Abstract
Influenza virus neuraminidase (NA), a type II transmembrane glycoprotein, possesses receptor-destroying activity and thereby facilitates virus release from the cell surface. Among the influenza A viruses, both the cytoplasmic tail (CT) and transmembrane domain (TMD) amino acid sequences of NA are highly conserved, yet their function(s) in virus biology remains unknown. To investigate the role of amino acid sequences of the CT and TMD on the virus life cycle, we systematically mutagenized the entire CT and TMD of NA by converting two to five contiguous amino acids to alanine. In addition, we also made two chimeric NA by replacing the CT proximal one-third amino acids of the NA TMD [NA(1T2N)NA] and the entire NA TMD (NATRNA) with that of human transferrin receptor (TR) (a type II transmembrane glycoprotein). We rescued transfectant mutant viruses by reverse genetics and examined their phenotypes. Our results show that all mutated and chimeric NAs could be rescued into transfectant viruses. Different mutants showed pleiotropic effects on virus growth and replication. Some mutants (NA2A5, NA3A7, and NA4A10) had little effect on virus growth while others (NA3A2, NA5A27, and NA5A31) produced about 50- to 100-fold-less infectious virus and still some others (NA5A14, NA4A19, and NA4A23) exhibited an intermediate phenotype. In general, mutations towards the ectodomain-proximal sequences of TMD progressively caused reduction in NA enzyme activity, affected lipid raft association, and attenuated virus growth. Electron microscopic analysis showed that these mutant viruses remained aggregated and bound to infected cell surfaces and could be released from the infected cells by bacterial NA treatment. Moreover, viruses containing mutations in the extreme N terminus of the CT (NA3A2) as well as chimeric NA containing the TMD replaced partially [NA(1T2N)NA] or fully (NATRNA) with TR TMD caused reduction in virus growth and exhibited the morphological phenotype of elongated particles. These results show that although the sequences of NA CT and TMD per se are not absolutely essential for the virus life cycle, specific amino acid sequences play a critical role in providing structural stability, enzyme activity, and lipid raft association of NA. In addition, aberrant morphogenesis including elongated particle formation of some mutant viruses indicates the involvement of NA in virus morphogenesis and budding.
Influenza viruses are enveloped, segmented negative-strand RNA viruses belonging to the Orthomyxoviridae family (27). Among the three known types of influenza viruses (A, B, and C), influenza A viruses are the most important from the standpoint of pathogenicity in humans and animals. Influenza A viruses possess two surface spike glycoproteins, hemagglutinin (HA) and neuraminidase (NA) (27). HA and NA represent about 85 and 15% of the surface proteins, respectively. HA is visible by electron microscopy as spikes outside the virus envelope. The HA protein, a type I transmembrane protein, is responsible for binding to the cell surface sialic acid (the receptor), eliciting neutralizing antibodies, and mediating virus entry into the cell by fusion of the viral membrane with the endosomal membrane (46). HA is synthesized as a single polypeptide (HA0) and then cleaved into HA1 and HA2 subunits by host proteases (22). Such a cleavage is essential for viral infectivity because in the acidic pH of the endosome it exposes a membrane fusion peptide at the amino terminus of the HA2 subunit required for membrane fusion (26).
NA, a type II transmembrane glycoprotein, possesses an enzyme activity that removes sialic acid from host cell glycocongugates as well as newly synthesized viral proteins. NA is a homotetramer and possesses three spatially distinct domains, the ectodomain, transmembrane domain (TMD), and cytoplasmic tail (CT). The globular ectodomain with a stalk possesses the site for enzyme activity. The structure of the globular domain including the active enzyme site has been solved by X-ray crystallography (1). The TMD, which functions as both translocation signal and anchor domain for membrane binding (8), interacts with lipid rafts and also possesses the determinants for apical sorting in polarized epithelial cells (5, 24). The short CT contains six highly conserved amino acids (7) with no known function(s) in virus biology.
It has been shown previously that NA is not required for virus replication or budding except in the final step of releasing the virus particles from the cell surface sialic acid receptor as well as preventing aggregation among the progeny virus particles (29). When the NA activity was eliminated either by antibodies (11), inhibitors (16, 35), ts mutation at the restricted temperature (37, 41), or deletion (16), sialic acids were not removed from the cell surface or viral envelope, and after budding, progeny virus particles remained attached to the cell surface and clumped together. This block in the virus replication cycle could be released by adding bacterial NA in the external medium (37). In addition, NA has been proposed to function in viral entry in infected animals by removing the sialic acids present in the mucin covering the epithelial surface of respiratory mucosa (10). Moreover, in some viruses such as neurovirulent A/WSN/33 virus, NA has been shown to bind and convert plasminogen to active plasmin, which cleaves HA0 to HA1 and HA2 and thereby enhances virus pathogenicity and neurovirulence (14, 15). Furthermore, NA has been shown to facilitate fusion between liposomes containing NA and erythrocytes (18, 19), but HA alone was found necessary and sufficient for pH-dependent fusion in CV1 cells, suggesting no role for NA in fusion (45).
Little information is available about the function of NA TMD and CT amino acid sequences in virus biology, although they are highly conserved among influenza A viruses. Using chimeric constructs and mutation, it was shown that NA TMD, but not the CT, possesses the determinants for apical sorting in polarized MDCK cells (24). Furthermore, the NA TMD was responsible for interaction with nonionic detergent-resistant lipid rafts (24) and that ectodomain-proximal amino acids of TMD were critical for lipid raft association (5). Using coexpression of M1 and NA or HA in HeLa cells, it was shown that both HA and NA interact with M1 and, further, CT and TMD of HA were responsible for interacting with M1 (2).
Although the 6 amino acids (aa) in the CT of NA are extremely conserved among the influenza A viruses (7), little is known about the requirement of the tail or the conservation of the amino acid sequences in virus biology. Earlier studies with the tail deletion NA mutant (NAt−) showed that NAt− virus can be rescued but was attenuated in growth (13) and that some virus particles were elongated in shape (30). These properties were further investigated by deletion of both the HA and NA tails in HAt−/NAt− mutant virus. Deletion of the CT of both HA and NA led to the formation of bizarre filamentous virus particles (21). Authors further observed that the deletion of CT caused a reduction in raft association, and they concluded that reduced raft association was responsible for budding defects (47). However, deletion of the complete CT could cause structural perturbation in the TMD, leading to instability in protein structure and reduced raft association. Therefore, it becomes important to define the functional significance of the amino acid sequences of CT and TMD of NA in the virus life cycle. We have therefore made site-specific mutations in both the TMD and CT of NA, rescued transfectant mutant viruses by reverse genetics, and investigated the properties of mutant viruses. Our results show that the CT and TMD of NA play an important role in virus budding. Some mutants affected the virus life cycle by reduced enzyme activity, others did so by affecting raft association, and still others affected virus budding independent of NA activity and raft association.
MATERIALS AND METHODS
Cell cultures.
MDCK cells, obtained from the American Type Culture Collection, were maintained in Dulbecco modified Eagle's medium (DMEM; Invitrogen Corporation, Grand Island, N.Y.) supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). Costar polycarbonate insert filter units (24-mm diameter) with a pore size of 3.0 μm (Corning Costar, Cambridge, Mass.) were used for growing polarized MDCK cells. Cells (1.2 × 106) were seeded on filters with 2.5 and 1.5 ml of growth medium in basolateral and apical chambers, respectively, and grown for about 60 h to form tight monolayers. The polarity of cell monolayers and the formation of tight junctions were determined by measuring the transepithelial resistance (3) with an EVOM voltmeter (World Precision Instruments, New Haven, Conn.).
Constructions of different mutant NA cDNAs.
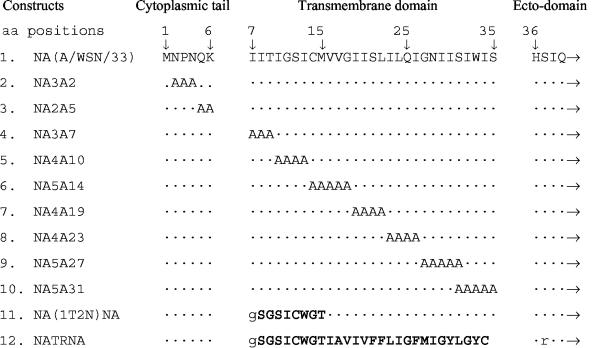
Alanine mutations in the CT and TMD of NA (A/WSN/33) were introduced either in the Pol I (34) or in the Pol I-Pol II (17) plasmid containing NA cDNA by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). Primers used in these reactions will be provided upon request. To construct NATRNA, the entire NA TMD was swapped (by using BamHI and EcoRI restriction sites) with the TR (human transferrin receptor). For NA(1T2N)NA, the N-terminal one-third portion of the NA TMD was swapped (by using BamHI and BstXII restriction sites) with the N-terminal one-third portion of the TR TMD (Fig. 1).
FIG. 1.
Schematic presentation of NA mutants. Sets of 2 to 5 aa in the NA CT or TMD were replaced with alanines. Chimeric NA TMDs [NA(1T2N)NA and NATRNA] were constructed by swapping either the first 9 aa or the entire NA TMD with that of the TR TMD. ·, amino acids from NA sequences. Amino acids shown in boldface type are from TR sequences. Amino acids in lowercase type are due to created restriction enzyme sites. Note that the TR TMD contains 28 aa instead of the 29 aa of the NA TMD. Numbers above amino acid sequences represent amino acid positions (indicated by ↓) of the WT NA peptide.
Generation of transfectant NA mutant viruses.
To generate NA5A14, NA4A19, NA5A27, NA(1T2N)NA, and NATRNA (Fig. 1) transfectant viruses, the mutant NA cDNAs in the Pol I plasmid containing the RNA polymerase I promoter and terminator were transfected in 293T cells with the other 16 plasmids as described previously (34). To generate NA3A2, NA2A5, NA3A7, NA4A10, NA4A23, and NA5A31 (Fig. 1) transfectant viruses, eight plasmids (seven wild-type [WT] [HA, M, NP, NS, PA, PB1, PB2] and one mutated NA Pol I-Pol II construct) were transfected in 293T cells as described previously (17, 20). The transfectant viruses were amplified in MDCK cells. The mutations in transfectant viruses were confirmed by sequencing the reverse transcription-PCR products of NA virus RNA.
Virus propagation and plaque assay.
For stock virus preparation, WT (A/WSN/33) or mutant virus was grown in MDCK cells as described previously (3). Briefly, MDCK cell monolayers were infected with viruses at a multiplicity of infection (MOI) of 0.001 by using virus dilution buffer (phosphate-buffered saline [PBS] supplemented with 0.5 mM MgCl2, 1.0 mM CaCl2, 50 μg of DEAE-dextran/ml and 0.2% bovine serum albumin [BSA]) and maintained at 33°C in virus growth medium (VGM; modified Eagle's medium containing 4% BME vitamins, 10 mM HEPES, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 0.2% BSA, 1.6 mg of NaHCO3/ml, and 15 μg of DEAE-dextran/ml) in the presence of 0.5 μg of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma Chemical Co., St. Louis, Mo.)/ml. Plaque assays were done with 24-h-old MDCK cell monolayers (35-mm-diameter dish) in the presence of 0.5 μg of TPCK-treated trypsin (unless stated otherwise)/ml in agarose overlay medium (VGM containing 0.6% low-melting-point agarose; Sigma) as reported previously (44).
Preparation of 35S-labeled virus.
Virus-infected MDCK cells were labeled at 4 h postinfection (p.i.) for 12 h with 35S-protein labeling mix (Perkin-Elmer Life Science, Inc., Boston, Mass.) in 2:8 VGM (VGM containing 20% modified Eagle's medium and 80% Met−-Cys− DMEM). At 16 h p.i., medium was harvested and clarified by low-speed centrifugation and virions were pelleted by ultracentrifugation (2.5 h, 35,000 rpm with an SW55 Ti rotor) through a 25% sucrose cushion and resuspended in TNE buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, and 1.0 mM EDTA).
endo H or PNGase digestion of NA.
Lysates of virus-infected cells or purified virions were immunoprecipitated with specific antibodies; immune complexes were recovered by protein A-Sepharose beads and eluted in denaturing buffer by boiling for 10 min. Aliquots of the samples were treated with either 1,000 U of endo-β-N-acetylglucosaminidase Hf (endo Hf; NEB, Beverly, Mass.) or 500 U of peptide N-glycosidase (PNGase F; NEB) or were mock treated for 3 h.
Fluorometric sialidase activity assay.
The sialidase activity of purified virus particles was assayed with 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (Sigma) as a substrate as described previously (23). Briefly, 5 μl of 0.2 mM 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid in 0.2 mM potassium phosphate (pH 5.9), was added to an equal volume of purified virus samples in TNE and incubated for 30 min at 37°C. The reaction was stopped by adding 200 μl of 0.1 M glycine-25% ethanol (pH 10.7), and the fluorescence was measured at an excitation wavelength of 360 nm and an emission wavelength of 465 nm. Each reaction was performed in triplicate.
Assays for Triton X-100 insolubility.
The Triton X-100 insolubility of proteins expressed at the cell surface was determined as described previously (3, 47) with the following modification. At 4 h p.i. (2.5 h p.i. for vesicular stomatitis virus [VSV]-infected cells), virus-infected cells were labeled with 35S-protein labeling mix in Met−/Cys− DMEM for 60 min followed by a 90-min chase. Cell surface proteins were assayed by surface-specific biotinylation as described previously (3). Briefly, the biotin reaction was done three times for 20 min (each) at 4°C with 0.6 μg of Sulfo-NHS-LC-Biotin (Pierce Chemical Co., Rockford, Ill.)/ml in PBS containing 0.1 mM CaCl2 and 1.0 mM MgCl2. Unreacted biotin was removed by two washes of DMEM containing 1% BSA. Cells were then extracted with 1% Triton X-100 (Roche, Mannheim, Germany) in TNE on ice for 10 min. The supernatant was collected as the soluble fraction. The insoluble cell layer left on the dish was lysed in radioimmunoprecipitation assay (RIPA) buffer, and the clarified lysates were collected as the Triton X-100-insoluble fraction. Both soluble and insoluble surface proteins were immunoprecipitated by respective antibodies, analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and autoradiographed. Radioactive protein bands were detected by storage phosphorimaging with a PhosphorImager (Typhoon 9410) and quantified by using the ImageQuant software program (Amersham Biosciences, Piscataway, N.J.).
To examine the Triton X-100 insolubility of different virion proteins, purified 35S-labeled virus particles were extracted with 0.1% Triton X-100 in 100 μl of TNE buffer for 10 min on ice (3) and diluted in 5 ml of TNE buffer, followed by ultracentrifugation (2 h, 40,000 rpm with an SW55 Ti rotor) to separate the soluble and insoluble fractions. Soluble and insoluble proteins were adjusted to 1× RIPA buffer, immunoprecipitated by respective antibodies, treated with PNGase F, analyzed by SDS-PAGE, autoradiographed, and quantified. Since 1% Trition X-100 made virion proteins highly soluble, 0.1% Triton X-100 was used, as reported earlier (3, 47).
Thin-section electron microscopy.
Thin-section electron microscopy was done as described previously (3). Briefly, MDCK cell monolayers grown on polycarbonate filters (3.0-μm pore size) were infected with different viruses at an MOI of 3.0 from the apical side. At 12 h p.i., virus-infected cells were cross-linked in 2% glutaraldehyde in PBS++ (PBS containing 0.5 mM MgCl2 and 1.0 mM CaCl2) and postfixed with 1% OsO4 in PBS++. Filters were dehydrated, cut out from filter units, and embedded in Epon. Sixty-nanometer-thick sections were stained with uranyl acetate and lead citrate and examined with a JEM-100CX electron microscope (JEOL Ltd., Tokyo, Japan).
RESULTS
Generation of transfectant NA mutant viruses.
Alanine mutations in the CT and TMD of NA protein (A/WSN/33) were introduced by site-directed mutagenesis (Fig. 1). Chimeric NA TMDs [NA(1T2N)NA and NATRNA] were constructed by swapping either the first 9 aa or the entire NA TMD with that of TR TMD, respectively (Fig. 1). The transfectant viruses containing the mutated NA were generated entirely from cloned cDNAs (A/WSN/33) by using either the 17-plasmid method (34) for NA5A14, NA4A19, NA5A27, NA(1T2N)NA, and NATRNA or the 8 plasmid-method (17) for NA3A2, NA2A5, NA3A7, NA4A10, NA4A23, and NA5A31. All mutant viruses were successfully rescued, and mutations in the NA gene of progeny viruses were confirmed by sequencing the reverse transcription-PCR products of virion NA RNAs.
Growth characteristics and plaque morphology of transfectant NA mutant viruses.
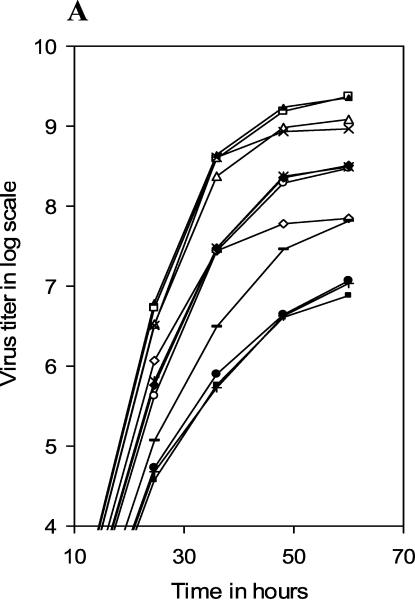
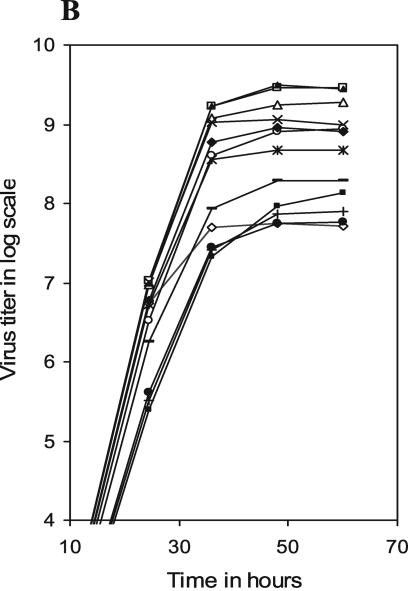
To investigate virus propagation under multiple cycles of infection, MDCK cell monolayers were infected with WT or different mutant viruses at an MOI of 0.001 and incubated in VGM (without trypsin) at 33°C. At different times p.i., progeny viruses in cell supernatants were titrated by PFU assay and plotted in log scale against time p.i. (Fig. 2A). Results show that according to their growth characteristics, mutant viruses could be divided into three major groups. The first group (mutant NA2A5, NA3A7, and NA4A10 viruses) grew normally, like the WT virus, to a final titer of about 1 × 109 to 3 × 109 PFU/ml (at 60 h p.i.). The second group (NA5A14, NA4A19, and NA4A23) produced virus titers about 1 log less (approximately 3 × 108 PFU/ml at 60 h p.i.). The third group (NA5A27, NA5A31, and NATRNA) grew the least and had a 100-fold-lower final titer (∼107 PFU/ml at 60 h p.i.) than the WT virus. The growth rates of the transfectant viruses NA3A2 and NA(1T2N)NA were in between those of the second and third groups, with a final titer of about 7 × 107 PFU/ml.
FIG. 2.
Growth curves of different mutant viruses. MDCK cell monolayers were infected with viruses at an MOI of 0.001 and maintained at 33°C in VGM without (A) or with (B) 0.5 μg of TPCK-treated trypsin/ml. At different time points p.i., aliquots of culture supernatant were collected and titrated for infectious viruses by PFU assay. ▴, WT; ⋄, NA3A2; □, NA2A5; Δ, NA3A7; ×, NA4A10; Ж, NA5A14; ○, NA4A19; ♦, NA4A23; •, NA5A27; +, NA5A31; −, NA(1T2N)TR; ▪, NATRNA. Results represent averages of the results from four independent experiments.
Under multiple cycles of infections at a very low initial MOI (0.001 PFU/cell), the reduced virus yield can be primarily due to two reasons. (i) It is known that WSN NA can enhance HA cleavage (14). Therefore, mutations in NA can cause less HA cleavage and thereby produce fewer infectious progeny viruses, which in turn will reduce the final virus titer during reinfection. (ii) Mutations in NA can affect NA activity (for removing sialic acid) and thereby reduce the release of the progeny viruses into the medium and reinfection.
To investigate the possibility that less HA cleavage was responsible for the lower titer of some mutant viruses, we used TPCK-treated trypsin (0.5 μg/ml) in VGM. For WT as well as NA3A2, NA2A5, NA3A7, and NA4A10 mutant viruses, trypsin did not have a significant effect in producing infectious viruses, although in the presence of trypsin some increase in virus titer was always observed (Fig. 2B). However, in the presence of trypsin, the titer of mutant NATRNA, NA5A27, and NA5A31 viruses (third group), which had the lowest titer in the absence of trypsin increased significantly (∼5 to 20 times). Still even in the presence of trypsin, the final titer of these mutant viruses remained 10 to 20 times lower than that of the WT virus. The effect of trypsin in producing infectious viruses was intermediate for NA(1T2N)NA, NA5A14, NA4A19, and NA4A23 mutant viruses, and titer increased only two- to threefold.
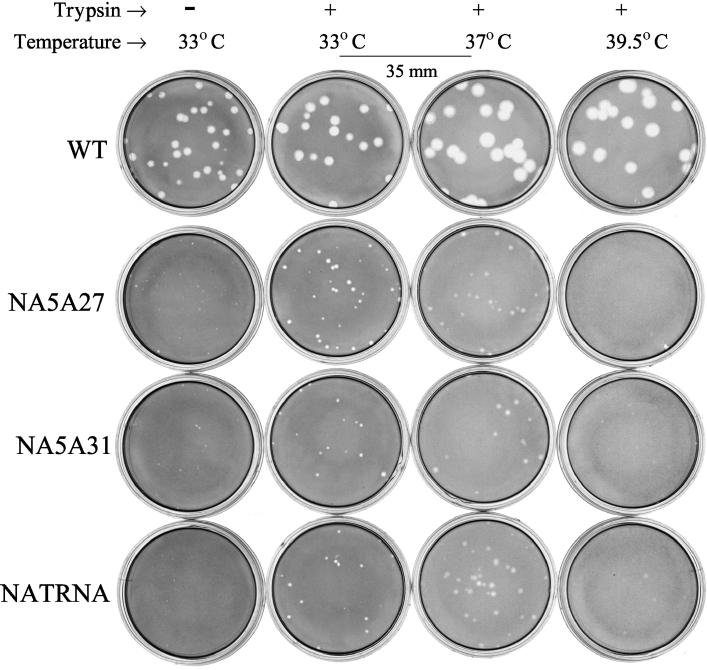
Plaque morphology of the WT and different mutant viruses was examined at 33°C either in the presence (0.5 μg/ml) or absence of trypsin in the agar overlay medium, and plaque sizes were measured at 60 h p.i. Results (Table 1; Fig. 3) show that compared to the WT virus, the plaque size and morphology were similar for some mutant viruses but different for others. However, all viruses, including the WT virus, exhibited an increase in plaque size in the presence of trypsin compared to that in the absence of trypsin. Mutant NA2A5, NA3A7, and NA4A10 viruses behaved like the WT virus and produced clear distinct plaques both in the absence (1.5- to 2.0-mm diameters) and presence (2.3- to 2.5-mm diameters) of trypsin. Mutant NA5A27, NA5A31, NA(1T2N)NA, and NATRNA viruses produced pinpoint plaques in the absence of trypsin which increased in size (0.7 to 0.9 mm) in the presence of trypsin. Plaques of mutant viruses NA5A14, NA4A19, and NA4A23 were intermediate in size and ranged from 0.7 to 0.8 mm in the absence and 1.2 to 1.5 mm in the presence of trypsin. These results show that plaque size of mutant viruses (Table 1) essentially correlated with their multiple-cycle growth characteristics (Fig. 2).
TABLE 1.
Plaque sizes of mutant viruses at different temperatures
| Virus | Avg plaque size (mm)a:
|
|||
|---|---|---|---|---|
| Without trypsin (33°C) | With trypsin at temp (°C)
|
|||
| 33 | 37 | 39.5 | ||
| WT | 2.0 | 2.5 | 4.0 | 3.8 |
| NA3A2 | 1.3 | 1.9 | 3.0 | 3.0 |
| NA2A5 | 2.0 | 2.5 | 4.0 | 3.8 |
| NA3A7 | 1.5 | 2.3 | 3.4 | 3.4 |
| NA4A10 | 1.9 | 2.4 | 3.8 | 3.6 |
| NA5A14 | 0.8 | 1.2 | 2.5 | 2.5 |
| NA4A19 | 0.8 | 1.2 | 2.0 | 2.0 |
| NA4A23 | 0.7 | 1.5 | 2.0 | 2.0 |
| NA5A27 | PPc | 0.9 | 1.0 (D) | (D)b |
| NA5A31 | PP | 0.7 | 1.0 (D) | (D)b |
| NA(1T2N)NA | PP | 0.9 | 1.4 | 1.2 |
| NATRNA | PP | 0.8 | 1.0 (D) | (D)b |
Plaque morphology and size (average of 10 plaques) were determined at 60 h p.i.
Diffuse (D) plaques were visible but not measurable.
PP, plaques were clear but pinpoint in size.
FIG. 3.
Plaque morphologies of different mutant viruses. MDCK cell monolayers (35-mm-diameter dish) were infected with serially diluted viruses and maintained at different temperatures in agarose overlay medium with (+) or without (−) TPCK-treated trypsin (0.5 μg/ml). At 60 h p.i., the agarose overlay medium was removed, stained for 1 min with crystal violet, and washed with PBS.
Next, we determined whether the mutations in NA caused any temperature-sensitive defect. Accordingly, the plaque morphology of WT and different mutant viruses was also determined at 37 and 39.5°C in the presence of trypsin (Table 1 and Fig. 3). At both the temperatures (37 and 39.5°C), the morphology of WT and mutant virus plaques were somewhat defuse compared to that at 33°C. WT as well as all mutant viruses except NA5A27, NA5A31, and NATRNA (Fig. 3) produced larger plaques at both the temperatures (37 and 39.5°C) than at 33°C (Table 1). However, these three mutant viruses (NA5A27, NA5A31, and NATRNA) exhibited a different plaque morphology. At 37°C they produced larger and diffuse plaques. At 39.5°C, plaques of NA5A27 and NA5A31 mutant viruses were highly diffuse and hardly visible. Also, at 39.5°C, plaques of NATRNA mutant virus were diffuse but some were visible. Therefore, mutant NA5A27, NA5A31, and NATRNA viruses exhibited some temperature-sensitive defect at 39.5°C.
Sialidase activity of mutant viruses.
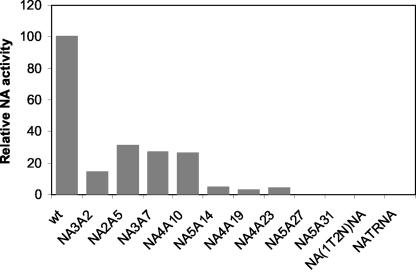
Our results (Fig. 2) showed that trypsin can only partially restore the attenuation of growth of mutant viruses in MDCK cells. Moreover, trypsin did not increase the PFU titer for attenuated mutant virus NA3A2 (about 50-fold-lower titer than WT in the presence or absence of trypsin). We therefore investigated the possibility that mutation in CT or TMD of NA can lead to lower NA enzyme activity of virus particles and thereby cause less-efficient release of virus particles from the cell surface and attenuation of virus growth after multiple cycles of infection. Enzyme activity was measured by incubating 5 μl of purified virus suspension with 5 μl of 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid substrate for 30 min at 37°C and assaying the amount of released fluorescent compound, 4-MU, as described in Materials and Methods. The NA enzyme activity of mutant viruses was normalized against M1 protein and presented as percentage of that of WT virus (Fig. 4). Results were essentially the same when HA was used for normalization (data not shown). M1 and HA proteins were estimated by immunoblotting with antibodies against M1 and HA followed by densitometric quantification. The results (Fig. 4) show that according to their NA enzyme activity, mutant viruses could be divided into three major groups. The first group (mutant NA3A2, NA2A5, NA3A7, and NA4A10 viruses) showed NA sialidase activities between 14 and 32% of that of the WT. For the mutant NA5A14, NA4A19, and NA4A23 viruses (second group), the NA enzyme activities were 4.8, 3.0, and 4.5% of that of the WT, respectively. The third group [NA5A27, NA5A31, NA(1T2N)NA, and NATRNA] had essentially little or no measurable NA activity. These data indicate that greatly reduced NA enzyme activity of some mutant viruses may be responsible for less-efficient release of virus particles from the cell surface and the attenuation of virus growth and that 26 to 32% of the NA activity was sufficient for WT virus release, as has been reported previously (36).
FIG. 4.
Sialidase activity of mutant viruses. Sialidase activity was determined with the 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid substrate as described in Materials and Methods. Specific activity was calculated by normalizing NA activity to the level of virus particles, as determined by immunoblotting viral proteins (separated by SDS-PAGE) with antibody against M1, followed by densitometric analysis. Relative NA activity, taking that of WT virus as 100%, was calculated from the results from three independent experiments, with an average variation of less than 10%.
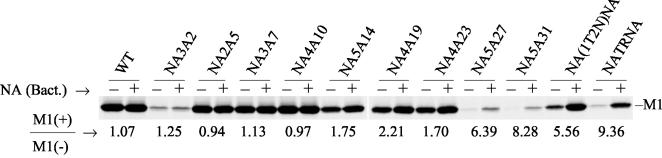
We therefore investigated whether the bacterial NA can enhance the release of virus particles from the cell surface. Accordingly, at 5 h p.i., virus-infected cells were labeled with 35S-protein labeling mix for 9 h. Two hours before collection, virus-infected cells were treated or mock treated with bacterial NA (10 mU/ml). At 14 h p.i., released virus particles in the supernatant were collected and clarified by low-speed centrifugation. Virions were purified and pelleted by ultracentrifugation through a 25% sucrose cushion. Viral proteins were separated by SDS-PAGE, autoradiographed, and quantified. The ratios of virus particles released in the presence and absence of bacterial NA were estimated by using the M1 protein band. As expected, there was no increase in virus release for WT virus in the presence of bacterial NA. Similarly, no significant increase in virus release was observed for NA3A2, NA2A5, NA3A7, and NA4A10 mutants (Fig. 5). However, in the presence of bacterial NA, the release of mutant NA5A27, NA5A31, NA(1T2N)NA, and NATRNA viruses was increased significantly (about five- to ninefold) (Fig. 5). On the other hand, in the presence of bacterial NA, only a small increase (1.7- to 2.2-fold) in virus release was observed for NA5A14, NA4A19, and NA4A23 mutants. Results were similar if HA or NP protein instead of M1 was quantified in released virus particles (data not shown). These results show that where NA activity was greatly reduced or undetectable, aggregation of virus particles on the cell surface was at least partially responsible for the reduced virus release and low PFU titer for mutant NA5A27, NA5A31, NA(1T2N)NA, and NATRNA viruses. However, it should be noted that even in the presence of bacterial NA, the amounts of released virus particles in the cells infected with mutant NA3A2, NA5A27, and NA5A31 viruses were not restored to the level of WT virus, suggesting that another defect(s) in addition to reduced NA enzyme activity was affecting virus growth.
FIG. 5.
Effect of bacterial NA on virus release. Virus-infected MDCK cells were metabolically labeled (at 5 h p.i.) with 300 μCi of 35S-protein labeling mix in 2:8 VGM for 9 h. Two hours before collection, virus-infected cells were treated (+) or mock treated (−) with bacterial NA (10 mU/ml). At 14 h p.i., released virus particles in the supernatant were collected and clarified by low-speed centrifugation and virions were purified and pelleted by ultracentrifugation through a 25% sucrose cushion. Pellets were dissolved in TNE buffer. Viral proteins were analyzed directly by SDS-PAGE, autoradiographed, and quantified. The increase in virus release after NA treatment was determined by using the M1 band. The average value of M1(+)/M1(−) was calculated from the results from three to five independent experiments, with a variation of less than 10%.
Expression and maturation of NA proteins in virus-infected cells and incorporation of NA proteins in virions.
To determine whether mutation caused structural perturbation affecting synthesis, stability, transport, or maturation of NA protein, virus-infected MDCK cells were metabolically labeled at 4 h p.i. with 35S-protein labeling mix for 20 min and chased with unlabeled methionine and cysteine in excess for 0 or 90 min. Cell lysates were immunoprecipitated with anti-NA and anti-M1 antibodies, proteins were eluted from Sepharose beads, treated with either endo H or PNGase F or mock treated, analyzed by SDS-PAGE and quantified. Since NA protein bands were diffuse because of glycosylation, the total amount of NA protein expressed was determined from the PNGase F-treated band and normalized against the M1 band. The results are presented as the percentage of WT NA protein expressed (Table 2). The amount of endo H-resistant protein after 90 min of chase was considered to be mature protein, was obtained by subtracting the endo H-sensitive protein band from the total protein band obtained after PNGase F treatment, and is presented as the percentage of total NA protein (Table 2). Results show that in NA2A5 and NA4A23 virus-infected cells, NA protein expression was similar to that of WT. In virus-infected cells, NA3A2 and NA5A14 protein expression were only 27 and 23% of the WT NA, respectively. For other mutant viruses, NA protein expression was intermediate, ranging from 41 to 82% (Table 2). These results show that in some mutant virus-infected cells, NA expression was reduced, and this reduction was not due to MOI effect, since the expression of other viral proteins such as HA, M1, and NP was essentially the same as in the WT virus-infected cells (data not shown). For up to 90 min of chase, all mutant NA proteins were stable like WT NA (data not shown). For all mutant viruses except NA4A19, NA4A23, and NA(1T2N)NA, the maturation of NA protein after 90 min of chase was essentially the same as the WT NA (Table 2). However, after 90 min of chase, only 28, 24, and 15% NA protein became mature for mutant NA4A19, NA4A23, and NA(1T2N)NA viruses, respectively, compared to 60% for WT virus (Table 2).
TABLE 2.
Expression and maturation of NA protein in virus-infected cells and percentage of NA protein incorporated in virion envelope
| Virus | Amt of NA protein expresseda (avg ± SD)d | Maturation of NA (% of endo H resistance)b (avg ± SD)d | NA incorporation in virion envelopec (avg ± SD)d |
|---|---|---|---|
| WT | 100 | 60 ± 4 | 100 |
| NA3A2 | 27 ± 3 | 58 ± 3 | 12.7 ± 2.1 |
| NA2A5 | 105 ± 4 | 57 ± 3 | 50.5 ± 3.2 |
| NA3A7 | 60 ± 3 | 57 ± 4 | 27.3 ± 2.6 |
| NA4A10 | 71 ± 4 | 59 ± 3 | 44.2 ± 3.6 |
| NA5A14 | 23 ± 3 | 49 ± 3 | 3.6 ± 0.7 |
| NA4A19 | 53 ± 3 | 28 ± 3 | 7.1 ± 1.2 |
| NA4A23 | 103 ± 4 | 24 ± 3 | 6.4 ± 0.9 |
| NA5A27 | 50 ± 5 | 65 ± 5 | 5.6 ± 0.8 |
| NA5A31 | 44 ± 4 | 55 ± 6 | 5.4 ± 0.7 |
| NA(1T2N)NA | 41 ± 4 | 15 ± 3 | 2.5 ± 0.6 |
| NATRNA | 82 ± 3 | 64 ± 4 | 10.3 ± 1.1 |
Virus-infected MDCK cells were metabolically labeled (at 4 h p.i.) for 20 min with 300 μCi of 35S-protein labeling mix/ml, labeled cells were lysed and immunoprecipitated with anti-NA and anti-M1 antibodies, and proteins were eluted, treated with PNGase F, analyzed by SDS-PAGE, autoradiographed, and quantified as described in Materials and Methods. The M1 band was used for normalization, and the results are shown with the WT result as 100%.
Virus-infected MDCK cells were metabolically labeled for 20 min as described above and chased for 90 min. Labeled cells were lysed and immunoprecipitated with anti-NA and anti-M1 antibodies, and proteins were eluted, divided into two parts, treated with either endo H or PNGase F, analyzed by SDS-PAGE, autoradiographed, and quantified. Percentages of endo H resistance proteins were calculated with the following formula: (PNGase F sensitive − Endo H sensitive) × 100/PNGase F sensitive.
Virus-infected MDCK cells were metabolically labeled (at 4 h p.i.) for 12 h with 200 μCi of 35S-protein labeling mix/ml with 2:8 media (see Materials and Methods). At 16 h p.i., medium was harvested and clarified by low-speed centrifugation, and virions were pelleted by ultracentrifugation through a 25% sucrose cushion. Labeled viruses were lysed in RIPA buffer and immunoprecipitated with anti-NA and anti-M1 antibodies, and proteins were eluted, treated with PNGase F, analyzed by SDS-PAGE, autoradiographed, and quantified (Fig. 6). Amounts of NA proteins were normalized with M1 proteins for each mutant virus and presented as percentages of that in WT virus.
Results represent averages of those from three to five independent experiments.
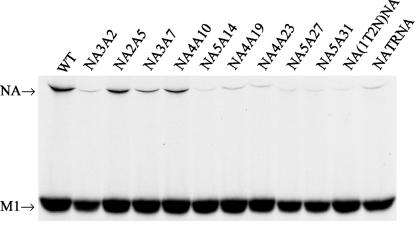
Since NA enzyme activity varied significantly among the mutant viruses, we wanted to determine whether this was due to differential incorporation of the NA protein in the virion envelope. Accordingly, at 4 h p.i., virus-infected cells were metabolically labeled with 35S-protein labeling mix for 12 h. At 16 h p.i., medium was harvested and clarified by low-speed centrifugation and virions were pelleted by ultracentrifugation through a 25% sucrose cushion. Viral proteins were separated by SDS-PAGE, autoradiographed, and quantified. Results show that M1/HA ratios were essentially the same for WT and all mutant viruses (about 1.1). However, M1/NP ratios varied. Mutant NA3A2 and NA(1T2N)NA viruses showed significantly higher ratios of M1 to NP (3.4 and 3.0, respectively, compared to 1.2 for WT) (data not shown). Due to the proximity of NP and highly defuse nature of NA protein, bands of these two proteins were difficult to separate and quantify in the same gel. Moreover, small amounts of NA present in some mutant viruses made it harder to quantify. To overcome this problem, labeled viruses were lysed in RIPA buffer and immunoprecipitated with anti-NA and anti-M1 antibodies and proteins were eluted from Sepharose beads, treated with PNGase F (to get a sharper NA band), analyzed by SDS-PAGE, autoradiographed, and quantified. Amounts of NA proteins were normalized against M1 proteins for each mutant virus and presented as percentages of that for WT virus. Results (Fig. 6) show that the percentage of NA protein incorporated in the virion envelope (represented by M1 protein) varied significantly for different mutant viruses (Table 2). NA2A5, NA3A7, and NA4A10 proteins were incorporated 50.5, 27.3, and 44.2% of WT NA, respectively. NA incorporation in virus particles was the least in NA(1T2N)NA (2.5% of WT). For others, NA incorporation was between 5.4 and 12.7% of WT. These results show that reduced incorporation of NA protein in virion particles in NA3A2, NA5A14, NA4A19, NA4A23, NA5A27, NA5A31, and NA(1T2N)NA mutant viruses was likely due to reduced NA expression or maturation or both. However, reduced NA incorporation in NATRNA virus was unlikely due to either defect in NA expression or maturation (Table 2).
FIG. 6.
NA protein incorporation in virions. Virus-infected MDCK cells were labeled at 4 h p.i. for 12 h with 200 μCi of 35S-protein labeling mix in 2:8 VGM. At 16 h p.i., medium was harvested and clarified by low-speed centrifugation and virions were pelleted by ultracentrifugation through a 25% sucrose cushion. Labeled virion lysates were immunoprecipitated with the respective antibodies; proteins were eluted, treated with PNGase F for 3 h, analyzed by SDS-PAGE, and autoradiographed.
Triton X-100 insolubility of NA proteins at virus-infected cell surface and in virions.
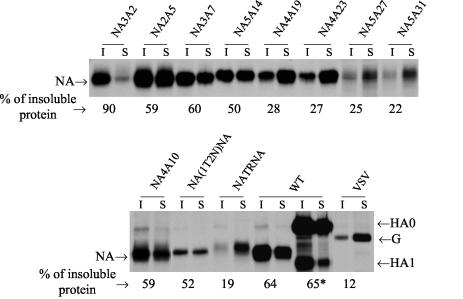
With influenza viruses, it has been reported that both NA and HA become raft associated, as determined by Triton X-100 insolubility, and that influenza viruses have been shown to bud from the lipid raft domain of the plasma membrane enriched in cholesterol and glycosphingolipids (31, 32, 39). Furthermore, lipid raft association of HA and NA is mediated by the TMD (4, 5, 24, 28, 40). However, CT deletions (47) have also been shown to affect lipid raft association of HA and NA and have been implicated in virus budding. Therefore, we wanted to determine Triton X-100 insolubility of mutant and chimeric NA proteins present at the surface of virus-infected cells. Accordingly, virus-infected MDCK cells were metabolically labeled (at 4 h p.i.) for 60 min with 35S-protein labeling mix and chased for 90 min. Cell surface proteins were biotinylated and extracted with 1% Triton X-100 on ice for 10 min. Triton X-100 soluble and insoluble fractions of surface-biotinylated proteins were immunoprecipitated by the respective antibodies, analyzed by SDS-PAGE, autoradiographed, and quantified. Cell surface influenza HA and VSV G were used as positive and negative controls and showed 65 and 12% Triton X-100 insolubility, respectively (Fig. 7). Results (Fig. 7) show that the Triton X-100 insolubility of mutant proteins NA3A2, NA2A5, NA3A7, NA4A10, and NA5A14 and the chimeric protein NA(1T2N)NA was similar to that of WT NA (i.e., 50% or higher compared to 64% insolubility for WT NA). On the other hand, for mutant NA4A19, NA4A23, NA5A27, NA5A31, and NATRNA viruses, only 28, 27, 25, 22, and 19% of cell surface NA proteins, respectively, were Triton X-100 isoluble. For WT and all mutant viruses, the Triton X-100 insolubility of HA at the virus-infected cell surface was essentially the same (data not shown).
FIG. 7.
Triton X-100 insolubility of cell surface proteins. At 4 h p.i., influenza virus-infected (2.5 h p.i. for VSV-infected cells) MDCK cells were metabolically labeled for 60 min with 300 μCi of 35S-protein labeling mix/ml and chased for 90 min. Cell surface proteins were biotinylated and extracted with 1% Triton X-100 on ice for 10 min. Triton X-100-soluble (S) and -insoluble (I) fractions of surface biotinylated proteins were immunoprecipitated with specific antibodies for NA, HA, or VSV G proteins, analyzed by SDS-PAGE, autoradiographed, and quantified. The percentage of Triton X-100-insoluble protein was calculated from the results from three to five independent experiments, with average variation of less than 10%. *, the Triton X-100 insolubility of HA was calculated with both HA0 and HA1 bands.
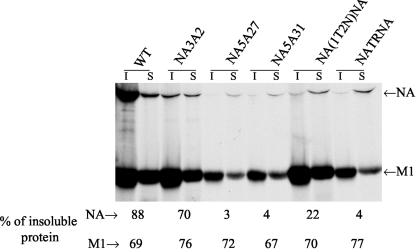
Since NA3A2, NA5A27, NA5A31, NA(1T2N)NA, and NATRNA mutant viruses were most attenuated in replication, and since reduced raft association of HA has been implicated in attenuated virus growth (43), we wanted to determine the Triton X-100 resistance of NA present in these virion envelopes. Accordingly, labeled virus particles were purified and treated with 0.1% Triton X-100, and soluble and insoluble proteins were immunoprecipitated, treated with PNGase F, analyzed by SDS-PAGE, autoradiographed, and quantified. The results (Fig. 8) show that virion NA was highly Triton X-100 soluble for NA5A27, NA5A31, NA(1T2N)NA, and NATRNA mutant viruses (3, 4, 22, and 4%, respectively) but not for NA3A2, which essentially exhibited the WT phenotype. The Triton X-100 insolubility of M1 (Fig. 8) and HA (data not shown) proteins of these mutant viruses was essentially similar to that of the WT virus.
FIG. 8.
Triton X-100 insolubility of different proteins in virions. 35S-labeled virions were prepared as described in the legend to Fig. 6 and treated with 0.1% Triton X-100 on ice for 10 min, soluble (S) and insoluble (I) fractions were immunoprecipitated with the respective antibodies, and proteins were eluted, treated with PNGase F for 3 h, analyzed by SDS-PAGE, autoradiographed, and quantified. The percentage of Triton X-100-insoluble protein was calculated from the results from three to five independent experiments, with an average variation of less than 10%.
Aggregation and morphological changes of mutant viruses on infected cell surfaces.
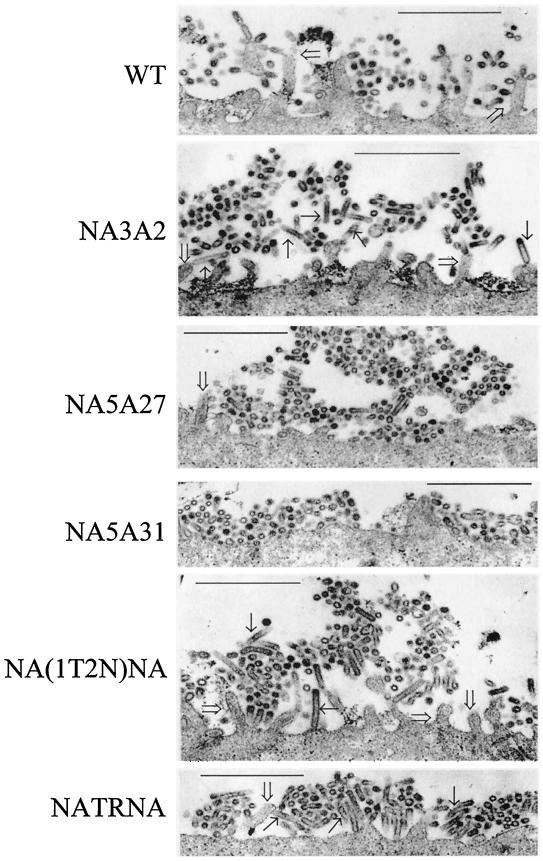
Since five mutant viruses [NA3A2, NA5A27, NA5A31, NA(1T2N)NA, and NATRNA] exhibited the greatest reduction in virus growth even in the presence of trypsin, we wanted to determine whether there was any defect in virus budding and release. Previously, it was shown that deletion of the CT of NA caused an elongated virus morphology (30) and that low NA enzyme activity caused aggregation of virus particles on the infected cell surface (29). In this report we have shown that for mutant NA5A27, NA5A31, NA(1T2N)NA, and NATRNA viruses enzyme activity was very low and, in the presence of bacterial NA, the release of virus particles increased significantly (about five- to ninefold), indicating aggregation of virus particles at the cell surface. We therefore examined for virus particles at the infected cell surface by thin-section electron microscopy. Accordingly, MDCK cells grown on polycarbonate filters that were infected with WT and different mutant viruses. At 12 h p.i., infected cells were fixed for thin-section electron microscopy. The results (Fig. 9) show that the WT virus particles were mostly spherical in shape and that fewer particles remained attached to the cell surface. On the other hand, NA5A27 and NA5A31 mutant viruses exhibited spherical shape but produced large aggregation of progeny virus on the cell surface. However, NA3A2, NA(1T2N)NA, and NATRNA mutant virus particles produced a different morphological phenotype. Many virus particles were elongated in shape. NATRNA and NA(1T2N)NA also exhibited aggregation on the infected cell surface.
FIG. 9.
Budding of viruses by thin-section electron microscopy. MDCK cells grown on polycarbonate filters were infected with different viruses at an MOI of 3.0. At 12 h p.i., infected cell monolayers were cross-linked in 2% glutaraldehyde and postfixed with 1% OsO4. Filters were embedded in Epon, and 60-nm-thick sections were stained with uranyl acetate and lead citrate and examined with a transmission electron microscope. Bar, 1 μm. →, elongated virus particles; ⇒, villi.
DISCUSSION
The amino acid sequences of the CT and TMD of NA are highly and moderately conserved, respectively, among the influenza A viruses. Yet the specific function and role of these amino acid sequences in virus biology remain unknown. Results presented in this report show that the specific amino acid residues are not absolutely required for the influenza virus life cycle, since either the complete or part of the NA TMD or CT can be replaced and modified, yet infectious viruses can be rescued and propagated. On the other hand, our data show that specific amino acids in some regions of the TMD and CT as well as a foreign TMD have a profound influence on virus biology, causing reduction in growth during multiple cycles of infection. Reduced yield of NA mutants can be attributed to decreased enzyme activity in the virion and a defect in budding at the cell surface.
Mutations in the TMD and CT of NA can affect protein expression, maturation, transport, incorporation into virions, and enzyme activity and thus can affect virus growth and replication. In addition, mutations in the NA can also affect virus budding and release. We examined the properties of each NA mutant virus in these categories. Among the mutants, NA2A5, NA3A7, and NA4A10 exhibited essentially WT virus replication, NA expression, maturation, and lipid raft association, indicating that these amino acid sequences (aa 5 to 13) were not critical for NA function. Within the TMD, mutations of the ectodomain-proximal amino acids had a progressively detrimental effect on virus production. The mutations in the viruses NA5A27 and NA5A31 reduced virus production the most, about 50 and 250 times less in the presence and absence of trypsin, respectively. The reduction in virus titer in these mutants was clearly due to the loss of NA activity in the virus particles causing virus aggregation (Fig. 9). Also, the presence of bacterial NA in the medium released more virus particles into the supernatant (Fig. 5). However, reduced NA enzyme activity of some mutant viruses could be only partly correlated with reduced NA incorporation. When relative NA enzyme activity was normalized against NA content in virions, calculated as 100 × (%NA activity)/(% NA content in virion), for mutant NA3A2, NA3A7, and NA4A14 viruses, the efficiency of NA in catalyzing its substrate was similar to WT NA. For mutant NA3A5, NA3A10, NA4A19, and NA4A23 viruses, those efficiencies were 62, 60, 42, and 70% of the WT efficiency, respectively. Furthermore, mutant NA5A27, NA5A31, NA(1T2N)NA, and NATRNA viruses containing 5.6, 5.4, 2.5, and 10.3% of WT virus level of NA protein, respectively, showed very little or no measurable NA activity (Fig. 4), indicating further reduction in the efficiency of NA to catalyze its substrate. Mutations may have caused structural perturbation to the NA, leading to a reduction in the efficiency with which it catalyzed its substrate, and TMDs are known to be involved in oligomerization and structural stability (25).
The cause of reduced NA incorporation in the virion envelope varied and was based on a combination of factors such as reduced NA expression, maturation, and raft association as exhibited by low Triton X-100 insolubility. These results essentially confirm the earlier observation about raft association of NA TMD mutants with stably expressed mutated NATR chimeric proteins containing the TMD of NA and the ectodomain of TR (5). In NA5A14 virus, reduced NA incorporation in the virion envelope (3.6% of WT) was most likely due to reduced expression (23% of WT), since protein maturation or raft association was not greatly affected (Fig. 6; Table 2). On the other hand, for NA4A19 and NA4A23 mutant viruses, reduced NA incorporations (7.1 and 6.4% of WT virus, respectively) were due to a combination of reduced maturation (28 and 24% of WT, respectively) as well as reduced raft association (28 and 27% of WT, respectively). Poor incorporation of NA in NA5A27 and NA5A31 viruses could be due to low raft association both on the cell surface (Fig. 7) and virus envelope (Fig. 8), since NA expression and maturation were not greatly affected. The mutant virus NA(1T2N)NA, containing the chimeric NA TMD, also exhibited attenuated virus growth due to greatly reduced NA activity and poor incorporation (2.5% of WT) in virion. This reduction in NA incorporation could be due to combinations of poor expression (41% of WT) and maturation (15%). Again, chimeric TMD in NA(1T2N)NA may be responsible for poor oligomerization leading to poor maturation (25). On the other hand, reduced growth of NATRNA, in which the entire TMD of NA was replaced by TR TMD, was likely due to reduced NA activity, which in turn was likely due to poor NA incorporation and Triton X-100 insolubility. Expression and maturation of this mutant protein were essentially normal and were presumably due to the presence of the entire TR TMD, which may not interfere with oligomerization and maturation. This would support the idea that a foreign TMD affects incorporation of NA in the viral envelope. However, reduced raft association may also be a contributing factor causing poor incorporation of the NA into the virion envelope, as suggested previously for some other mutant viruses (47). Among the two CT mutants, NA2A5 (the TMD-proximal mutant), behaved essentially similar to the WT virus. On the other hand, the NA3A2, the extreme N-terminal mutant, had a 50-fold reduction in virus titer with reduced NA expression (27%) in infected cells and incorporation in the virion (12.7%), but it had no effect on maturation and Triton X-100 insolubility of the NA protein. This mutation apparently caused a defect in virus budding (Fig. 9). In addition, this mutation may have caused interference in NA-M1 interaction, since the CT is involved in interacting with M1. Earlier attempts to rescue infectious virus from mutation in this region (P3→A) failed, suggesting some critical function of this region (6).
Three mutant NA3A2, NA(1T2N)NA, and NATRNA viruses also exhibited an increased number of elongated virus particles, indicating a defect in the budding process. These mutant virus particles exhibited a budding defect with a morphological phenotype similar to that recently reported for an M1 mutant (R101A) (20). The processes and factors involved in bud formation and release of virus buds are complex and yet undetermined (32, 33). Bud formation requires bending of the membrane and the planar membrane structure to a curved structure. Assembly of lipid bilayers, including formation of lipid microdomains such as lopid rafts as well as clustering of matrix (M1) proteins on the inner bilayer, can cause membrane bending and initiation of budding. Finally, pinching off of the virus buds requires the fusion of the apposing viral and cellular membranes, leading to fission and separation of the bud from the cell (33). Factors affecting the fusion and fission of the bud will affect the size and the shape of the virus particles. Among the viral components, matrix proteins have been shown to be the key component in both bud formation and pinching off (32, 33). It was previously shown that influenza virus M1 plays an important role in filamentous particle formation (9, 38). In three mutants [NA3A2, NA(1T2N)NA, and NATRNA], a defect in the fusion of the apposing membranes and fission of the buds appears to be the major contributing factor in elongated rather than spherical particle formation. Apparently, there appears to be a defect in bud release rather than bud formation with these mutants. The role of NA in the budding process is further supported by the presence of more elongated progeny virus particles in mutant viruses lacking the NA gene (29) and NAt− viruses containing a reduced amount of NA on the virus envelope (30).
However, how NA and transmembrane protein can effect budding remains unclear. Earlier it was proposed that the reduced raft association of HAt− and NAt− proteins was responsible for the formation of filamentous progeny virus particles (46). However, lack of raft association of mutant NA cannot explain the budding defect (i.e., elongated particle formation) in NA3A2 mutant virus, since this mutant NA showed Triton X-100 insolubility similar to that of WT NA on the virus-infected cell surface as well as on the virion envelope. Therefore, it is likely that NA either directly or indirectly may have a role in the budding process independent of raft association. A number of host factors have been shown to be involved in the release of virus buds for a number of viruses (12, 42). Since these have been primarily shown to interact with matrix proteins, we propose that NA-M1 interaction may directly or indirectly facilitate bud release by bringing these host factors to the budding site.
In summary, we have shown that the conserved TMD or CT sequences of NA are not absolutely required for the virus life cycle, since they can be partially or totally replaced by other sequences, yet infectious virus can be obtained and propagated. However, the data presented here support the idea that specific sequences of TMD and CT of NA are required for optimal virus replication and that some of these residues have a direct role in virus budding.
Acknowledgments
This work was supported by USPHS grants AI 16348 and AI 41681.
We thank Elizabeth F. Neufeld and Ke-Wei Zhao for help with the fluorometric sialidase activity assay and B. M. Sjostrand (BRI EM Core Facility, University of California—Los Angeles) for assistance in electron microscopy.
REFERENCES
- 1.Air, G. M., and W. G. Laver. 1989. The neuraminidase of influenza virus. Proteins 6:341-356. [DOI] [PubMed] [Google Scholar]
- 2.Ali, A., R. T. Avalos, E. Ponimaskin, and D. P. Nayak. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 74:8709-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barman, S., L. Adhikary, Y. Kawaoka, and D. P. Nayak. 2003. Influenza A virus hemagglutinin containing basolateral localization signal does not alter the apical budding of a recombinant influenza A virus in polarized MDCK cells. Virology 305:138-152. [DOI] [PubMed] [Google Scholar]
- 4.Barman, S., A. Ali, E. K.-W. Hui, L. Adhikary, and D. P. Nayak. 2001. Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses. Virus Res. 77:61-69. [DOI] [PubMed] [Google Scholar]
- 5.Barman, S., and D. P. Nayak. 2000. Analysis of the transmembrane domain of influenza virus neuraminidase, a type II transmembrane glycoprotein, for apical sorting and raft association. J. Virol. 74:6538-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilsel, P., M. R. Castrucci, and Y. Kawaoka. 1993. Mutations in the cytoplasmic tail of influenza A virus neuraminidase affect incorporation into virions. J. Virol. 67:6762-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blok, J., and G. M. Air. 1982. Sequence variation at the 3′ end of the neuraminidase gene from 39 influenza type A viruses. Virology 121:211-229. [DOI] [PubMed] [Google Scholar]
- 8.Bos, T. J., A. R. Davis, and D. P. Nayak. 1984. NH2-terminal hydrophobic region of influenza virus neuraminidase provides the signal function in translocation. Proc. Natl. Acad. Sci. USA 81:2327-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourmakina, S. V., and A. Garcia-Sastre. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517-527. [DOI] [PubMed] [Google Scholar]
- 10.Burnet, F. M., and J. D. Stone. 1947. The receptor-destroying enzyme of V. cholerae. Aust. J. Exp. Biol. Med. Sci. 25:227-233. [DOI] [PubMed] [Google Scholar]
- 11.Compans, R. W., N. J. Dimmock, and H. Meier-Ewert. 1969. Effect of antibody to neuraminidase on the maturation and hemagglutinating activity of an influenza A2 virus. J. Virol. 4:528-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Sastre, A., and P. Palese. 1995. The cytoplasmic tail of the neuraminidase protein of influenza A virus does not play an important role in the packaging of this protein into viral envelopes. Virus Res. 37:37-47. [DOI] [PubMed] [Google Scholar]
- 14.Goto, H., and Y. Kawaoka. 1998. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc. Natl. Acad. Sci. USA 95:10224-10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto, H., K. Wells, A. Takada, and Y. Kawaoka. 2001. Plasminogen-binding activity of neuraminidase determines the pathogenicity of influenza A virus. J. Virol. 75:9297-9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gubareva, L. V., M. S. Nedyalkova, D. V. Novikov, K. G. Murti, E. Hoffmann, and F. G. Hayden. 2002. A release-competent influenza A virus mutant lacking the coding capacity for the neuraminidase active site. J. Gen. Virol. 83:2683-2692. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, R. T., E. Dietsch, and R. Rott. 1985. Further studies on the role of neuraminidase and the mechanism of low pH dependence in influenza virus-induced membrane fusion. J. Gen. Virol. 66:295-301. [DOI] [PubMed] [Google Scholar]
- 19.Huang, R. T., R. Rott, K. Wahn, H. D. Klenk, and T. Kohama. 1980. The function of the neuraminidase in membrane fusion induced by myxoviruses. Virology 107:313-319. [DOI] [PubMed] [Google Scholar]
- 20.Hui, E. K.-W., S. Barman, T. Y. Yang, and D. P. Nayak. 2003. Basic residues of the helix six domain of influenza virus M1 involved in nuclear translocation of M1 can be replaced by PTAP and YPDL late assembly domain motifs. J. Virol. 77:7078-7092. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Jin, H., G. P. Leser, J. Zhang, and R. A. Lamb. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klenk, H. D., and W. Garten. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2:39-43. [DOI] [PubMed] [Google Scholar]
- 23.Kobasa, D., S. Kodihalli, M. Luo, M. R. Castrucci, I. Donatelli, Y. Suzuki, T. Suzuki, and Y. Kawaoka. 1999. Amino acid residues contributing to the substrate specificity of the influenza A virus neuraminidase. J. Virol. 73:6743-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kundu, A., R. T. Avalos, C. M. Sanderson, and D. P. Nayak. 1996. Transmembrane domain of influenza virus neuraminidase, a type II protein, possesses an apical sorting signal in polarized MDCK cells. J. Virol. 70:6508-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kundu, A., M. A. Jabbar, and D. P. Nayak. 1991. Cell surface transport, oligomerization, and endocytosis of chimeric type II glycoproteins: role of cytoplasmic and anchor domains. Mol. Cell. Biol. 11:2675-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamb, R. A. 1983. The influenza virus RNA segments and their encoded proteins, p. 26-69. In P. Palese and D. W. Kingsbury (ed.), Genetics of influenza viruses. Springer-Verlag, Vienna, Austria.
- 27.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1531. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams, and Wilkins, Philadelphia, Pa.
- 28.Lin, S., H. Y. Naim, A. C. Rodriguez, and M. G. Roth. 1998. Mutations in the middle of the transmembrane domain reverse the polarity of transport of the influenza virus hemagglutinin in MDCK epithelial cells. J. Cell Biol. 142:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, C., M. C. Eichelberger, R. W. Compans, and G. M. Air. 1995. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J. Virol. 69:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitnaul, L. J., M. R. Castrucci, K. G. Murti, and Y. Kawaoka. 1996. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J. Virol. 70:873-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nayak, D. P., and S. Barman. 2002. Role of lipid rafts in virus assembly and budding. Adv. Virus Res. 58:1-28. [DOI] [PubMed] [Google Scholar]
- 32.Nayak, D. P., and E. K.-W. Hui. 2002. Assembly and morphogenesis of influenza viruses. Recent Res. Dev. Virol. 4:35-54. [Google Scholar]
- 33.Nayak, D. P., and E. K.-W. Hui. The role of lipid microdomains in virus biology. In P. Quinn (ed.), Subcellular biochemistry, vol. 37, in press. Kluwer Academic/Plenum Publishers, London, United Kingdom.
- 34.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palese, P., and R. W. Compans. 1976. Inhibition of influenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): mechanism of action. J. Gen. Virol. 33:159-163. [DOI] [PubMed] [Google Scholar]
- 36.Palese, P., and J. Schulman. 1975. Susceptibility of different strains of influenza A virus to the inhibitory effects of 2-deoxy-2,3-dehydro-n-trifluoracetylneuraminic acid (FANA). Virology 63:98-104. [DOI] [PubMed] [Google Scholar]
- 37.Palese, P., K. Tobita, M. Ueda, and R. W. Compans. 1974. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61:397-410. [DOI] [PubMed] [Google Scholar]
- 38.Roberts, P. C., R. A. Lamb, and R. W. Compans. 1998. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 240:127-137. [DOI] [PubMed] [Google Scholar]
- 39.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038-2044. [DOI] [PubMed] [Google Scholar]
- 40.Scheiffele, P., M. G. Roth, and K. Simons. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibata, S., F. Yamamoto-Goshima, K. Maeno, T. Hanaichi, Y. Fujita, K. Nakajima, M. Imai, T. Komatsu, and S. Sugiura. 1993. Characterization of a temperature-sensitive influenza B virus mutant defective in neuraminidase. J. Virol. 67:3264-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 43.Takeda, M., G. P. Leser, C. J. Russell, and R. A. Lamb. 2003. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA 100:14610-14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tobita, K., A. Sugiura, C. Enomote, and M. Furuyama. 1975. Plaque assay and primary isolation of influenza A viruses in an established line of canine kidney cells (MDCK) in the presence of trypsin. Med. Microbiol. Immunol. (Berlin) 162:9-14. [DOI] [PubMed] [Google Scholar]
- 45.White, J., A. Helenius, and M. J. Gething. 1982. Haemagglutinin of influenza virus expressed from a cloned gene promotes membrane fusion. Nature 300:658-659. [DOI] [PubMed] [Google Scholar]
- 46.Wiley, D. C., and J. J. Skehel. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56:365-394. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]