Abstract
Bovine herpesvirus 1 (BHV-1), like other Alphaherpesvirinae subfamily members, establishes latency in sensory neurons. The latency-related (LR) RNA is abundantly expressed during latency, and expression of an LR protein is required for the latency reactivation cycle in cattle. Within LR promoter sequences, a 135-amino-acid open reading frame (ORF) was identified, ORF-E, that is antisense to the LR RNA. ORF-E is also downstream of the gene encoding the major viral transcriptional activator, bICP0. Strand-specific reverse transcription-PCR demonstrated that a transcript containing ORF-E was consistently expressed in trigeminal ganglia (TG) of latently infected calves, productively infected cultured cells, and acutely infected calves. As expected, a late transcript encoding glycoprotein C was not detected in TG of latently infected calves. The ORF-E transcript is polyadenylated and is expressed early when cultured bovine cells are productively infected. Protein coding sequences containing ORF-E were fused to green fluorescent protein (GFP) to examine the cellular localization of the putative protein. In transiently transfected mouse neuroblastoma (neuro-2A) and human neuroblastoma (SK-N-SH) cells, the ORF-E/GFP fusion protein was detected in discreet domains within the nucleus. In contrast, the ORF-E/GFP fusion protein was detected in the cytoplasm and nucleus of rabbit skin cells and bovine kidney cells. As expected, the GFP protein was expressed in the cytoplasm and nucleus of transfected cells. These studies indicate that the ORF-E transcript is consistently expressed during latency. We suggest that the ORF-E gene regulates some aspect of the latency reactivation cycle.
Bovine herpesvirus 1 (BHV-1) establishes lifelong latency in ganglionic neurons of the peripheral nervous system after initial replication in mucosal epithelium. Virus reactivation and spread to other susceptible animals occur after natural and corticosteroid-induced stress (23, 27). Infection can cause conjunctivitis, pneumonia, genital disorders, abortions, and an upper respiratory infection referred to as shipping fever (28). Since BHV-1-associated pathogenesis and shipping fever cost the U.S. cattle industry at least $500 million per year (2), modified live vaccines that protect cattle from disease but do not reactivate virus from latency are desirable.
The latency-related (LR) RNA is the only abundant viral transcript that has been detected in latently infected neurons (20, 23, 24). A fraction of LR RNA is polyadenylated and alternatively spliced in trigeminal ganglia (TG), suggesting that this RNA is translated into more than one protein (10, 13). LR gene products inhibit S-phase entry, and an LR protein is associated with cyclin-dependent kinase 2 (cdk2) and cdc2/cyclin complexes (13, 17). LR gene products also inhibit apoptosis (7), suggesting that one important function of the LR gene is to promote neuronal survival. A mutant BHV-1 strain that contains three stop codons near the beginning of the LR RNA was constructed to test whether LR proteins play a role in virus growth in cattle (14). Calves infected with the LR mutant exhibited diminished clinical symptoms and ocular shedding of the virus relative to calves infected with the wild-type and LR-rescued virus (14). Diminished levels of virus were also detected in TG of calves acutely infected with the LR mutant (15). Wild-type BHV-1 and the LR-rescued virus but not the LR mutant virus reactivated from latency following treatment with dexamethasone. During the transition from acute infection to latency (establishment of latency), higher levels of apoptosis occur in TG neurons of calves infected with the LR mutant compared to calves infected with wild-type BHV-1 (21). Taken together, these studies indicate that wild-type expression of LR gene products is required for the latency reactivation cycle in cattle.
A small open reading frame (ORF) was identified within the LR promoter and named ORF-E. ORF-E is antisense to the LR transcript and downstream of the bICP0 ORF. By using strand-specific reverse transcription (RT)-PCR, a transcript that encompasses ORF-E in TG of latently infected calves was detected. In productively infected bovine cells, a polyadenylated ORF-E transcript was also detected. Sequences containing ORF-E were cloned into a mammalian expression vector and expressed as a green fluorescent protein (GFP) fusion protein. The ORF-E/GFP fusion protein was primarily detected in the nucleus of transfected neuronal cells, whereas it was detected in the cytoplasm and nucleus of nonneuronal cells. These results suggest that ORF-E may play a role in the latency reactivation cycle.
MATERIALS AND METHODS
Cells and virus.
Cells were plated at a density of 5 × 105 cells/100-mm plastic dish in Earl's modified Eagle's medium supplemented with 5% fetal bovine serum (FBS). Bovine kidney cells (MDBK; ATCC CCL-22) were grown in 5% FBS, split 1:6 every 4 to 5 days, and used to propagate BHV-1.
Neuro-2A cells (ATCC CCL131) are mouse neuroblastoma cells, which were grown in Earle's minimal essential medium supplemented with 10% FBS. RS cells are rabbit epidermal skin cells (ATCC CCL68) and were supplemented with 5% FBS. SK-N-SH cells (ATCC HTB 11) are human neuroblastoma cells and were supplemented with 10% FBS. All media contained penicillin (10 U/ml) and streptomycin (100 μg/ml).
The Cooper strain of BHV-1 (wild-type virus) was obtained from the National Veterinary Services Laboratory, Animal and Plant Health Inspection Services, Ames, Iowa. Viral stocks were prepared by infecting MBDK cells at a multiplicity of infection of 0.001 from a plaque-purified virus and subsequently titrated on MDBK cells.
Animal experiments.
BHV-1-free crossbred calves (≈250 kg) were randomly assigned and housed in isolation rooms to prevent cross contamination. Calves were anaesthetized with xylazine (approximately 50 mg/50 kg of body weight; Bayer Corp., Shawnee Mission, Kans.). Calves were then inoculated with 1 ml of a solution containing 107 PFU of virus per ml in each nostril and eye, dropwise without scarification, for a total of 4 × 107 PFU per animal, as described previously (14, 15, 26, 29). Experiments with animals were performed in accordance with the American Association of Laboratory Animal Care guidelines. Calves were housed under strict isolation containment and given antibiotics before and after BHV-1 infection to prevent secondary bacterial infection. Calves were euthanized at the indicated times after infection. All calves designated as being latently infected were euthanized at 60 days after infection, and these calves were not shedding infectious virus from the nasal cavity or the eye.
Preparation of RNA and RT-PCR.
RNA from cultured cells and TG was prepared as described previously (6, 10, 13). Briefly, a guanine isothiocynate-acidic phenol chloroform solution was used to prepare total RNA. RNA was quantified spectrophotometrically (optical density at 260 nm) and stored at −80°C in 3 volumes of ethanol.
RNA samples were treated with 2 U of DNase I (RNase free) and 10 U of RNasin (Promega, Madison, Wis.) per μg of total RNA for 30 min at 20°C. DNase I was removed by chloroform extraction. For random-primed cDNA synthesis, the RNA was incubated with 0.5 μg of random hexamers (Boehringer Mannheim) and 0.5 mM deoxynucleoside triphosphate mix for 7 min at 65°C and immediately chilled on ice. Then 16 μl of reverse transcriptase mix (20 mM Tris-Cl [pH 8.3], 50 mM KCl, 2.5 mM MgCl2, 100 mg of bovine serum albumin per ml, 1 mM dithiothreitol, 10 U of RNasin, and 100 U of Superscript reverse transcriptase II [Stratagene]) was added to the solution. The reaction was incubated for 10 min at 20°C and then at 42°C for 45 min. Reverse transcriptase activity was eliminated by incubation at 95°C for 5 min.
For strand-specific and oligo(dT)-primed cDNA synthesis, the RNA was incubated with the specific primer and oligo(dT) and 0.5 mM deoxynucleoside triphosphate mix for 10 min at 65°C and then placed at 50°C; 16 μl of reverse transcriptase mix was preheated to 50°C, added to the RNA, and incubated at 50°C for 1 h. Reverse transcriptase activity was eliminated by incubation at 95°C for 5 min. PCR was performed with 2.0 μl of cDNA in a 50-μl solution containing 1× commercial PCR buffer (Advantage-GC cDNA PCR kit; BD Biosciences, Palo Alto, Calif.), 200 mM deoxynucleoside triphosphate mix, 200 μM each primer, 1 μl GC-Melt, and 1 μl of polymerase mix (KlenTaq-1 and Deep Vent polymerases and TaqStart antibody for the hot start). All PCR amplifications were performed on the Omn-E Hybaid thermal cycler with the following cycle conditions: 95°C for 3 min. (hot start), 95°C for 30 s, 57 to 65°C for 45 s, and 72°C for 1.5 min for 30 cycles and a final step of 72°C for 5 min. Amplified products were detected by 2% agarose gel electrophoresis.
Primers.
See Fig. 1C and 4 for ORF-E primer positions and sequences. Strand-specific primer 308, 5′-CTCGAGGCAGGCGCAACGGAC-3′, specifically amplifies ORF-E RNA. Primers that amplify the LR gene, IETU1, and gC have been described elsewhere (15, 26). β-Actin primers to screen reverse transcriptase reactions were derived from rat sequences. The β-actin primers are 5′-GTGGGGCGCCCCAGGCACCA-3′ and 5′-CTCCTTAATGTCACGCACGATTTC-3′. All primers were purchased from Integrated DNA Technologies (Coralville, Iowa).
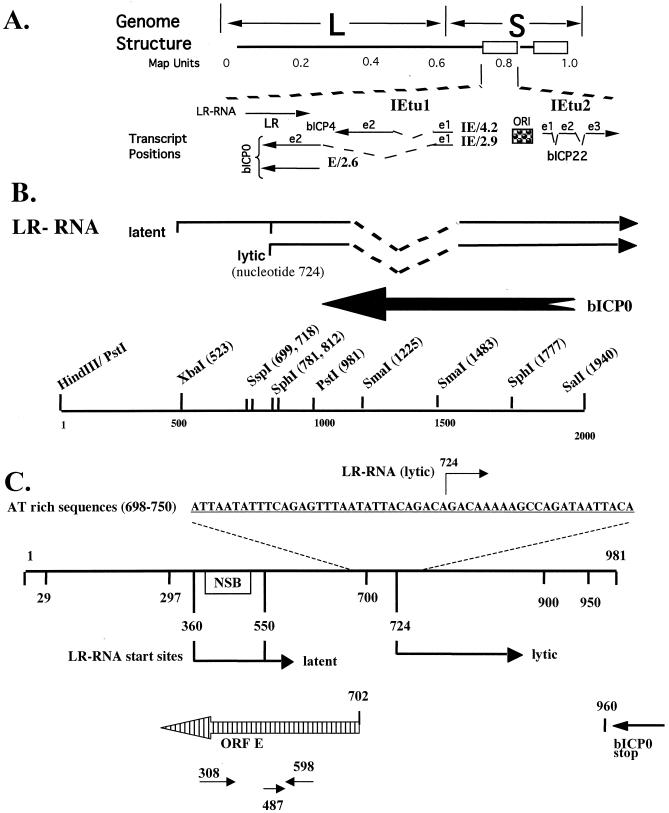
FIG. 1.
Location of the ORF-E gene within the BHV-1 genome. (A) Schematic representation of the BHV-1 genome. L and S indicate the unique long and short regions, respectively. The boxes represent the inverted and terminal repeats. The map units are shown below the schematic of the genome. The positions of immediate-early transcripts and LR transcript are presented. IE/4.2 is the immediate-early transcript that encodes bICP4. IE/2.9 is the immediate-early transcript that encodes bICP0. One immediate-early promoter activates expression of IE/4.2 and IE/2.9, and this immediate-early transcription unit is designated IETU1. E/2.6 is the early transcript that activates expression of E2.6, which encodes bICP0. Exon 2 (e2) of bICP0 contains all of the protein-coding sequences of bICP0. The origin of replication (ORI) separates IETU1 from IETU2. IETU2 encodes a protein, bICP22. Solid lines in the transcript position map represent exons 1, 2, and 3. (B) Partial restriction enzyme map of the sequences encompassing the LR gene. The locations of the LR transcripts, approximate sites for splicing of the LR RNA (dashed lines), and the 3′ terminus of bICP0 are marked. (C) Expanded map of the PstI fragment that contains the LR promoter, 5′ terminus of the LR transcript (13), and the putative ORF-E transcript. The AT-rich sequences located near the start site of the LR RNA are postulated to promote ORF-E transcription, and the arrow denotes the start sites for LR RNA transcription (nucleotide 724). The numbers indicate the mapped start sites for LR transcription during latent and lytic infections (4, 13). The open box represents the neuron-specific binding (NSB) site within the LR promoter (8). Also shown for reference is the termination site for bICP0 translation (bp 960) (31). ORF-E is antisense with respect to the LR gene (striped arrow). The primers used for strand-specific RT-PCR and detection of the ORF-E transcript are listed below ORF-E.
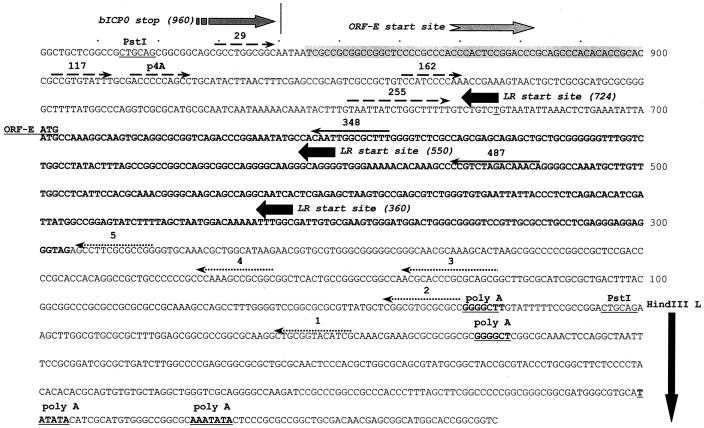
FIG. 4.
Strategy for localizing the 5′ and 3′ ends of the ORF-E transcript. The DNA sequence of the ORF-E/LR gene region (PstI fragment shown in Fig. 1) and positions of the primers used for the studies in Fig. 5 and 6 are shown. A partial sequence of the adjacent HindIII-L is also presented because it contains putative polyadenylation sites for ORF-E. The ORF-E sense strand is presented. The 3′ terminus of bICP0 and the LR start site (productive infection and latency) are presented to provide landmarks. Solid lines with arrow denote the primers used for strand-specific cDNA synthesis (primers 348 and 487). The dashed lines with arrows denote the positions of the primers used for PCR amplification to localize the 5′ terminus of the ORF-E transcript. The arrows show the directions of the primers. The shaded box indicates the region where the 5′ end is localized. The start codon for ORF-E and the putative protein-coding region of ORF-E are in bold. The positions of the primers used to localize the 3′ end of the ORF-E transcript (1 to 5) are dotted lines with an arrow and are in the directions of the respective primers. Potential polyadenylation signals are in bold and underlined. The nucleotide numbers are those proposed for the LR gene (20).
Cloning ORF-E into plasmid vectors.
For identification of polyadenylation of ORF-E, the PstI fragment that contains ORF-E was cloned into pUC19 and pCDNA3.1(+) (Invitrogen, catalogue no. V790-20). Briefly, a plasmid containing the entire LR region (Fig. 1) and each vector was digested to completion with PstI. Each fragment was agarose gel purified and used for ligation. The correct orientation of the PstI insert was determined by restriction digestion. To study the cellular localization of ORF-E expression, ORF-E was cloned into the mammalian expression vector Monster Green Fluorescent Protein (phMGFP vector; Promega, catalogue no. E6421) to generate an ORF-E/GFP fusion protein. PCR was performed on viral genomic DNA with primers that contained unique restriction sites for amplification of ORF-E. The Forward primer is 5′-GCGCGCTAGCCTGAAATATTAATGCCAAAGGC-3′, and the reverse primer is 5′-TAACCCGGGAGGAGGAGGTCCTCCCTCGAGGCA-3′. The forward primer contains the first in-frame ATG for ORF-E (bold ATG) and a unique NheI site. The reverse primer contains ORF-E sequences that encompass the stop site of ORF-E with the actual stop codon (Fig. 4) replaced with three glycines (TAG->GGA GGA GGA) and a unique SmaI site.
PCR was performed as described above. PCR products were digested with SmaI at 25°C for 1 h and then NheI was added, and the reaction was incubated at 37°C for 1 h. phMGFP was digested with SmaI, and then calf intestinal phosphatase (New England Biolabs) and NheI were added. The digested products were isolated by agarose gel electrophoresis, and the PCR product was ligated into the digested phMGFP to generate a plasmid in which ORF-E is 5′ of the GFP ORF (pORF-E-GFP). Sequencing of pORF-E-GFP verified that there were no PCR-induced mutations.
Confocal microscopy of the ORF-E fusion protein.
Confocal microscopy was used to examine the subcellular localization of ORF-E as previously described (16). Cells were plated at a density of 2 × 105/well in 60-mm plastic plates (60 mm/well) 12 to 16 h prior to transfection. Then 2 μg of the designated plasmids was transfected into cells with Transit-LT1 solution as described by the manufacturer (Mirus, Madison, Wis.; catalog no. MIR2300). Briefly, 3 μl of Transit per μg of plasmid was equilibrated with 200 μl of cell medium (minus serum and antibiotics) for 20 min. Then 2 μg of the indicated plasmid was added and incubated for 20 min. The solution was added directly to the cells and incubated for 24 or 48 h. Live cell images were collected with a water immersion lens (40 by 60) under an Olympus FW500/BX60 confocal laser scanning microscope with a simultaneous three-channel display mode, including the 488-nm excitation laser and 522-nm emission filter set for GFP expression, differential interference contrast/transmitted light (grey scale), and overplayed and merged images of the first two.
RESULTS
Identification of a novel ORF upstream of the LR gene.
The LR gene encodes the only viral transcript (LR RNA) that is abundantly expressed in latently infected neurons (20, 23, 24). The LR gene partially overlaps bICP0 and is antisense to the bICP0 transcript (Fig. 1A and B). A fraction of LR RNA is polyadenylated and alternatively spliced in TG, suggesting that this family of transcripts could be translated into more than one protein (10, 13). While performing experiments with the LR mutant virus and plasmids expressing LR gene products, we occasionally obtained results suggesting that other functional genes were located within the LR gene locus. Consequently, we performed computer analysis on the region downstream of the bICP0 gene and in the opposite orientation of the LR gene.
A small ORF was identified within the LR promoter, which was designated ORF-E (Fig. 1C). ORF-E is antisense to the LR transcript and downstream of the bICP0 ORF, but ORF-E does not overlap bICP0 (Fig. 1C). The initiating methionine codon for ORF-E is located at nucleotide 697, and the terminating codon resides at bp 297. The LR promoter contains multiple cis-acting motifs and has a neuron-specific binding domain (NSB; Fig. 1C) (3, 4, 8, 18). The sequences that activate expression of the LR RNA during productive infection contain a long AT-rich motif (40 of 53 nucleotides are A or T). The AT-rich motif is located in a position that could promote expression of a transcript encompassing ORF-E (Fig. 1C). Two putative polyadenylation sites were also detected near the 3′ terminus of ORF-E. HSV-1 expresses a transcript (AL) during productive infection that is antisense to the latency-associated transcript (LAT) (22). Taken together, these observations suggested that a transcript containing ORF-E was expressed in infected cells.
Detection of a transcript that encompasses ORF-E.
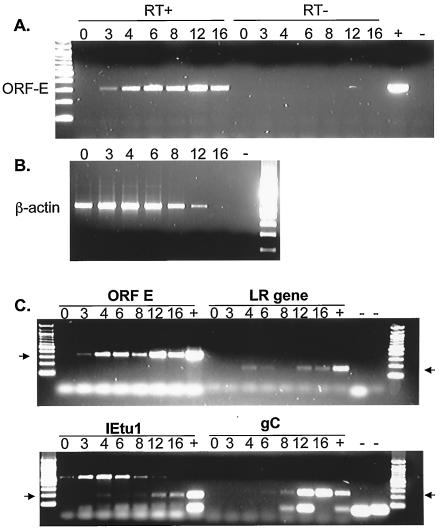
We predicted that if an ORF-E transcript was expressed, it would be small and not very abundant and thus might be difficult to detect by standard Northern blot analysis. Consequently, we developed oligonucleotide primers corresponding to nucleotides 308 to 327 and 598 to 578 (primers 308 and 598 in Fig. 1C). Primer 308 was used for first-strand cDNA synthesis because this would yield a cDNA product that was antisense to LR RNA but would not prime bICP0 cDNA synthesis. MDBK (bovine kidney) cells were infected with BHV-1 for 1 h at 4°C to synchronize virus infection. After washing and addition of fresh medium, the cultures were incubated at 37°C, and total RNA was extracted at different times after infection. An RT-PCR product of the predicted size was detected as early as 3 h after infection (Fig. 2A). The expected size of the RT-PCR product is shown in the + lane. The PCR products were a result of cDNA amplification because reactions without reverse transcriptase did not contain an amplified product (Fig. 2A, RT- lanes). As a control, random-primed RT-PCR was performed with primers that detect β-actin. Figure 2B shows that β-actin was detected from time zero through 12 h postinfection. Because of the viral host shut-off function (11, 12), β-actin RNA expression was not readily detected at 16 h after infection.
FIG. 2.
Detection of ORF-E RNA by RT-PCR. MDBK cells were infected with the Cooper isolate of BHV-1 (multiplicity of infection of 5). Total RNA was extracted at the indicated times postinfection. (A) Strand-specific RT-PCR was conducted with primer 308 (Fig. 1C) to synthesize cDNA. The primers for PCR were 308 and 598. (B) With the same RNA as shown in panel A, cDNA synthesis was primed with random primers, and β-actin cDNA was amplified with the primers described in Materials and Methods. Reactions without reverse transcriptase did not yield specific amplified products with the β-actin primers (data not shown). (C) Random primers were used to synthesize cDNA. PCR was then performed with specific primers for ORF-E (308 and 598), the LR gene (L3B), IETU1, and gC. Lanes +, positive controls consisting of DNA from infected cells; lanes −, no-template controls; RT−, no reverse transcriptase enzyme in the reaction. PCR products were separated on a 2% agarose gel, and the products were visualized with ethidium bromide. The numbers above the lanes indicate hours after infection. The arrows denote products of the expected sizes from the PCR.
The time course of ORF-E RNA expression was compared to that of immediate-early transcription unit 1 (IETU1), the LR gene, and a late transcript encoding glycoprotein C (gC) (Fig. 2C). In agreement with the results in Fig. 2A, the ORF-E transcript was detected 3 h after infection, which was similar to the IETU1 and LR gene transcripts. In contrast, the ORF-E transcript was detected earlier then the late gC transcript. Cycloheximide blocking studies indicated that the ORF-E transcript was not an immediate-early transcript (data not shown). In summary, these results demonstrated that a transcript encompassing ORF-E was detected in productively infected MDBK cells.
Detection of ORF-E RNA in BHV-1-infected calves.
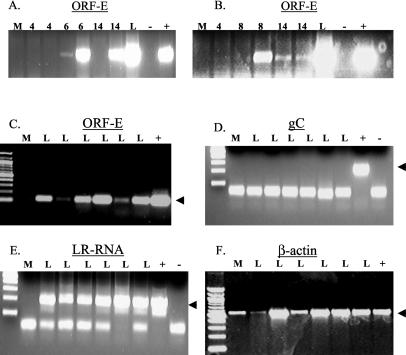
We then tested whether ORF-E RNA was expressed in TG of BHV-1-infected calves. Calves were infected with BHV-1 as previously described (14, 26, 30). TG were collected during acute infection (days 2 to 14) and latency (60 days after infection). Total RNA was prepared as described in Materials and Methods, and RT-PCR was performed with strand-specific primers. Low levels of ORF-E RNA expression were detected in TG of calves during acute infection (Fig. 3A and B). We were unable to amplify ORF-E cDNA prior to day 6 of infection. However the ORF-E transcript was detected at days 6, 8, and 14 postinfection in two of four, three of four, and three of four TG samples, respectively. During latency, ORF-E RNA expression was readily detected in TG of six of six calves latently infected with BHV-1 (Fig. 3C, lanes L) with strand-specific priming for first-strand synthesis. As expected, ORF-E RNA expression was not detected in a mock-infected calf (lane m). All calves in this study were defined as being latently infected because they were not shedding infectious virus from the eyes or nose at 60 days after infection (15).
FIG. 3.
Detection of ORF-E in TG of BHV-1-infected calves. Calves were infected with BHV-1 as previously described (14, 30). At 4, 6, 8, 14, and 60 days postinfection, TG were collected and total RNA was extracted as previously described (6, 13). (A, B, and C) Strand-specific RT-PCR was performed to detect expression of ORF-E RNA with primer 308. ORF-E-specific transcripts were detected by PCR with primers 478 and 598. Panels A and B show a representation of two different experiments. The number of days that the calves were infected is denoted on the top of the panels. L, RNA extracted from TG of latently infected calves (60 days after infection), included to give a relative level of ORF-E RNA detection between acutely and latently infected tissue. (D and F) Random-primed RT-PCR was performed to detect gC and β-actin RNA expression, respectively. (E) Strand-specific RT-PCR was performed to detect expression of the LR RNA with the L3B reverse primer, and PCR was performed with the L3B primers. The arrow denotes the expected PCR product. The bands below the expected products are primer dimers. Lanes: M, mock-infected calf; L, latently infected calves; +, DNA from BHV-1-infected MDBK cells; −, no-template control.
To confirm that these six calves were latently infected with BHV-1, RT-PCR was performed to detect BHV-1 gC. As expected, gC RNA expression was not detected in TG of the six latently infected calves used for this study (Fig. 3D). In contrast, LR RNA expression was detected in all latently infected calves but not in the mock-infected calf (Fig. 3E, lane M). β-Actin primers detected an RT-PCR product in all samples, including that from the mock-infected calf (Fig. 3F). In all of the reactions, no PCR product was obtained when reverse transcriptase was omitted from the first-strand synthesis (data not shown). In summary, these results demonstrated that the ORF-E transcript was consistently expressed in latently infected calves. The ORF-E transcript was not detected as consistently in TG of acutely infected calves relative to latently infected calves.
Localization of the 5′ terminus of ORF-E RNA.
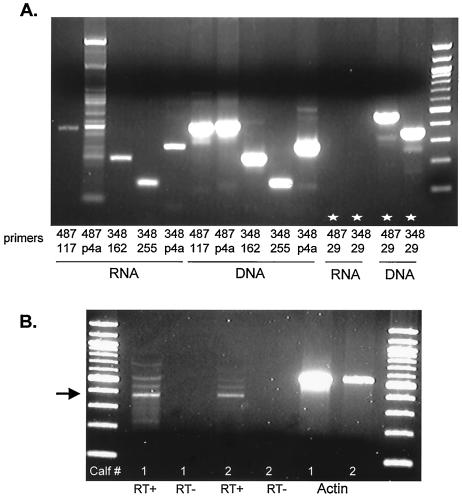
5′ Rapid amplification of cDNA ends was initially used to map the 5′ terminus. Due to the extensive GC content of ORF-E sequences, we had difficulties obtaining unambiguous results. As an alternative approach, a single reverse primer that was within 500 bp of the expected 5′ terminus was used to generate first-strand cDNA. Different primers were then synthesized to “walk” down to the 5′ terminus of the ORF-E transcript (for the locations of the primers used for this study, see Fig. 4). We were confident that amplified products were derived from the ORF-E transcript because a strand-specific primer was used. MDBK cells were infected with BHV-1, and total RNA was extracted at 6 h postinfection (Fig. 5A). Figure 5A shows that both primers used for the first-strand synthesis (primers 348 and 487) yielded the expected amplified products. When DNA was the template for PCR, all primers produced the expected product. However, when RNA was used as the template, this was not the case. For example, when primer 29 (Fig. 4) was used in the PCR, an amplified product was not detected when RNA was the original template, but the expected product was detected with DNA as the template (last four lanes in Fig. 5A, denoted by stars).
FIG. 5.
Mapping the 5′ region of the ORF-E RNA. (A) MDBK cells were infected with BHV-1, and total RNA or DNA was extracted at 6 h postinfection. Strand-specific cDNA synthesis was performed with 1 μg of total RNA with either primer 487 or 348. PCR was performed with the indicated primers. (B) Calves were infected with BHV-1, and TG were extracted at either 6 days postinfection (calf 1) or during latency at 60 days postinfection (calf 2). Strand-specific cDNA synthesis was performed with 2 μg of total RNA and primer 487. PCR amplification of cDNA products was performed with primers 487 and 348. The arrow denotes the expected band.
To determine if the 5′ terminus was the same in TG of latently infected calves, RNA was extracted from TG of infected calves and then subjected to a modified primer extension assay. Primer 487 was used for first-strand synthesis (Fig. 5B). For PCR, primer 487 and another primer of the same sense (primer 348) were used to generate a PCR product. A PCR product of about 390 bp was detected in TG of calves that were latently (calf 1) and acutely (calf 2) infected. These products were derived from cDNA because the products were not detected in reactions without reverse transcriptase. In summary, these studies demonstrated that the 5′ terminus of ORF-E RNA was localized to sequences that are −256 to −204 bp from the ORF-E initiating ATG codon (Fig. 4).
Localization of the 3′ terminus of ORF-E RNA.
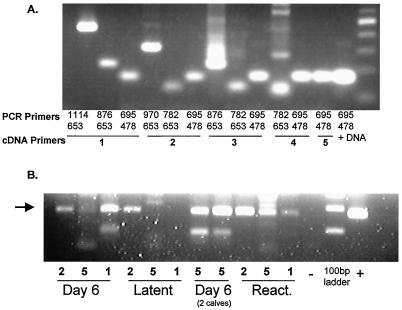
Initially, rapid amplification of cDNA ends was used to map the 3′ terminus of the ORF-E RNA. However, inconsistent results were obtained, and as described below, the discrepancy occurred in part because of different 3′ termini. A similar method was then employed for localizing the 5′ terminus of the ORF-E RNA (see Fig. 4 for the locations of the primers). First-strand cDNA synthesis was conducted with antisense primers. In BHV-1-infected MDBK cells, all primers generated an RT-PCR product (Fig. 6A). Although there were sequences (GGGCTT, Fig. 4) that could serve as polyadenylation signals (1), this result suggested that the polyadenylation sites were further downstream.
FIG. 6.
Localization of the 3′ terminus of the ORF-E RNA. (A) MDBK cells were infected with BHV-1, and total RNA was extracted at 6 h postinfection. Strand-specific cDNA synthesis was performed with 1 μg of total RNA with the indicated primers (1, 2, 3, 4, or 5) that are shown in Fig. 4. PCR was performed with the indicated primers. (B) Calves were infected with BHV-1 as described in Materials and Methods, and TG were extracted at the indicated times postinfection. Latently infected calves (60 days postinfection) were treated with 100 mg of dexamethasone as described previously to induce reactivation from latency (15), and RNA was extracted 21 h later (React.). The numbers above the line indicate which primer was used for strand-specific cDNA synthesis. PCR was performed with primers 478 and 695. The arrow denotes the expected size of the PCR product.
Additional 3′ terminus mapping studies were performed with total RNA prepared from TG of BHV-1-infected calves. The procedure described for Fig. 6A was used to generate cDNA and then PCR was performed. The numbers under each lane in Fig. 6B represent the primers used in the reverse transcriptase reaction. Regardless of whether RNA was prepared from TG of acutely infected calves or latently infected calves, similar results were obtained with primer 2 (Fig. 6B). Primer 1 was unable to amplify the ORF-E-specific cDNA that was generated from total RNA prepared from TG of latently infected calves (a representative sample is shown). However, primer 1 did amplify ORF-E-specific cDNA in acutely infected calves (day 6) and in latently infected calves that were treated with dexamethasone for 21 h to initiate reactivation from latency. In summary, these results suggested that the 3′ terminus of ORF-E RNA in TG was different at different times after BHV-1 infection. During acute infection and when reactivation was initiated with dexamethasone, the ORF-E RNA was longer at its 3′ terminus than it was during latency.
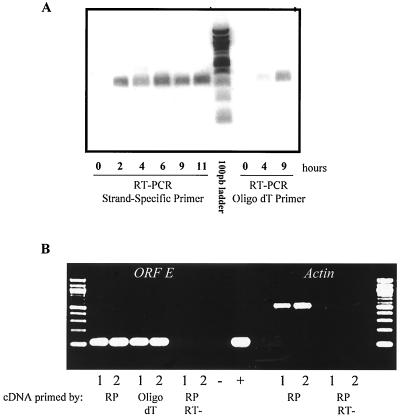
Additional studies were subsequently performed to determine whether the ORF-E RNA was polyadenylated and which polyadenylation signal was used. To this end, MDBK cells were infected with BHV-1, and total RNA was extracted. First-strand synthesis was oligo(dT) primed, and PCR was performed with primers 308 and 598. A PCR product was detected at 4 and 9 h after infection (Fig. 7A, last two lanes). With the same RNA, strand-specific RT-PCR was performed and results similar to those shown in Fig. 2 were obtained. The polyadenylated transcript detected in the oligo(dT)-primed reaction was ORF-E and not the LR gene because the 5′ terminus of the LR RNA is downstream of primer 308 during productive infection (13) (Fig. 1B). The PCR product was not likely bICP0 because the polyadenylation signal for bICP0 is located at nucleotide 960 (see Fig. 1 and 4) (31).
FIG. 7.
Is the ORF-E transcript polyadenylated in transfected cells? (A) MDBK cells were infected with BHV-1, and total RNA was prepared at the indicated times. RT-PCR was performed with strand-specific or oligo(dT) primers to synthesize cDNA. PCR was performed with the same primers (308 and 598) for all reactions. (B) The PstI fragment shown in Fig. 1 was cloned into either pUC19 or pCDNA3.1. Neuro-2A cells were transfected with the designated plasmid, and at 48 h posttransfection total RNA was prepared. cDNA was synthesized with random primers or oligo(dT) primers. PCR was then performed with the same primers for all reactions (478 and 598). Lanes 1, cells transfected with pUC19-ORF-E. Lanes 2, cells transfected with pCDNA3.1-ORF-E. RT-, reverse transcriptase was not added to the reaction; −, no-template control; +, BHV-1 DNA. As another control, β-actin primers were used in the PCR on the same reactions.
Transient-transfection assays were used to further test whether the ORF-E transcript could actually terminate at the first polyadenylation signal at the GGGCTT motif near nucleotides 20 to 30 (Fig. 4). The PstI fragment (Fig. 1C) was cloned into pUC19 and the mammalian vector pCDNA3.1. pUC19 does not have a known mammalian polyadenylation signal, whereas pCDNA3.1 contains the simian virus 40 polyadenylation signals. We predicted that if a pUC19 plasmid containing the ORF-E gene was transfected into mammalian cells, an RT-PCR product would be amplified from oligo(dT)-primed RT-PCR if polyadenylation occurred at the GGGCTT motif. We also expected that the ORF-E transcript would be polyadenylated when cloned into a pcDNA3.1-based vector. These two plasmids were transfected into neuro-2A cells, and total RNA was extracted 48 h later. Random primers and oligo(dT) were used in the first-strand synthesis reactions. Both vectors expressed an ORF-E transcript that was detected by either random priming or oligo(dT) (Fig. 6B). These products were derived from cDNA because when reverse transcriptase was omitted from the reaction, the PCR product was not detected. In summary, the ORF-E transcript appears to utilize different polyadenylation sites during latency versus productive infection.
Localization of the ORF-E protein in transfected cells.
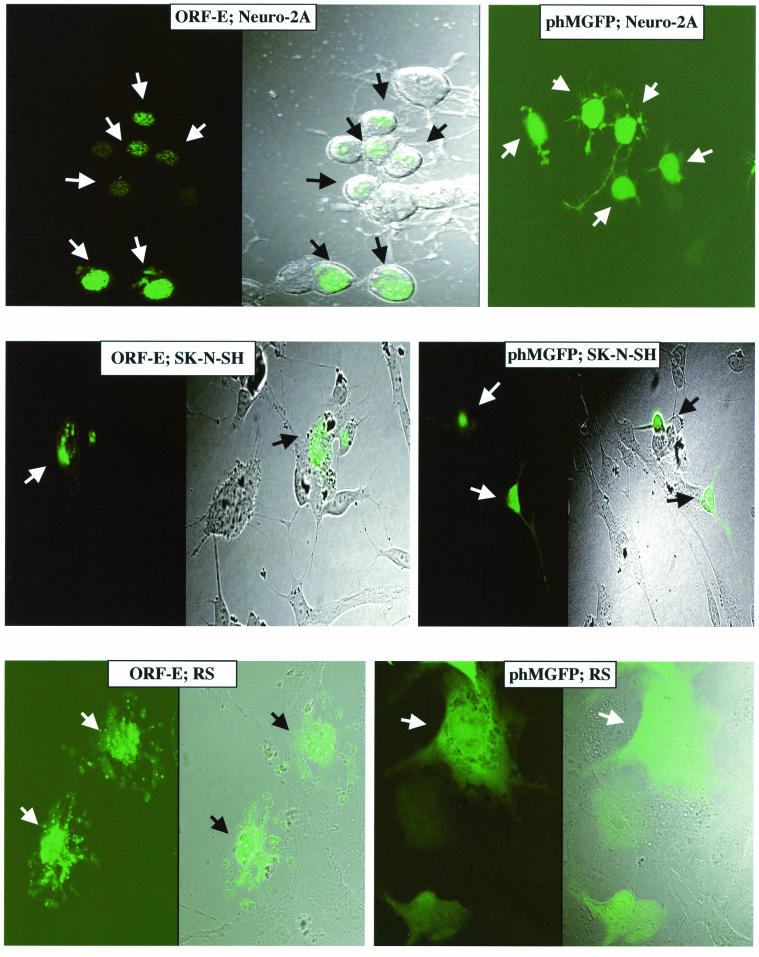
Studies were performed to test whether ORF-E was produced in transfected cells and, if so, to examine its subcellular localization. The putative protein-coding sequences of ORF-E (Fig. 4) were cloned upstream of the GFP coding sequences in phMGFP. The ORF-E/GFP plasmid and the blank GFP vector were transfected into two nonneuronal (MDBK and rabbit skin) and two neuronal (neuro-2A and SK-N) cell lines. The ORF-E/GFP fusion protein was localized to the nucleus when expressed in neuro-2A cells and SK-N cells (Fig. 8). The expression of the fusion protein was punctate and appeared to be localized to distinct nuclear regions. GFP expression was not detected in the projections (dendrites) of the neuron-like cells when transfected with ORF-E/GFP. In contrast, neuro-2A and SK-N-SH cells transfected with the blank GFP expression vector (phMGFP) contained GFP throughout the cell, including the dendrites. When the ORF-E/GFP fusion was transfected into RS cells, the expression pattern was still punctate, but the fusion protein was detected in the nucleus and cytoplasm. A similar expression pattern was observed when MDBK cells were transfected with the ORF-E/GFP fusion vector (data not shown). In summary, this study suggested that ORF-E was localized to the nucleus of neuronal cells.
FIG. 8.
Localization of ORF-E expression. The designated cells were transfected with 2 μg of the ORF-E/GFP plasmid or the GFP blank expression vector phMGFP. At 24 h after transfection, cells were viewed by confocal microscopy. The panels on the left are fluorescent images, and those on the right are differential interference/phase contrast images (see Materials and Methods). The magnification was 160× for SK-N-SH cells and 240× for neuro-2A and RS cells. The arrows denote GFP-positive cells. These images are representative of many cells that were examined.
DISCUSSION
This report describes the identification of a transcript that contains a 135-amino-acid open reading frame, ORF-E. The ORF-E transcript is antisense to the LR gene and was consistently expressed during latency. Although we were unable to detect the ORF-E transcript by Northern analysis, the ORF-E transcript was readily detected by RT-PCR with strand-specific primers used to initiate cDNA synthesis. This is similar to the herpes simplex virus type 1 AL gene and the UL43 gene, which were not detected by Northern blotting but were detected by RT-PCR (5, 22). When ORF-E was fused to GFP, this fusion protein was expressed in the nucleus of transfected neuronal cells.
The herpes simplex virus type 1 AL transcript (antisense to LAT) contains an ORF that is expressed upstream and antisense of herpes simplex virus type 1 LAT (22). The AL transcript overlaps 198 nucleotides of the LAT promoter and 158 nucleotides of the 5′ end of the primary 8.3-kb LAT transcript. The sequence of the AL ORF is highly conserved among different herpes simplex virus type 1 isolates that have been sequenced, suggesting that this protein is expressed. The finding that sera from infected rabbits recognize peptides from the AL ORF (22) supports the prediction that this protein is expressed. Although the AL gene and the ORF-E gene occupy similar locations on their respective genomes, there is no obvious amino acid sequence similarity between the putative ORF-E protein and the AL protein (data not shown). Among the BHV-1 isolates that have been sequenced, the ORF-E amino acid sequences are highly conserved. In contrast, an ORF-E gene was not detected at the same location on the BHV-5 genome (9). Although the genomes of BHV-1 and BHV-5 are very similar, BHV-5 causes severe central nervous disorders in cattle, whereas BHV-1 primarily induces upper respiratory tract disorders and conjunctivitis. The most dramatic difference in the genomes of BHV-1 and BHV-5 was the LR gene locus, including the ORF-E coding sequences. Since it was hypothesized that differences in the biological properties of BHV-1 and BHV-5 are linked to differences in the LR gene locus (9), it is logical to speculate that ORF-E may contribute to these differences.
The 5′ terminus of the ORF-E transcript was localized to sequences that were adjacent to the 3′ terminus of bICP0 (Fig. 4). The putative ATG for ORF-E is more than 200 bp downstream from this region. This result was unexpected because the AT-rich motif that is present within the LR promoter (Fig. 1C) was downstream of the 5′ terminus of the ORF-E transcript, suggesting that this was the ORF-E promoter. However, our results suggested that ORF-E promoter sequences are located within the bICP0 coding sequences. The finding that more than one 3′ end exists for the ORF-E transcript was also not expected. We suggest that virus-encoded and induced functions and neuron-specific factors regulate the selection of polyadenylation sites. It is also possible that a family of ORF-E transcripts are synthesized, which would have complicated mapping the 5′ and 3′ termini. This possibility is supported by the fact that the LR gene encodes a family of transcripts, some of which are alternatively spliced (10, 13). Since ORF-E and the LR gene are both consistently expressed during latency, we suggest that these genes regulate each other's expression and/or work in conjunction to promote latency. Studies to address what role the ORF-E gene plays in the latency reactivation cycle of BHV-1 are in progress.
We predict that ORF-E encodes a regulatory protein in neurons because the ORF-E/GFP fusion protein was primarily detected in the nucleus of transfected neuronal cells (Fig. 8). The cells transfected with ORF-E/GFP continued to grow (data not shown). In contrast, the LR gene induced cell cycle arrest (25), and the bICP0 gene is cytotoxic (16). Although we do not know the function of the ORF-E gene, it seems clear that it has novel functions relative to the LR gene and bICP0. The LR gene has antiapoptotic activity (7, 21). In contrast, our studies indicate that the ORF-E gene does not protect cells against an apoptotic insult in vitro (data not shown). Sequence analysis of the putative protein encoded by ORF-E suggested that it can be myristoylated, glycosylated, and phosphorylated. Amino acid sequences near the N terminus of ORF-E can be found in other proteins that have anti-inflammatory properties. Infiltration of immune cells into TG plays an active role in maintaining latency (19), suggesting that the ORF-E protein may dampen the effects of infiltrating immune cells on infected neurons. We recently expressed the ORF-E protein in a baculovirus system, and thus we should be able to generate antibodies that will be useful for detecting ORF-E protein expression after infection.
Acknowledgments
This study was supported by two USDA grants (2002-02450 and 2003-02213) and a Public Health Service grant (1P20RR15635).
We thank Terri Fangman for help in performing confocal microscopy studies.
REFERENCES
- 1.Beaudoing, E., S. Freier, J. Wyatt, J. Claverie, and D. Gautheret. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10:1001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowland, S. L., and P. E. Shewen. 2000. Bovine respiratory disease: commercial vaccines currently available in Canada. Can. Vet. J. 41:33-48. [PMC free article] [PubMed] [Google Scholar]
- 3.Bratanich, A. C., N. D. Hanson, and C. J. Jones. 1992. The latency-related gene of bovine herpesvirus 1 inhibits the activity of immediate-early transcription unit 1. Virology 191:988-991. [DOI] [PubMed] [Google Scholar]
- 4.Bratanich, A. C., and C. J. Jones. 1992. Localization of cis-acting sequences in the latency-related promoter of bovine herpesvirus 1 which are regulated by neuronal cell type factors and immediate-early genes. J. Virol. 66:6099-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter, K., P. Ward, and B. Roizman. 1996. Charaterization of the products of the U(L)43 gene of herpes simplex virus 1: potential implications for regulation of gene expression by antisense transcription. J. Virol. 70:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chomczynski, P., and N. Sacchi. 1987. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 7.Ciacci-Zanella, J., M. Stone, G. Henderson, and C. Jones. 1999. The latency-related gene of bovine herpesvirus 1 inhibits programmed cell death. J. Virol. 73:9734-9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delhon, G., and C. Jones. 1997. Identification of DNA sequences in the latency related promoter of bovine herpes virus type 1 which are bound by neuronal specific factors. Virus Res. 51:93-103. [DOI] [PubMed] [Google Scholar]
- 9.Delhon, G., M. P. Moraes, Z. Lu, C. L. Afonso, E. F. Flores, R. Weiblen, G. F. Kutish, and D. L. Rock. 2003. Genome of bovine herpesvirus 5. J. Virol. 77:10339-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devireddy, L. R., and C. Jones. 1998. Alternative splicing of the latency-related transcript of bovine herpesvirus 1 yields RNAs containing unique open reading frames. J. Virol. 72:7294-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everly, D., and G. Read. 1999. Site-directed mutagenesis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): analysis of functional differences between HSV type 1 (HSV-1) and HSV-2 alleles. J. Virol. 73:9117-9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gopinath, R., A. Ambagala, S. Hinkley, and S. Srikumaran. 2002. Effects of virion host shut-off activity of bovine herpesvirus 1 on MHC class I expression. Viral Immunol. 15:595-608. [DOI] [PubMed] [Google Scholar]
- 13.Hossain, A., L. M. Schang, and C. Jones. 1995. Identification of gene products encoded by the latency-related gene of bovine herpesvirus 1. J. Virol. 69:5345-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inman, M., L. Lovato, A. Doster, and C. Jones. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J. Virol. 75:8507-8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inman, M., L. Lovato, A. Doster, and C. Jones. 2002. A mutation in the latency related gene of bovine herpesvirus 1 interferes with the latency reactivation cycle of latency in calves. J. Virol. 76:6771-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inman, M., Y. Zhang, V. Geiser, and C. Jones. 2001. The zinc ring finger in the bICP0 protein encoded by bovine herpes virus-1 mediates toxicity and activates productive infection. J. Gen. Virol. 82:483-492. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, Y., A. Hossain, M. T. Winkler, T. Holt, A. Doster, and C. Jones. 1998. A protein encoded by the latency-related gene of bovine herpesvirus 1 is expressed in trigeminal ganglionic neurons of latently infected cattle and interacts with cyclin-dependent kinase 2 during productive infection. J. Virol. 72:8133-8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, C., G. Delhon, A. Bratanich, G. Kutish, and D. Rock. 1990. Analysis of the transcriptional promoter which regulates the latency-related transcript of bovine herpesvirus 1. J. Virol. 64:1164-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khanna, K., R. Bonneau, P. Kinchington, and R. Hendricks. 2003. Herpes Simplex virus-specific memory CD8+ T cells are selectively activated and retained in latently infected sensory ganglia. Immunity 18:593-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutish, G., T. Mainprize, and D. Rock. 1990. Characterization of the latency-related transcriptionally active region of the bovine herpesvirus 1 genome. J. Virol. 64:5730-5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovato, L., M. Inman, G. Henderson, A. Doster, and C. Jones. 2003. Infection of cattle with a bovine herpesvirus 1 strain that contains a mutation in the latency-related gene leading to increased apoptosis in trigeminal ganglia during the transition from acute infection to latency. J. Virol. 77:4848-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perng, G.-C., B. Maguen, L. Jing, K. R. Mott, N. Osorio, S. M. Slanina, A. Yukht, H. Ghiasi, A. B. Nesburn, M. Inman, G. Henderson, C. Jones, and S. L. Wechsler. 2002. A novel herpes simplex virus type 1 transcript antisense to the 5′ end of latency-associated transcript produces a protein in infected rabbits. J. Virol. 76:8003-8010. [DOI] [PMC free article] [PubMed]
- 23.Rock, D., J. Lokensgard, T. Lewis, and G. Kutish. 1992. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J. Virol. 66:2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rock, D. L., S. L. Beam, and J. E. Mayfield. 1987. Mapping bovine herpesvirus type 1 latency-related RNA in trigeminal ganglia of latently infected rabbits. J. Virol. 61:3827-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schang, L. M., A. Hossain, and C. Jones. 1996. The latency-related gene of bovine herpesvirus 1 encodes a product which inhibits cell cycle progression. J. Virol. 70:3807-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schang, L. M., and C. Jones. 1997. Analysis of bovine herpesvirus 1 transcripts during a primary infection of trigeminal ganglia of cattle. J. Virol. 71:6786-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheffy, B. E., and D. H. Davies. 1972. Reactivation of a bovine herpesvirus after corticosteroid treatment. Proc. Soc. Exp. Biol. Med. 140:974-976. [DOI] [PubMed] [Google Scholar]
- 28.Tikoo, S. K., M. Campos, and L. A. Babiuk. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv. Virus Res. 45:191-223. [DOI] [PubMed] [Google Scholar]
- 29.Winkler, M. T., A. Doster, J. H. Sur, and C. Jones. 2002. Analysis of bovine trigeminal ganglia following infection with bovine herpesvirus 1. Vet. Microbiol. 86:139-155. [DOI] [PubMed] [Google Scholar]
- 30.Winkler, M. T. C., A. Doster, and C. Jones. 2000. Persistence and reactivation of bovine herpesvirus 1 in the tonsils of latently infected calves. J. Virol. 74:5337-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wirth, U. V., C. Fraefel, B. Vogt, C. Vlcek, V. Paces, and M. Schwyzer. 1992. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J. Virol. 66:2763-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]