 5). The 55-kDa Gag polypeptide contains matrix (MA), capsid (CA), NC, and the viral late domain (p6), in addition to the spacer peptides p1 and p2 (MA-CA-p2-NC-p1-p6). The 160-kDa GagPol precursor consists of MA, CA, p2, and NC, followed by the pol-encoded transframe protein (p6*) and protease (PR), reverse transcriptase (RT), and integrase (IN) (see references 3 and 44 and references therein). PR cleaves Gag and GagPol into their structural and enzymatic subunits during the late stages of viral budding, thereby ensuring maturation to infectious particles (49).
Nef from HIV-1 is a 27- to 32-kDa viral accessory protein that participates in numerous processes during the viral replicative cycle. It is also incorporated into virions, where Nef is cleaved by PR (39, 51). Initially, Nef was thought to decrease viral replication in vitro. Subsequently, because it supports high levels of viremia and progression to AIDS as well as induces an AIDS-like disease in transgenic mice, Nef was defined as a key factor for the pathogenesis of primate lentiviruses (9, 10, 22, 24).
Functions of Nef include the removal of CD4 and the major histocompatibility complex class I molecules from the surface of infected cells, thereby facilitating viral replication and immune evasion, respectively (20). Moreover, Nef activates signaling cascades that increase levels of viral replication in primary T cells (11, 14, 44). Finally, Nef increases levels of Gag in detergent-resistant microdomains (lipid rafts) at the plasma membrane to enhance the infectivity of progeny virions (53). Another important clue for a role of Nef during virion production came from the finding that the dominant negative Nef protein from HIV-1F12 (NefF12) interferes with Gag processing, production, and infectivity of other HIV-1 strains in trans (8, 12, 15, 16, 35). HIV-1F12 was cloned as a provirus from chronically infected HUT78 cells that produced no virions (16). Not only was NefF12 primarily responsible for this phenotype of HIV-1F12, but it could inhibit the production of other strains of HIV-1 (8, 12, 15, 35). In those studies, an unusual localization of NefF12 to a perinuclear compartment correlated with its interfering phenotype. Together, those studies suggested that Nef might interact with viral structural components during the assembly of HIV-1.
In this study, by placing an endoplasmic reticulum (ER) retention signal at its C terminus, we forced the retention of a common Nef protein within the biosynthetic pathway. This perinuclear retention was instrumental for subsequent inhibitory effects of Nef on Gag processing and virion production, which correlated with the binding between the C-terminal flexible loop of Nef and p6* from GagPol. Additionally, the mutant Nef protein lacking the flexible loop was no longer incorporated into viral particles. Finally, a trans-complementation assay for virus release provided genetic evidence for the interaction between Nef and GagPol. We conclude that Nef binds viral structural components during the biosynthesis of progeny virions in the producer cell.
MATERIALS AND METHODS
Antibodies.
Monoclonal antihemagglutinin (anti-HA) epitope (F7) (Santa Cruz Biotechnology, Santa Cruz, Calif.), monoclonal anti-Nef (52), mouse anti-p24Gag (AG3.0) (referred to as anti-CA throughout), monoclonal anti-RT (NEA-9304) (New England Nuclear, Boston, Mass.), and polyclonal anti-Rab6 (C-19) (Santa Cruz Biotechnology) antibodies were used as first antibodies to detect HA epitope-tagged proteins, Nef, Gag, GagPol, and Rab6, respectively. Secondary horseradish peroxidase-conjugated anti-mouse antibody (Santa Cruz Biotechnology) was detected by enhanced chemiluminescence (Amersham Biosciences, Evanston, Ill.). Secondary antibodies for immunofluorescence were from Molecular Probes (Eugene, Oreg.). For immunoprecipitation assays, rabbit anti-p24Gag polyclonal (ABI) (referred as anti-CApoly throughout), and rabbit polyclonal anti-HA (Y11) (Santa Cruz Biotechnology) antibodies were used. The fluorescein isothiocyanate-conjugated anti-CD8 antibody used for immunofluorescence was purchased from Becton Dickinson (San Jose, Calif.).
Plasmid constructions.
Plasmid DNAs encoding replication-competent HIV-1 proviruses were from HIV-1NL4-3 (1). The variant NL4-3ΔNef with nef deleted was generously provided by John Guatelli, University of California, San Diego. Expression plasmids for mutant Nef proteins were generated by inserting the nef genes into the pEF-BOS vector. The nefF12 gene was derived from the HIV-1F12 isolate (16), while NefKKXX was generated by amplifying the nef gene from the NL4-3 provirus with a reverse primer encoding the KKMP sequence at the 3′ end. The Nef expression plasmid was generated from the NL4-3 provirus and inserted into the pcDNA3.1D (Invitrogen, Carlsbad, Calif.) plasmid at the TOPO site. This plasmid was used to derive an expression construct for NefΔFL (Nef with a deletion in the flexible loop, amino acids 148 to 180) by standard mutagenesis techniques. The expression plasmid for NefΔFLKKXX was generated by amplifying the nef gene from pcDNA3.1D NefΔFL with a reverse primer encoding the KKMP sequence at the 3′ end. Expression plasmids for hybrid CD8.Nef proteins were generated by inserting nef genes into the EF-BOS plasmid with the extracellular and transmembrane portion of CD8 (28). The expression plasmid for CD8.NefKKXX was derived from HIV-1NL4-3. GST.Nef and mutant GST.Nef1-60, GST.Nef55-210, and GST.HIVNefΔFL hybrid proteins were derived from HIV-1SF2. nef sequences were amplified by PCR with BamHI (5′) and EcoRI (3′) restriction sites and inserted into the pGEX-4T1 vector (Pharmacia, Piscataway, N.J.). For in vitro translation, GagPol and its truncated derivatives were generated by PCR from HIV-1HxB2 proviral DNA with a frameshift mutation between Gag and GagPol, to ensure the synthesis of GagPol and no Gag, and a point mutation in the active site of PR (40). Gag was amplified from HIV-1NL4-3. All sequences were introduced into the T7-plink2 vector (28).
To generate the GagPol provirus, a single point mutation (aspartic acid at position 25 to alanine) was introduced in the PR active site of HIV-1NL4-3 with a frameshift mutation that bypassed the p1 and p6. The GagPR provirus was generated as before, with the addition of a termination codon at the end of the PR gene. For the generation of Gag/GagPol provirus, the same PR-inactivating mutation was inserted in HIV-1NL4-3. Gag and GagΔNef proviruses were generated by introducing an early termination codon into the pol frames of HIV-1NL4-3 and HIV-1NL4-3Δnef, respectively. All mutations were introduced into the HIV-1NL4-3 backbone by standard PCR mutagenesis. Expression plasmids for HA epitope-tagged p6* and p6 were generated by inserting the p6* and p6 genes from HIV-1NL4-3 into the pEF-BOS/HA plasmid.
The plasmids pNL−, pNLHXB, pENX, pENX.p6, and pENX.Tsg101 were generous gifts from Paul Bieniasz (The Rockefeller University, New York, N.Y.) and are described elsewhere (32). To generate the expression plasmid for the hybrid Nef.HA.Tsg101, Nef.Tsg101.V5, and NefΔFL.Tsg101.V5 proteins, nef genes from HIV-1NL4-3 were inserted into the pEF-BOS or the pcDNA3.1D vector. The HA epitope tag was placed at the 3′ end of the nef gene, followed by the cellular gene Tsg101. In another scenario, the nef gene was followed by the cellular gene Tsg101 and the V5 tag was placed at the 3′ end.
Cells and transfections.
293T, HeLa, and NIH 3T3 cells were grown in Dulbecco modified minimal essential medium supplemented with 10% fetal calf serum and streptomycin-penicillin. Transfections were performed with Lipofectamine (Gibco BRL, Rockville, Md.) according to the manufacturer's instructions.
Virus production and Gag processing.
To assess the effects of Nef during virion production, 293T cells were transfected with proviral DNA and Nef expression plasmids at 1:1 molar ratio. At 48 h posttransfection, cells and cell culture supernatants were harvested. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris [pH 7.2], 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]), and cleared supernatants were analyzed by Western blotting for Nef and Gag. Culture supernatants were cleared through a 45-μm-pore-size filter (Millipore, Bedford, Mass.) and stored at −80°C or used directly in infectivity assays. Quantification of virion production was performed by p24Gag (CA) enzyme-linked immunosorbent assay (ELISA) capture assays (NEN Life Science Products-Perkin-Elmer, Boston, Mass.) according to the manufacturer's instructions.
Isolation of virions and VLPs.
Culture supernatants, collected at 48 h after transfection, were clarified by low-speed centrifugation, followed by ultracentrifugation through a 20% sucrose cushion at 100,000 × g for 1.5 h. Pellets were suspended in 1× phosphate-buffered saline (PBS) overnight at 4°C. Viruses and virus-like particles (VLPs) were lysed in SDS loading buffer, and viral protein content was analyzed by Western blotting.
Subcellular localization.
The subcellular localization of CD8.Nef fusion proteins was analyzed as described previously (12). Briefly, NIH 3T3 cells were plated on coverslips overnight and subsequently transfected with 1 μg of plasmid DNA. At 36 h posttransfection, cells were fixed in 3.7% paraformaldehyde for 5 min at room temperature, washed in PBS, and then permeabilized with 0.1% Triton X-100 in PBS for 3 min at room temperature. After the cells were washed in PBS, they were blocked in PBS-1% bovine serum albumin for 30 min at room temperature and then incubated with the appropriate first and secondary antibodies. After extensive washing in PBS, the coverslips were mounted in HistoGel (Linaris, Bettingen, Germany) and analyzed by using a Leica DM IRBE confocal laser scanning microscope with a 63× oil objective. Pictures of individual optical sections were processed with Adobe PhotoShop software.
Protein purification, in vitro translation, and glutathione S-transferase (GST) pull-down assays.
GST.Nef hybrid proteins were expressed in Escherichia coli strain DH5α and purified by using glutathione-Sepharose beads (Pharmacia) with a modified lysis buffer (50 mM HEPES [pH 7.8], 100 mM KCl, 1% Triton X-100, 2 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, and 1 mg of lysozyme per ml). The purity of GST.Nef hybrid proteins was verified by Coomassie blue staining after SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and their concentration was determined with a protein assay kit (Bio-Rad, Hercules, Calif.). 35S-labeled Gag, GagPol, and its truncated derivatives were transcribed and translated according to the manufacturer's instructions, using the rabbit reticulocyte system (TNT; Promega, Madison, Wis.). The quality of translated proteins was verified by SDS-PAGE and autoradiography.
For in vitro binding assays, 0.5 μg of immobilized GST or hybrid GST.Nef proteins was incubated with 5 μl of 35S-labeled proteins for 4 h at 4°C in 750 μlof CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}buffer (50 mM Tris-HCl [pH 7.4], 0.05 mM EDTA, 10 mM CHAPS, and protease inhibitors). The beads were then washed five times in the same buffer and subjected to SDS-PAGE and autoradiography.
Coimmunoprecipitations.
293T cells were transfected with 1 μg of each proviral clone, i.e., Gag, Gag/GagPol, GagPol, or GagPR. At 48 h after the transfection, cells were washed in PBS and lysed in RIPA buffer for 30 min at 4°C. Clarified lysates from Gag- or GagPR-expressing cells were incubated for 4 h at 4°C with anti-CApoly antibody, followed by incubation with protein A-Sepharose (Amersham-Pharmacia, Uppsala, Sweden) for 90 min at 4°C. For Gag/GagPol and GagPol clarified lysates, anti-RT antibody was used. Immunocomplexes were then precipitated and washed three times in RIPA buffer containing 150 mM NaCl and two times in RIPA buffer containing 500 mM NaCl. Precipitates were resuspended in SDS sample buffer, boiled for 5 min, and resolved by SDS-PAGE, followed by Western blotting with anti-Nef antibody. For 293T cells coexpressing p6* or p6 together with Nef, immunoprecipitations were performed with the anti-HA polyclonal antibody.
trans-complementation assay for virus release.
HeLa cells were transfected with 1.6 μg of pL− along with 0.4 μg of pENX and pENX-derived plasmids. For cotransfection of pL− and Nef.Tsg101 hybrid proteins, a 2:1 molar ratio was used. Culture supernatants were collected at 48 h after transfection and clarified by low-speed centrifugation, followed by ultracentrifugation through a 20% sucrose cushion at 100,000 × g for 1.5 h. Viral protein contents in cell and particle lysates were analyzed by Western blotting. Aliquots from viral particle preparations were assayed for RT activity, as described previously (12).
RESULTS
Mutant CD8.NefKKXX and NefF12 chimeras localize to the intermediate compartment.
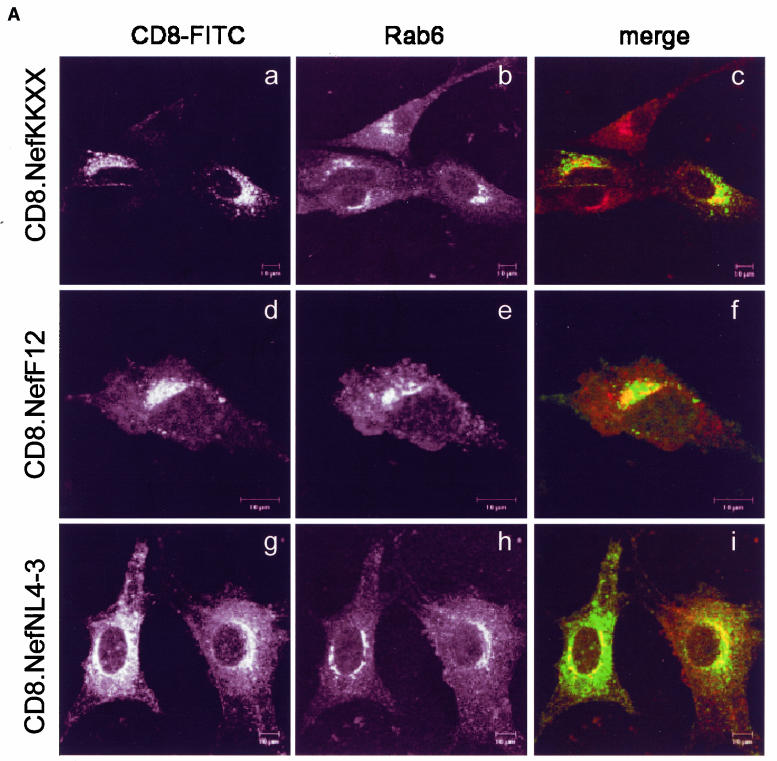
Previously, we and others have shown that the interfering action of NefF12 on Gag processing, virion production, and infectivity correlated with its unusual perinuclear localization (12, 35). To test the hypothesis that this localization is mechanistically involved in the F12 phenotype, a KKXX sorting motif (lysine, lysine, methionine, and proline [KKMP]) was inserted at the C terminus of a CD8.NefNL4-3 hybrid protein (CD8.NefKKXX). KKXX motifs bind the β subunit of the COP-I coatomer and mediate the retrieval of cargo from later compartments back to the ER during biosynthetic transport (7, 26). We analyzed whether the addition of the KKXX motif to Nef induced a subcellular localization similar to that of the CD8.NefF12 chimera in NIH 3T3 cells by using confocal microscopy. As expected (12, 35), the hybrid CD8.F12Nef protein was concentrated in perinuclear areas of the cell (Fig. 1A, panel d), while the wild-type CD8.NefNL4-3 chimera was found throughout the cytoplasm and at the cell membrane (Fig. 1A, panel g). Importantly, the subcellular distribution of the mutant CD8.NefKKXX chimera resembled closely that of the CD8.NefF12 fusion protein and was clearly distinct from wild-type hybrid CD8.NefNL4-3 protein (Fig. 1A, panel a), suggesting that the addition of the KKXX motif resulted in the targeting of the hybrid CD8.Nef and CD8.NefF12 proteins to the same compartment. To characterize this compartment, costaining with markers for the ER (not shown) and the trans-Golgi network (TGN) (Rab6) was performed. As expected, a perinuclear subpopulation of the CD8.NefNL4-3 chimera localized partially to the TGN (Fig. 1A, panel h [merged in panel i]), with no significant overlap with markers for the ER in the cytoplasm (data not shown). Surprisingly, CD8.NefF12 and mutant CD8.NefKKXX chimeras colocalized only partially with Rab6 (Fig. 1A, panels e [merged in panel f] and b [merged in panel c], respectively) and were not detected at ER structures (data not shown). This pattern is reminiscent of that described in a recent report, where a CD8.E19 chimera that contained the KKMP motif at the C terminus was localized mostly to the intermediate compartment and the TGN (27, 45). We conclude that the mutant CD8.NefKKXX and the CD8.NefF12 chimeras both localized to the intermediate compartment in cells.
FIG. 1.
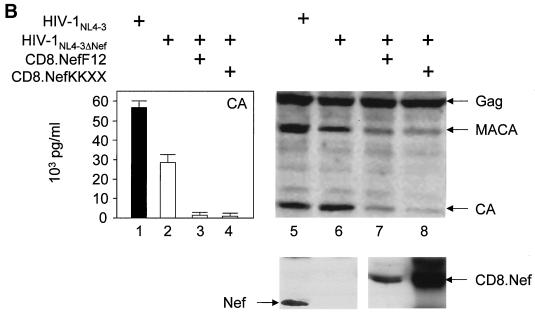
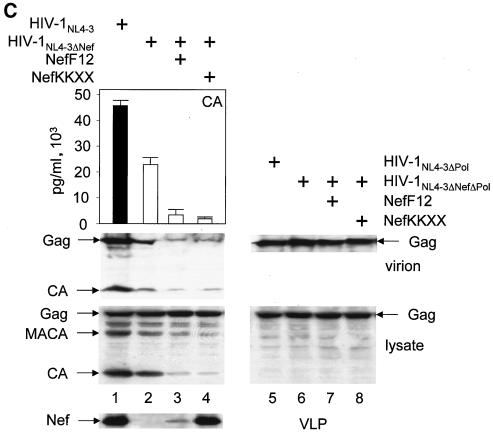
(A) The mutant CD8.NefKKXX chimera localizes to the intermediate compartment between the ER and TGN as determined by confocal microscopy. Shown is the immunostaining of NIH 3T3 cells, which expressed the indicated proteins. The subcellular distribution of expressed proteins and of the TGN was visualized with fluorescein isothiocyanate-conjugated anti-CD8 (green staining but left black and white for better resolution) and anti-Rab6 (red staining but left black and white for better resolution) antibodies, respectively. The right panels show the merge of the two pictures on the left. Cells expressed the hybrid mutant CD8.NefKKXX (a to c), the hybrid CD8.NefF12 (d to f), and the hybrid CD8.NefNL4-3 (g to i) proteins. (B) The hybrid mutant CD8.NefKKXX protein interferes with virion production and Gag processing in 293T cells. The mutant CD8.NefKKXX chimera inhibits the production of HIV-1. Left panel, virion production from 293T cells, which expressed HIV-1NL4-3 (bar 1) or HIV-1NL4-3ΔNef (bar 2) or coexpressed HIV-1NL4-3ΔNef and CD8.NefF12 (bar 3) or mutant CD8.NefKKXX (bar 4) chimeras, was measured by CA capture ELISA. Means and standard deviations from three independent experiments are shown. Right panels, Western blotting of these cell lysates was performed with anti-CA (top panel) and anti-Nef (bottom panel) antibodies. (C). Mutant NefKKXX protein inhibits Gag processing and virion production. 293T cells expressed HIV-1NL4-3 (bar 1) or mutant HIV-1NL4-3ΔNef (bar 2) viruses or coexpressed the mutant HIV-1NL4-3ΔNef virus and NefF12 (bar 3) or mutant NefKKXX (bar 4) proteins. Viral supernatants and cell lysates (lysate) were monitored for CA as described for panel B (left panels). Additionally, anti-CA Western blotting was performed on viral supernatants (virion). As controls, effects of NefF12 and NefKKXX on VLPs were analyzed (lanes 5 to 8). 293T cells expressed mutant HIV-1NL4-3ΔPol (lane 5) or HIV-1NL4-3ΔNef ΔPol (lane 6) viruses or coexpressed mutant HIV-1NL4-3ΔNef ΔPol virus and NefF12 (lane 7) or mutant NefKKXX (lane 8) proteins. Arrows indicate the Gag (55 kDa) and MACA (capsid, matrix) (41 kDa) precursors, the CA (24 kDa) and Nef (27 kDa) proteins, and the 57-kDa CD8.Nef chimera.
Addition of an ER retention signal creates an interfering Nef protein.
Next, we investigated whether the addition of the KKXX motif to Nef was sufficient for an F12 phenotype on virion production. Hybrid mutant CD8.NefKKXX or CD8.NefF12 proteins were coexpressed with HIV-1NL4-3 proviral DNA with nef deleted (HIV-1NL4-3ΔNef) in 293T cells, and virion production was monitored 2 days later. Quantification of virion production by CA capture ELISA revealed that the coexpression of both CD8.Nef chimeras resulted in more than a 10- to 30-fold reduction of virion production compared with the expression of HIV-1NL4-3ΔNef alone (Fig. 1B, left panel; compare bar 2 with bars 3 and 4). The expression of Gag was equivalent in these cells (Fig. 1B, right panel, compare lanes 5 to 8). This effect was specific for CD8.NefF12 and mutant CD8.NefKKXX chimeras and was not observed with the wild-type CD8.NefNL4-3 fusion protein (reference 12 and data not shown). Additionally, Western blotting of lysates of the virus-producing cells revealed that processing of the Gag precursor was severely affected in the presence of these mutant chimeras (Fig. 1B, right panel; compare MACA- and CA-related bands in lanes 5 and 6 with lanes 7 and 8). Thus, effects on Gag processing and virion production were equivalent. As a control for the KKXX motif, an AAMP sequence, which no longer binds to β-COP, was also placed at the C terminus of the CD8.Nef hybrid protein (CD8.NefAAXX). It had no effect on Gag processing and virion production (data not shown). Importantly, this block of Gag processing by the CD8.NefF12 chimera was identical to results reported by Fackler et al. (12). However, another study demonstrated that the presence of NefF12 in the backbone of HIV-1NL4-3 (HIV-1NL4-3NefF12) interfered only with processing of MACA (35). Since a single point mutation can reverse the phenotype of NefF12 (11) and since neither HIV-1F12 nor mutant HIV-1NL4-3NefF12 viruses replicate in cells (16, 35), this difference could be explained by the occurrence of back mutations in the nefF12 gene during viral propagation. We conclude that a mutant CD8.NefKKXX chimera can interfere with Gag processing and consequently virion production in a fashion similar to that of the CD8.NefF12 fusion protein.
Next, we examined whether mutant NefKKXX and NefF12 proteins without the N-terminal CD8 moiety could reproduce the same phenotype. The identical KKMP sequence was introduced at the C terminus of Nef from HIV-1NL4-3. To measure the effect of mutant Nef proteins on virion production more precisely, we first isolated viral particles and analyzed them by Western blotting. Indeed, both the mutant NefKKXX and NefF12 proteins inhibited virion production from the mutant HIV-1NL4-3ΔNef provirus (Fig. 1C, top panel; compare lane 2 with lanes 3 and 4). This effect correlated with the marked reduction of processing of the Gag precursor in the producer cells (Fig. 1C, left lower panels, lysate). Quantification of virion production by CA capture ELISA and anti-CA Western blotting demonstrated that the efficiency of this inhibition was only slightly decreased compared to that with the CD8.Nef chimeras (compare Fig. 1B and C, bars 2 with bars 3 and 4). This inhibition was from 5- to 15-fold as determined by ELISA and up to 20-fold as determined by Western blotting (Fig. 1C; compare lane 2 to lanes 3 and 4 in CA capture ELISA and Western blotting, virion). Again, the expression of Gag was equivalent in these cells (Fig. 1C, left lower panels, lysate). In sharp contrast, amounts of Gag were reduced in supernatants of cells expressing NefF12 and mutant NefKKXX proteins (Fig. 1C, right upper panel; compare lanes 1 and 2 to lanes 3 and 4, Gag, virion). Next, we confirmed that the mutant Nef proteins were interfering specifically with Gag processing and had no effect on Gag gene expression (Fig. 1C, left middle panel; compare Gag, MACA, and CA bands in lanes 1 and 2 with lanes 3 and 4, lysate). For this experiment, a mutant virus expressing only Gag and not GagPol, and therefore lacking the viral PR responsible for Gag processing, was coexpressed in 293T cells with the NefF12 or NefKKXX mutant proteins. Our results demonstrate that both NefF12 and mutant NefKKXX proteins had no effect on the expression of Gag in transfected cells or on the production of VLPs containing only Gag (Fig. 1C, right panels; compare lane 6 to lanes 7 and 8 for virion and lysate). In this experiment, a mutant HIV-1NL4-3 virus expressing only Gag (VLP) was used as the positive control (Fig. 1C, right panels, lane 5). Levels of Nef were similar to those in the previous experiment with HIV-1NL4-3 or mutant HIV-1NL4-3ΔNef with mutant CD8.NefF12 or CD8.NefKKXX chimeras (Fig. 1C, bottom panel). We conclude that effects of NefF12 and mutant NefKKXX proteins are specific for Gag processing and relate directly to the expression of GagPol.
Importantly, the effects of the mutant CD8.NefKKXX chimera and mutant NefKKXX proteins were even more pronounced than those of the CD8.NefF12 chimera or NefF12 (Fig. 1B [compare lanes 3 and 4 in the left panel to lanes 7 and 8 in the right panel] and C [compare bars 3 and 4 in the graph]), possibly due to its greater stability in 293T cells (Fig. 1B, bottom panel; compare lanes 7 and 8 to Fig. 1C, bottom panel, lanes 3 and 4, Western blotting for Nef). As we reported previously (12), NefF12 is very unstable in cells (Fig. 1C, bottom panel, lane 3). We conclude that the addition of a KKXX signal to the C terminus of Nef fully reconstitutes the NefF12 phenotype on Gag processing and virion production and that the retention of Nef during biosynthetic transport is critical for its inhibitory effects.
Nef binds GagPol in GST pull-down assays.
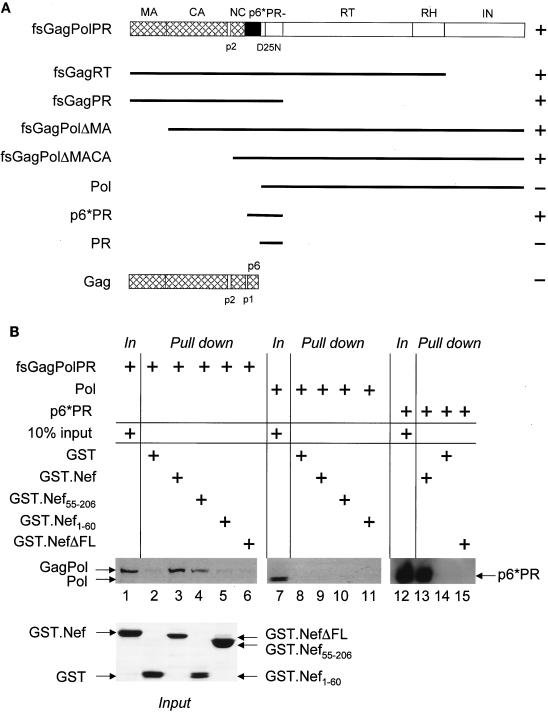
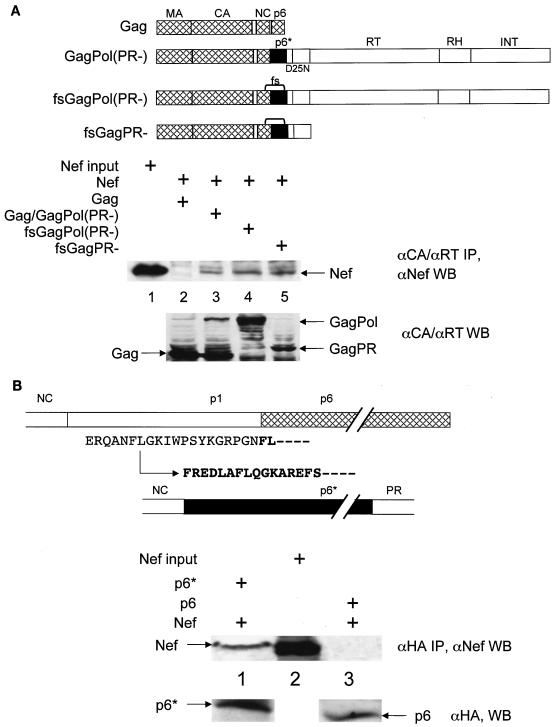
Since retention of the mutant CD8.NefKKXX and NefKKXX chimeras along the biosynthetic pathway was sufficient to inhibit processing of Gag and GagPol, we reasoned that the most likely mechanism for this inhibition would be physical targeting of one of these viral structural components by Nef. In initial experiments, a 160-kDa band was immunoprecipitated with the CD8.Nef fusion protein from metabolically labeled cells coexpressing it and the HIV-1NL4-3ΔNef proviral DNA. This band associated with Nef also in the absence of Env gp160, suggesting that it might represent the 160-kDa GagPol precursor (data not shown). To investigate this possibility, studies of binding between Nef and Gag or GagPol were performed. First, GST pull-down assays were performed with a GST.Nef fusion protein and 35S-labeled Gag, GagPol, and their truncated derivatives, which were transcribed and translated in vitro by using the rabbit reticulocyte lysate (Fig. 2A). As shown in Fig. 2B, none of the proteins bound GST alone (compare lanes 2, 8, and 14). Although we could not observe any binding between Gag and the GST.Nef chimera (Fig. 2A), it was clear that GagPol bound Nef (Fig. 2B, lane 3). Similar binding between GagPol and the hybrid GST.NefF12 protein was also observed (data not shown). To determine which regions in GagPol are important for its binding to Nef, several truncated GagPol proteins were synthesized. Figure 2A shows a summary of the mapping done with GagPol. First, IN and RT were dispensable for this binding to Nef. Likewise, Nef bound GagPol lacking MA and CA. In sharp contrast, Pol lacking the p6* region no longer bound Nef (Fig. 2B; compare lanes 9, 10, and 11 with lane 7). Finally, Nef retained the p6*PR fragment derived from GagPol (Fig. 2B; compare lanes 12 and 13). Since Nef did not bind PR alone, we conclude that Nef binds p6* from GagPol in vitro.
FIG. 2.
Nef binds GagPol and p6*PR polypeptides in GST pull-down assays. (A) Shown are the frameshifted, protease-negative GagPol (fsGagPolPR−) and Gag polyproteins and their truncated derivatives. MA, CA, NC, p6, PR, RT, RT, RNase H (RH), the spacer peptides p2 and p1, and p6* are shown. Note that p6 and p6* (frameshifted p6), which share no similarity in their sequences, are present only in Gag and GagPol, respectively. Black lines represent the truncated GagPol and Gag precursors. GagPol and its truncated derivatives contained an inactive protease (PR−). The plus and minus signs next to each polyprotein reflect its binding to the GST.Nef chimera (data not shown). (B) GST and GST.Nef fusion proteins were expressed in E. coli and purified from GST beads (bottom panel, Coomassie blue staining). They were incubated with 35S-labeled Gag, GagPol, or their truncated derivatives, which were transcribed and translated in vitro by using the rabbit reticulocyte lysate. Bound proteins were resolved by SDS-10% PAGE followed by autoradiography. Shown are three examples of GST pull-down assays. Arrows indicate the 160-kDa GagPol, the 115-kDa Pol, and the 17-kDa p6*PR polyproteins. GST was used as the negative control (lanes 2, 8, and 14).
To determine which region in Nef was required for this binding, its N- and C-terminal portions were expressed individually as GST fusion proteins (Fig. 2B, bottom panel, Coomassie blue staining of GST and GST.Nef fusion proteins). While there was no binding between GagPol and the N terminus of Nef (residues 1 to 60), the C terminus of Nef (residues 55 to 210) still bound GagPol (Fig. 2B; compare lanes 4 and 5 with lane 3 for the binding of GagPol to the full-length hybrid GST.Nef protein). Moreover, a mutant GST.Nef chimera lacking the C-terminal flexible loop (NefΔFL from positions 148 to 180) did not bind GagPol or the p6*PR fragment (Fig. 2B, lanes 6 and 15, respectively). We conclude that Nef specifically binds the p6* region of GagPol via its C-terminal flexible loop in vitro.
Nef binds GagPol and p6* in cells.
Since we demonstrated that Nef binds the p6* region of GagPol in vitro, we also wanted to determine if this binding occurs in cells. Coimmunoprecipitation experiments were performed with 293T cells that expressed various HIV-1NL4-3 proviral DNAs. Based on our previous results, we expected that only GagPol would interact with Nef. Moreover, p6* should be required for this binding (Fig. 2). Due to the relatively low levels of expression of GagPol and its cleavage by PR, proviruses harboring a protease-inactivating point mutation at the active site (D25N) were used. Moderate amounts of Nef could be detected in anti-Gag immunoprecipitations from cells coexpressing Gag, GagPol, and Nef (Fig. 3A, middle panel, lane 3). We then used a mutant HIV-1NL4-3 provirus that in addition to the D25N also contained a series of mutations in the transframe region, which led to the exclusive expression of GagPol but not Gag (Fig. 3A, bottom panel; compare lanes 3 and 4). In this scenario, binding of Nef was observed by its coimmunoprecipitation with GagPol with an anti-RT antibody (Fig. 3A, top panel, lane 4). Furthermore, by using a HIV-1NL4-3 provirus with RT and IN deleted, we confirmed that GagPR within GagPol is sufficient for binding to Nef (Fig. 3A, middle panel, lane 5). Indeed, no interaction was detected between Nef and Gag (Fig. 3A, middle panel, lane 2). From these results, we conclude that Nef also binds GagPol in cells.
FIG. 3.
Nef binds GagPol and p6* in cells. (A) Genomic regions of Gag and GagPol from HIV-1NL4-3 are represented as in Fig. 2. Gag which contained a deletion in the pol open reading frame was used for the expression of only Gag. Gag/GagPol directed the expression of Gag and GagPol. GagPol bears mutations in the ribosomal frameshifting site (TTTTTT→CTCCTC) and expressed only GagPol (indicated by brackets between NC and p6*). GagPR contained a stop codon at the end of the pr open reading frame and encodes only GagPR. PR in all these polyproteins was inactive due to the mutation of aspartate to alanine at position 25 in its active site. For coimmunoprecipitations, these proviruses were expressed in 293T cells. Cells were lysed in RIPA buffer, followed by incubation with anti-CA (αCA) polyclonal (for Gag, Gag/GagPol, and GagPro) or anti-RT monoclonal (for Gag/GagPol and GagPol) antibodies. After the immunoprecipitation, Western blotting (WB) was performed with the anti-Nef antibody (middle panel). A control Western blotting for protein inputs was performed with anti-CA and anti-RT antibodies and represents 50% of the input proteins (bottom panel). Immunoprecipitation of Gag was used as the negative control for the binding between Nef and GagPol (lane 2). (B). Shown are p6 and p6*. p1 is the coding region where the frameshifting sites are located. Note that the p6* open reading frame starts in p1 (the arrow indicates the frameshifting position) and is flanked by NC at the N terminus and by PR at the C terminus. The first amino acid residues of p6 and p6* are represented by boldface letters. HA epitope-tagged p6 and p6* proteins were expressed alone or coexpressed with wild-type NefNL4-3 protein in 293T cells. Cells were lysed and immunoprecipitated with the anti-HA polyclonal antibody. Immune complexes were processed as described above, and Western blotting was performed with the anti-Nef antibody (middle panel). A control Western blot for protein input was performed with the anti-HA antibody (bottom panel). Immunoprecipitation of p6 was used as the negative control for the binding between Nef and p6* (lane 3). Arrows indicate the 27-kDa Nef protein, the 6-kDa p6 and p6* proteins, the 55-kDa Gag protein, and the 160-kDa GagPol and 72-kDa GagPR precursors.
Next, we confirmed that p6* mediates the binding between GagPol and Nef in cells. Figure 3B shows the coimmunoprecipitation with the anti-HA antibody of an HA epitope-tagged p6* protein and Nef from 293T cells (Fig. 3B, middle panel, lane 1). In parallel, we were unable to detect any binding between Nef and an HA epitope-tagged p6 protein (Fig. 3B, middle panel, lane 3). Given the differences in inputs of Nef between Fig. 3A and B, the binding between Nef and p6* or GagPol was equivalent in vitro and in vivo. Taken together, these results confirm that Nef specifically binds GagPol from HIV-1 and that p6* is required for this binding.
Mutant NefΔFLKKXX protein no longer interferes with Gag processing or virion production, and mutant NefΔFL protein is not incorporated into viral particles.
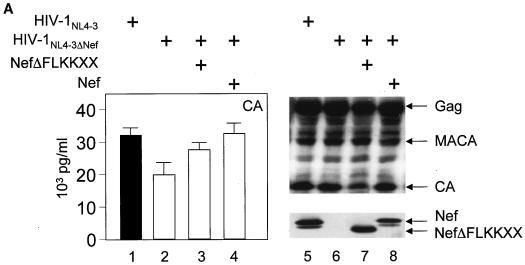
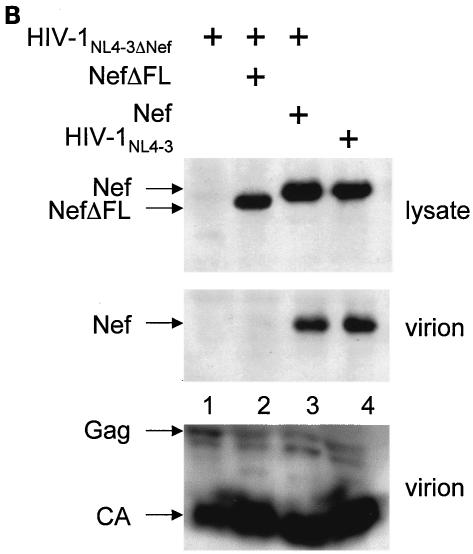
Next, we investigated whether the binding between Nef and GagPol plays a functional role in the action of interfering Nef proteins. To this end, a deletion of the C-terminal flexible loop was introduced in the mutant NefKKXX protein, and its ability to interfere with Gag processing and virion production was examined. Indeed, the mutant NefΔFLKKXX protein was unable to block virion production from cells coexpressing HIV-1NL4-3ΔNef (Fig. 4A, left panel; compare bar 3 with bars 2 and 4 and with Fig. 1C, lane 4), despite comparable levels of the mutant and wild-type Nef proteins (Fig. 4A, right panel; compare lanes 7 and 8). Additionally, normal processing of Gag was observed in the presence of this mutant protein (Fig. 4A; compare lane 7 with lanes 6 and 8 and with Fig. 1C, middle panel, lane 4). Finally, the mutant NefΔFL protein was no longer incorporated into progeny virions (Fig. 4B; compare lane 2 to lanes 3 and 4), despite equivalent levels of virion production (Fig. 4B, bottom panel, virion). Thus, the binding between Nef and GagPol is required for the interfering effects of the mutant NefKKXX protein. Moreover, mutant Nef protein that no longer binds GagPol is also not incorporated into new viral particles.
FIG. 4.
Mutant NefΔFLKKXX protein no longer interferes with Gag processing or virion production, and mutant NefΔFL protein is not incorporated into viral particles. (A) Virion production from 293T cells, which expressed HIV-1NL4-3 (bar 1) or HIV-1NL4-3ΔNef (bar 2) or coexpressed HIV-1NL4-3ΔNef and the mutant NefΔFLKKXX (bar 3) or the wild-type Nef (bar 4) proteins, was measured by CA capture ELISA. Means and standard deviations from three independent experiments are shown. Western blotting of these cell lysates was performed with anti-CA and anti-Nef antibodies. Arrows indicate the Gag (55 kDa) and MACA (41 kDa) precursors and the CA (24 kDa) and the Nef (27 kDa) proteins. (B) Nef lacking the flexible loop (NefΔFL) is not incorporated into progeny virions. From culture supernatants, virions were separated by centrifugation through a sucrose cushion and examined for the presence of Gag and Nef by Western blotting. Arrows point to different Nef proteins in cell lysates (top panel) and virions (middle panel) as well as structural proteins in virions (bottom panel).
Nef.Tsg101 but not mutant NefΔFL.Tsg101 chimeras rescue the budding and release of mutant proviruses lacking the PTAP motif in p6.
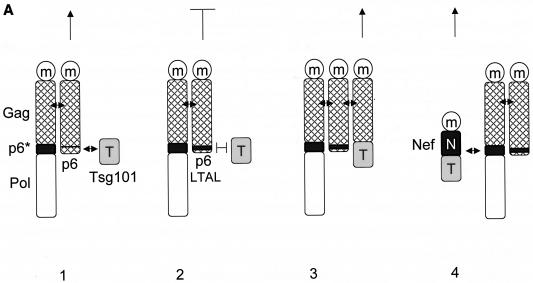
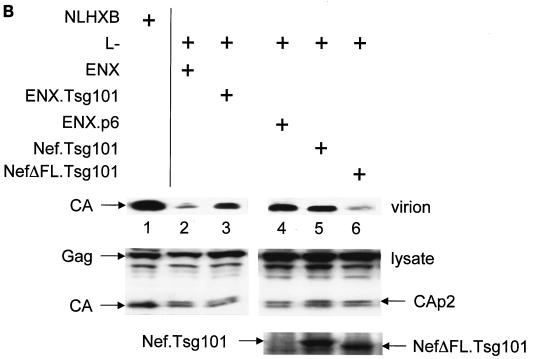
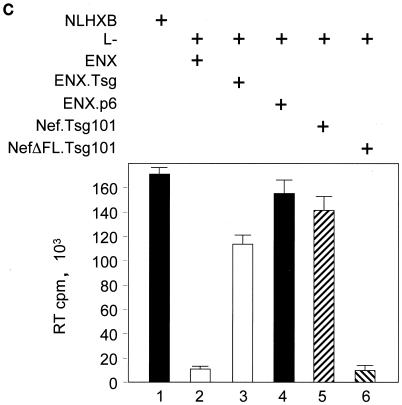
Finally, we addressed the question of whether the interaction with GagPol is also relevant for the physiological function of noninterfering Nef proteins. We took advantage of a trans-complementation assay for viral budding developed by Martin-Serrano and colleagues (32). In this assay, budding of new virions from a late-domain-inactive (L−) provirus harboring two mutations at p6 (P7L and P10L [both mutations were silent in p6*]) can be rescued by adding minimal late-domain motifs to a mutant proviral DNA that lacks p6 and pol (ENX). The requirement for the rescue of virion budding is the recruitment of the host protein Tsg101 to the site of virion assembly (32) (Fig. 5A, panels 1 to 3). Instead of using the ENX plasmids with the minimal late-domain motifs (ENXp6) to rescue viral budding, we used a Nef.Tsg101 fusion protein. In this scenario, via its interaction with GagPol, Nef should target Tsg101 to the site of virion assembly, thereby restoring the budding of new virions (Fig. 5A, panel 4). As expected, expression of the late-domain-inactive L− viruses along with the Nef.Tsg101 fusion protein rescued the release of viral particles to the levels of HIV-1NLHXB in HeLa cells (Fig. 5B; compare lanes 2, 5, and 7 to Fig. 5C, lanes 1, 2, and 5). Importantly, levels of virion production were equivalent to those achieved upon the expression of ENXp6 (Fig. 5B and C; compare lanes 4 and 5). Thus, Nef can target Tsg101 to the sites of viral budding.
FIG. 5.
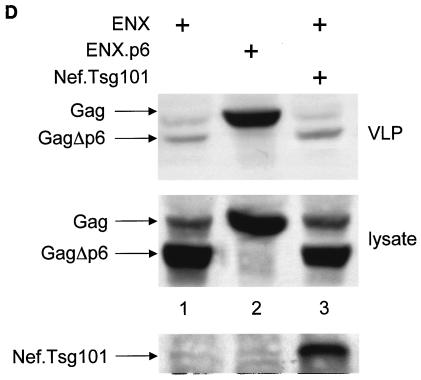
The Nef.Tsg101 chimera but not the mutant NefΔFL.Tsg101 chimera rescues mutant HIV-1 in the PTAP motif of the late domain. (A) Schematic representation of the trans-complementation assay for virus release. By recruiting Tsg101 to the PTAP domain of viral protein p6, cells expressing the HIV-1NLHXB provirus are able to release new virions to the supernatant (panel 1). To create the L− proviral clone, the two prolines in the PTAP motif in p6 of HIV-1NLHXB were mutated to leucines (PTAP→LTAL). Cells expressing the L− provirus no longer release virions to the supernatant (panel 2). To rescue this release, this L− mutant provirus was coexpressed with compensatory late domains (added back to a HIV-1NLHXB mutant, named ENX, with p6, pol, vif, vpr, and nef deleted) in 293T cells (panel 3). In another scenario, the L− provirus was coexpressed with a plasmid expressing the Nef.Tsg101 chimera (panel 4). (B) Cellular supernatants were harvested, and virions were precipitated by ultracentrifugation. Western blotting of virion preparations and cell lysates were performed with the anti-CA antibody. Lysates were also analyzed by Western blotting with the anti-Nef antibody for the presence of the Nef.Tsg101 chimera. Arrows indicate the Gag (55 kDa) and MACA (41 kDa) precursors, the CA (24 kDa) protein, and the Nef.Tsg101 (∼83 kDa) and the NefΔFL.Tsg101 (∼80 kDa) chimeras. (C) An aliquot of each virion preparation was used to determine amounts of virus present in supernatants of transfected cells (RT). The wild-type HIV-1NLHXB (bar 1) and the L− proviruses plus ENX.p6 (bar 4) were used as positive controls for virion production. The L− provirus plus the plasmid ENX were used as negative control for the trans-complementation assay (bar 2). The mutant NefΔFL.Tsg101 fusion protein (bar 6) was used as the negative control for the binding between HIVNef and GagPol (bar 5). (D) The Nef.Tsg101 chimera does not rescue the release of VLPs from ENX. VLPs from supernatants of cells expressing ENX, expressing ENX.p6 as a positive control, or coexpressing ENX with the Nef.Tsg101 chimera were collected and purified. Viral and cell-associated proteins were analyzed by Western blotting with the anti-CA antibody. Arrows indicate the Gag (55 kDa) and mutant GagΔp6 (49 kDa) proteins. The presence of the Nef.Tsg101 chimera was also confirmed.
To demonstrate that this rescue was due to the binding between Nef and GagPol and not only to the targeting of Nef to lipid rafts at the plasma membrane (53), a Nef protein with a deletion in the C-terminal flexible loop and therefore unable to bind to GagPol (Fig. 2B, lanes 6 and 15) was fused with the Tsg101 protein to generate the mutant NefΔFL.Tsg101 chimera. Figure 5B and C demonstrate that this hybrid protein did not rescue viral budding from the late-domain-inactive L− virus (compare lanes 4, 5 and 6). Levels of expression of Nef.Tsg101 and mutant NefΔFL.Tsg101 chimeras were identical (Fig. 5B, bottom panel, lanes 5 and 6). To confirm more precisely that the trans-complementation of L− by the Nef.Tsg101 chimera was due to direct binding to GagPol and not only recruitment of Nef to lipid rafts at the membrane, ENX was coexpressed with the Nef.Tsg101 chimera and the production of VLPs was analyzed 48 h later. In this experiment, ENXp6 was used as a positive control for VLP production. Although the same levels of expression of ENXp6 and ENX were achieved in cells (Fig. 5D, bottom panel; compare lanes 1 and 3 to lane 2), the production of VLPs from cells expressing only ENX or coexpressing ENX and the Nef.Tsg101 chimera was decreased equivalently (Fig. 5D, top panel; compare lanes 1 and 3 to lane 2). From these results, we conclude that the Nef.Tsg101 chimera rescues late-domain-defective proviruses only via interactions with GagPol.
DISCUSSION
This study demonstrates that the retention of Nef during its biosynthetic pathway in the intermediate compartment inhibits Gag processing and virion production. Importantly, by introducing an ER targeting signal, the wild-type NefNL4-3 protein could be made into an interfering protein for viral replication. This interference correlated with the binding between the flexible loop in Nef and p6* within GagPol in vitro and in vivo. Furthermore, since only the wild-type Nef.Tsg101, and not a mutant NefΔFL.Tsg101 chimera, rescued the release of mutant HIV-1 proviruses lacking a functional late domain and since the mutant NefΔFL protein was no longer incorporated into virions, this binding was biologically relevant. We conclude that Nef binds GagPol during the production of new virions.
In previous studies, the perinuclear localization of NefF12 correlated with its interfering phenotype (12, 35). Thus, we asked whether the addition of an ER retention signal to the C terminus of a Nef protein from a common HIV-1 strain could recapitulate this phenotype. Indeed, we demonstrated that the hybrid mutant CD8.NefKKXX or NefKKXX proteins interfered with Gag processing and virion production of a mutant HIV-1NL4-3ΔNef provirus (Fig. 1B and C). Although we chose a mutant HIV-1NL4-3ΔNef provirus that lacked the nef gene to eliminate the competition with Nef encoded by the provirus, our results could be reproduced with the wild-type HIV-1NL4-3 provirus (data not shown). Importantly, the mutant NefKKXX protein inhibited efficiently Gag processing and virion production in 293T cells (Fig. 1C), demonstrating that the CD8 transmembrane domain did not contribute to this phenotype. Notably, these effects were even more pronounced than those of the hybrid CD8.NefF12 protein. Most likely, this finding reflects the greater stability of mutant NefKKXX proteins in cells. Recently, a truncated nerve growth factor receptor (ΔNGFr).NefF12 hybrid protein was found to provide intracellular immunity against HIV-1 (33). However, the expression levels of this chimera were also relatively low, and its ability to inhibit the replication of diverse clinical isolates was variable. Because of its greater stability, the use of the mutant NefKKXX rather than NefF12 proteins might be advantageous for such intracellular immunization.
Because hybrid mutant CD8.NefKKXX and Nef.KKXX proteins blocked Gag processing, this finding implied that GagPol is a target of Nef. Indeed, Nef bound GagPol, but not Gag, via its C-terminal flexible loop (Fig. 2). The specificity of this binding is explained because Nef bound p6* in GagPol, which is not present in Gag. Importantly, our data in vitro were confirmed in cells, in which both GagPol and p6* coimmunoprecipitated Nef (Fig. 3). Furthermore, the interaction of Nef and GagPol was observed in the context of normal levels of proteins expressed from the provirus. Importantly, the Nef.Tsg101 chimera rescued the release of viral particles from a provirus lacking the late domain. This finding indicates that Nef binds GagPol at sites of virion assembly. The specificity of this assay was confirmed by the inability of the Nef.Tsg101 fusion protein to affect VLPs containing only Gag and the mutant NefΔFL.Tsg101 chimera to promote the release of complete viral particles. Thus, specific binding between Nef and GagPol rather than incidental coaggregation of both proteins in lipid rafts during viral budding was required in this trans-complementation assay. Therefore, these results are in agreement with a functional role for Nef and its interaction with GagPol during virion assembly.
The interaction of the C-terminal flexible loop in Nef with p6* in GagPol might also explain the phenotype of NefF12. The data presented demonstrate that only the retention of GagPol is needed for NefF12 to significantly reduce Gag processing and virion production. Therefore, the p72/p75 proteins that specifically interact with NefF12 (12) likely mediate the retention and possibly degradation of this complex within the intermediate compartment in cells. These results reveal that the NefF12 phenotype does not result from atypical gain of function but rather reflects the property of Nef binding GagPol in infected cells. The inhibitory effects of ER-retained Nef proteins then result from retention and degradation of complexes between Nef and GagPol. This effect is reminiscent of that of mutations in the PR (23) and IN (4) regions of GagPol that generate dominant negative proteins for the production and release of progeny virions. Retention of GagPol may synergize with the functional interaction of NefF12 with the cytoplasmic tail of gp41 to the complete block of HIV-1 replication in CD4-positive cells (35).
The misrouting of GagPol by NefF12 is determined by the three amino acid changes specific to this Nef variant (8, 12). Importantly, all mutations in NefF12 lie outside the flexible loop (8, 16), which still binds GagPol with the same affinity as that of NefNL4-3 in vitro (data not shown). The C-terminal flexible loop in Nef has been implicated in binding of many cellular sorting proteins containing armadillo repeats, such as the β-chains of AP complexes (3, 19), the β subunit of the COP-I coatomer (41), and subunit H of the vacuolar ATPase (V1H) to internalize CD4 from the cell surface (18, 28, 31). The interaction of Nef with p6* may be of a similar nature on the molecular level. Since Nef augments virion infectivity in a manner that depends on the flexible loop in the absence of CD4 (29), it is possible that the binding of the flexible loop of Nef to GagPol is critical for this effect. Whether Nef still internalizes CD4 when associated with p6* will have to be determined. Thus, our findings suggest that Nef and its C-terminal flexible loop in particular might have different roles during early and late phases of the viral replicative cycle.
Several observations support a role for the interaction of Nef with GagPol in the replication of HIV-1. First, Ono et al. (38) demonstrated that a deletion of Nef had different effects in two highly related proviruses (HIV-1LAIBru and HIV-1NL4-3), in which viral replication for only one provirus (HIV-1NL4-3) depended on Nef. Importantly, GagPol (minimally a region containing the NC.PR) was responsible for this difference. Analyses of GagPol from these two proviruses confirmed that although their sequences are highly conserved, with the nef genes sharing 98% similarity, p6* from HIV-1LAIBru had an insertion of two residues followed by a duplication of 10 residues near the N terminus. This finding suggests genetically a link between Nef and p6* in GagPol. Second, when Nef from HIV-1 is introduced into the simian immunodeficiency virus SIVmac239, the resulting chimeric virus replicates poorly in rhesus macaques until mutations in Nef are observed (30). One study describes the accumulation of such compensatory changes in the C-terminal flexible loop of Nef. Although further mutations were observed in the transmembrane portion of Env, GagPol was not examined (2). The flexible loop in SIV Nef is also a key determinant for SIV infectivity in human lymphocytes (31), and the ability of Nef to internalize CD4 (via the flexible loop) was correlated with its role in the pathogenesis of simian AIDS (reference 31 and references therein). Finally, with a truncated Nef protein that resulted from larger deletions of the nef gene in SIVmac239, the restoration of virion infectivity correlated with the return of the flexible loop (6, 43). Of interest is that p6* sequences are highly divergent between HIV-1 and SIV, and our preliminary studies reveal a species-specific restriction to interactions between Nef and p6* from HIV-1 and SIV (data not shown). Therefore, these findings suggest that Nef and p6* are matched within the primate lentiviruses HIV-1 and SIV and possibly HIV-2 and that the interaction between Nef and GagPol facilitates viral replication.
Our data suggest a model for this new role of Nef in the viral replicative cycle. Nef binds p6* in GagPol within the cytoplasm, probably in the context of complexes between Gag and GagPol (see references 34 and 48 and references therein). Therefore, GagPol likely represents the viral interaction partner that allows Nef to target HIV-1 budding to these detergent-resistant microdomains (53). Indeed, in the absence of GagPol, only a small fraction of complexes containing only Gag move to lipid rafts (21). Putative assembly intermediates between Gag, GagPol, and Nef may be efficiently incorporated into lipid rafts at the plasma membrane due to the presentation of multiple membrane-targeting motifs on these proteins (21, 36, 37, 50, 53). Oligomerization of Nef (17) could help to multimerize these assembly intermediates and thus facilitate this incorporation. Nef also causes cytoskeletal rearrangements that could further trap these lipid rafts and/or help to form and release immature virions (13, 42). Moreover, as it induces intracellular accumulation of multivesicular bodies, where these detergent-resistant microdomains are formed and from which HIV buds, Nef could also facilitate the maturation and release of progeny virions (47). In this regard, it is of some interest that a lentivirus that lacks Nef, the equine infectious anemia virus, also contains a late domain in the GagPol precursor (46). Finally, Nef not only activates downstream signaling cascades, which increase the biosynthesis of cholesterol, but also carries this new cholesterol to sites of viral budding (52). At the same time, the interaction of Nef with GagPol ensures its incorporation in the virion. In this scenario, interactions between Nef and GagPol could facilitate the production of optimal virions for subsequent rounds of infection.
Acknowledgments
We are grateful to Barbara Müller and Eva Gottwein for critical comments on the manuscript and to Hans-Georg Kräusslich for discussion and continuous support. We thank Walter Muranyi for help with confocal microscopy and Christine Chang and Nadine Tibroni for excellent technical assistance.
This research was supported by an Emmy-Noether Fellowship of the Deutsche Forschungsgemeinschaft (Fa 378/2-2) to O.T.F., a postdoctoral fellowship from the Brazilian Council for Research (CNPq-200834/01-0) to L.J.C., and a grant from the NIH (RO1 A151165-01) to B.M.P.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, L., Z. Du, A. Y. Howe, S. Czajak, and R. C. Desrosiers. 1999. Induction of AIDS in rhesus monkeys by a recombinant simian immunodeficiency virus expressing Nef of human immunodeficiency virus type 1. J. Virol. 73:5814-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bresnahan, P. A., W. Yonemoto, S. Ferrell, D. Williams-Herman, R. Geleziunas, and W. C. Greene. 1998. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr. Biol. 8:1235-1238. [DOI] [PubMed] [Google Scholar]
- 4.Bukovsky, A., and H. Gottlinger. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 70:6820-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassan, M., V. Berteaux, P. O. Angrand, and J. P. Rousset. 1990. Expression vectors for quantitating in vivo translational ambiguity: their potential use to analyse frameshifting at the HIV gag-pol junction. Res. Virol. 141:597-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakrabarti, L. A., K. J. Metzner, T. Ivanovic, H. Cheng, J. Louis-Virelizier, R. I. Connor, and C. Cheng-Mayer. 2003. A truncated form of Nef selected during pathogenic reversion of simian immunodeficiency virus SIVmac239Deltanef increases viral replication. J. Virol. 77:1245-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosson, P., and F. Letourneur. 1994. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science 263:1629-1631. [DOI] [PubMed] [Google Scholar]
- 8.D'Aloja, P., E. Olivetta, R. Bona, F. Nappi, D. Pedacchia, K. Pugliese, G. Ferrari, P. Verani, and M. Federico. 1998. gag, vif, and nef genes contribute to the homologous viral interference induced by a nonproducer human immunodeficiency virus type 1 (HIV-1) variant: identification of novel HIV-1-inhibiting viral protein mutants. J. Virol. 72:4308-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel, M. D., F. Kirchhoff, S. C. Czajak, P. K. Sehgal, and R. C. Desrosiers. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 258:1938-1941. [DOI] [PubMed] [Google Scholar]
- 10.Deacon, N. J., A. Tsykin, A. Solomon, K. Smith, M. Ludford-Menting, D. J. Hooker, D. A. McPhee, A. L. Greenway, A. Ellett, C. Chatfield, et al. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270:988-991. [DOI] [PubMed] [Google Scholar]
- 11.Fackler, O. T., and A. S. Baur. 2002. Live and let die: Nef functions beyond HIV replication. Immunity 16:493-497. [DOI] [PubMed] [Google Scholar]
- 12.Fackler, O. T., P. d'Aloja, A. S. Baur, M. Federico, and B. M. Peterlin. 2001. Nef from human immunodeficiency virus type 1(F12) inhibits viral production and infectivity. J. Virol. 75:6601-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fackler, O. T., W. Luo, M. Geyer, A. S. Alberts, and B. M. Peterlin. 1999. Activation of Vav by Nef induces cytoskeletal rearrangements and downstream effector functions. Mol. Cell 3:729-739. [DOI] [PubMed] [Google Scholar]
- 14.Fackler, O. T., D. Wolf, H. O. Weber, B. Laffert, P. D'Aloja, B. Schuler-Thurner, R. Geffin, K. Saksela, M. Geyer, B. M. Peterlin, G. Schuler, and A. S. Baur. 2001. A natural variability in the proline-rich motif of Nef modulates HIV-1 replication in primary T cells. Curr. Biol. 11:1294-1299. [DOI] [PubMed] [Google Scholar]
- 15.Federico, M., R. Bona, P. D'Aloja, M. Baiocchi, K. Pugliese, F. Nappi, C. Chelucci, F. Mavilio, and P. Verani. 1996. Anti-HIV viral interference induced by retroviral vectors expressing a nonproducer HIV-1 variant. Acta Haematol. 95:199-203. [DOI] [PubMed] [Google Scholar]
- 16.Federico, M., F. Nappi, G. Ferrari, C. Chelucci, F. Mavilio, and P. Verani. 1995. A nonproducer, interfering human immunodeficiency virus (HIV) type 1 provirus can be transduced through a murine leukemia virus-based retroviral vector: recovery of an anti-HIV mouse/human pseudotype retrovirus. J. Virol. 69:6618-6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geyer, M., O. T. Fackler, and B. M. Peterlin. 2001. Structure-function relationships in HIV-1 Nef. EMBO Rep. 2:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geyer, M., H. Yu, R. Mandic, T. Linnemann, Y. H. Zheng, O. T. Fackler, and B. M. Peterlin. 2002. Subunit H of the V-ATPase binds to the medium chain of adaptor protein complex 2 and connects Nef to the endocytic machinery. J. Biol. Chem. 277:28521-28529. [DOI] [PubMed] [Google Scholar]
- 19.Greenberg, M., L. DeTulleo, I. Rapoport, J. Skowronski, and T. Kirchhausen. 1998. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr. Biol. 8:1239-1242. [DOI] [PubMed] [Google Scholar]
- 20.Greene, W. C., and B. M. Peterlin. 2002. Charting HIV's remarkable voyage through the cell: basic science as a passport to future therapy. Nat. Med. 8:673-680. [DOI] [PubMed] [Google Scholar]
- 21.Halwani, R., A. Khorchid, S. Cen, and L. Kleiman. 2003. Rapid localization of Gag/GagPol complexes to detergent-resistant membrane during the assembly of human immunodeficiency virus type 1. J. Virol. 77:3973-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanna, Z., D. G. Kay, N. Rebai, A. Guimond, S. Jothy, and P. Jolicoeur. 1998. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell 95:163-175. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan, A. H., M. Manchester, and R. Swanstrom. 1994. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J. Virol. 68:6782-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchhoff, F., T. C. Greenough, D. B. Brettler, J. L. Sullivan, and R. C. Desrosiers. 1995. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 332:228-232. [DOI] [PubMed] [Google Scholar]
- 25.Kollmus, H., M. W. Hentze, and H. Hauser. 1996. Regulated ribosomal frameshifting by an RNA-protein interaction. RNA 2:316-323. [PMC free article] [PubMed] [Google Scholar]
- 26.Letourneur, F., E. C. Gaynor, S. Hennecke, C. Demolliere, R. Duden, S. D. Emr, H. Riezman, and P. Cosson. 1994. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 79:1199-1207. [DOI] [PubMed] [Google Scholar]
- 27.Lotti, L. V., G. Mottola, M. R. Torrisi, and S. Bonatti. 1999. A different intracellular distribution of a single reporter protein is determined at steady state by KKXX or KDEL retrieval signals. J. Biol. Chem. 274:10413-10420. [DOI] [PubMed] [Google Scholar]
- 28.Lu, X., H. Yu, S. H. Liu, F. M. Brodsky, and B. M. Peterlin. 1998. Interactions between HIV1 Nef and vacuolar ATPase facilitate the internalization of CD4. Immunity 8:647-656. [DOI] [PubMed] [Google Scholar]
- 29.Lundquist, C. A., M. Tobiume, J. Zhou, D. Unutmaz, and C. Aiken. 2002. Nef-mediated downregulation of CD4 enhances human immunodeficiency virus type 1 replication in primary T lymphocytes. J. Virol. 76:4625-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandell, C. P., R. A. Reyes, K. Cho, E. T. Sawai, A. L. Fang, K. A. Schmidt, and P. A. Luciw. 1999. SIV/HIV Nef recombinant virus (SHIVnef) produces simian AIDS in rhesus macaques. Virology 265:235-251. [DOI] [PubMed] [Google Scholar]
- 31.Mandic, R., O. T. Fackler, M. Geyer, T. Linnemann, Y. H. Zheng, and B. M. Peterlin. 2001. Negative factor from SIV binds to the catalytic subunit of the V-ATPase to internalize CD4 and to increase viral infectivity. Mol. Biol. Cell 12:463-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 33.Muratori, C., I. Schiavoni, G. Melucci-Vigo, E. Olivetta, A. C. Santarcangelo, K. Pugliese, P. Verani, and M. Federico. 2002. Inducible expression of the deltaNGFr/F12Nef fusion protein as a new tool for anti-human immunodeficiency virus type 1 gene therapy. Hum. Gene Ther. 13:1751-1766. [DOI] [PubMed] [Google Scholar]
- 34.Nermut, M. V., W. H. Zhang, G. Francis, F. Ciampor, Y. Morikawa, and I. M. Jones. 2003. Time course of Gag protein assembly in HIV-1-infected cells: a study by immunoelectron microscopy. Virology 305:219-227. [DOI] [PubMed] [Google Scholar]
- 35.Olivetta, E., K. Pugliese, R. Bona, P. D'Aloja, F. Ferrantelli, A. C. Santarcangelo, G. Mattia, P. Verani, and M. Federico. 2000. cis expression of the F12 human immunodeficiency virus (HIV) Nef allele transforms the highly productive NL4-3 HIV type 1 to a replication-defective strain: involvement of both Env gp41 and CD4 intracytoplasmic tails. J. Virol. 74:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono, A., D. Demirov, and E. O. Freed. 2000. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J. Virol. 74:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono, A., J. M. Orenstein, and E. O. Freed. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono, T., Y. Iwatani, A. Nishimura, A. Ishimoto, and H. Sakai. 2000. Functional association between the nef gene product and gag-pol region of HIV-1. FEBS Lett. 466:233-238. [DOI] [PubMed] [Google Scholar]
- 39.Pandori, M. W., N. J. Fitch, H. M. Craig, D. D. Richman, C. A. Spina, and J. C. Guatelli. 1996. Producer-cell modification of human immunodeficiency virus type 1: Nef is a virion protein. J. Virol. 70:4283-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, J., and C. D. Morrow. 1992. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J. Virol. 66:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piguet, V., F. Gu, M. Foti, N. Demaurex, J. Gruenberg, J. L. Carpentier, and D. Trono. 1999. Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of beta-COP in endosomes. Cell 97:63-73. [DOI] [PubMed] [Google Scholar]
- 42.Plemenitas, A., X. Lu, M. Geyer, P. Veranic, M. N. Simon, and B. M. Peterlin. 1999. Activation of Ste20 by Nef from human immunodeficiency virus induces cytoskeletal rearrangements and downstream effector functions in Saccharomyces cerevisiae. Virology 258:271-281. [DOI] [PubMed] [Google Scholar]
- 43.Sawai, E. T., M. S. Hamza, M. Ye, K. E. Shaw, and P. A. Luciw. 2000. Pathogenic conversion of live attenuated simian immunodeficiency virus vaccines is associated with expression of truncated Nef. J. Virol. 74:2038-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simmons, A., V. Aluvihare, and A. McMichael. 2001. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity 14:763-777. [DOI] [PubMed] [Google Scholar]
- 45.Stornaiuolo, M., L. V. Lotti, N. Borgese, M. R. Torrisi, G. Mottola, G. Martire, and S. Bonatti. 2003. KDEL and KKXX retrieval signals appended to the same reporter protein determine different trafficking between endoplasmic reticulum, intermediate compartment, and Golgi complex. Mol. Biol. Cell 14:889-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 47.Stumptner-Cuvelette, P., M. Jouve, J. Helft, M. Dugast, A. S. Glouzman, K. Jooss, G. Raposo, and P. Benaroch. 2003. Human immunodeficiency virus-1 Nef expression induces intracellular accumulation of multivesicular bodies and major histocompatibility complex class II complexes: potential role of phosphatidylinositol 3-kinase. Mol. Biol. Cell 14:4857-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 74:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogt, V. M. 1996. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 214:95-131. [DOI] [PubMed] [Google Scholar]
- 50.Wang, J. K., E. Kiyokawa, E. Verdin, and D. Trono. 2000. The Nef protein of HIV-1 associates with rafts and primes T cells for activation. Proc. Natl. Acad. Sci. USA 97:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welker, R., H. Kottler, H. R. Kalbitzer, and H. G. Krausslich. 1996. Human immunodeficiency virus type 1 Nef protein is incorporated into virus particles and specifically cleaved by the viral proteinase. Virology 219:228-236. [DOI] [PubMed] [Google Scholar]
- 52.Zheng, Y. H., A. Plemenitas, C. J. Fielding, and B. M. Peterlin. 2003. Nef increases the synthesis of and transports cholesterol to lipid rafts and HIV-1 progeny virions. Proc. Natl. Acad. Sci. USA 100:8460-8465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng, Y. H., A. Plemenitas, T. Linnemann, O. T. Fackler, and B. M. Peterlin. 2001. Nef increases infectivity of HIV via lipid rafts. Curr. Biol. 11:875-879. [DOI] [PubMed] [Google Scholar]