Abstract
Hepatitis B virus (HBV) and woolly monkey hepatitis B virus (WMHBV) have natural host ranges that are limited to closely related species. The barrier for infection of primates seems to be at the adsorption and/or entry steps of the viral replication cycle, since a human hepatoma cell line is permissive for HBV and WMHBV replication following transfection of cloned DNA. We hypothesized that the HBV and WMHBV envelope proteins contain the principal viral determinants of host range. As previously shown by using the hepatitis D virus (HDV) system, recombinant HBV-HDV particles were infectious in chimpanzee as well as human hepatocytes. We extended the HDV system to include HDV particles pseudotyped with the WMHBV envelope. In agreement with the natural host ranges of HBV and WMHBV, in vitro infections demonstrated that HBV-HDV and WM-HDV particles preferentially infected human and spider monkey cells, respectively. Previous studies have implicated the pre-S1 region of the large (L) envelope protein in receptor binding and host range; therefore, recombinant HDV particles were pseudotyped with the hepadnaviral envelopes containing chimeric L proteins with the first 40 amino acids from the pre-S1 domain exchanged between HBV and WMHBV. Surprisingly, addition of the human amino terminus to the WMHBV L protein increased infectivity on spider monkey hepatocytes but did not increase infectivity for human hepatocytes. Based upon these data, we discuss the possibility that the L protein may be comprised of two domains that affect infectivity and that sequences downstream of residue 40 may influence host range and receptor binding or entry.
Many virus-receptor interactions contribute to viral host range and therefore constitute an interspecies barrier, as is the most apparent circumstance for hepatitis B virus (HBV). Hepadnaviruses, like HBV, characteristically display very narrow host ranges that extend only to closely related species. The discovery and characterization of a nonhuman primate hepadnavirus from woolly monkeys confirmed the restrictions on the transmissibility of the virus between species, as chimpanzees were not susceptible to an efficient infection with woolly monkey hepatitis B virus (WMHBV) (18). However, analyses of the host range of WMHBV indicated that the host range extends to the close relative of the woolly monkey, the black-handed spider monkey; both primates are within the Atelidae family and the Atelinae subfamily. Transfection of the cloned genomes for both HBV and WMHBV into the human liver cell line Huh7 demonstrated assembly of infectious particles following unimpeded replication of both viruses (17), suggesting that the barrier for interspecies transmission is at the initial steps of infection, which include virus adsorption and uptake.
The viral glycoproteins contained in the HBV envelope are encoded by a single open reading frame and are translated from different in-frame start codons to a common stop codon to generate the small (S), middle (M), and large (L) proteins. All three proteins contain the surface domain (S domain), while the M and L proteins have a 55-amino-acid (aa) extension from the S domain that is known as the pre-S2 domain. The L protein has a further 108-aa region that extends from the pre-S2 domain to compose the pre-S1 domain. Synthesis of these proteins occurs at the endoplasmic reticulum membrane where lumenal translocation results in the addition of N-linked carbohydrates at Asn-146 in the S domains of half of the population of S, M, and L proteins. An N-linked carbohydrate is attached to Asn-4 of the pre-S2 domain of the M protein as well, which contains an additional modification of an O-linked glycosylation at Thr-37 in the pre-S2 domain. The L protein is modified at Gly-2 of the pre-S1 domain by myristylation (26), which is required for infectivity (6, 9, 25). Aside from being constrained to interact with a functional receptor, sequences in the HBV envelope proteins are inherently constrained by the small size of the genome. The genes encoding the envelope proteins overlap with the viral polymerase gene; therefore, the HBV envelope proteins are restricted by the maintenance of a sequence expressing a functional polymerase. The sequence variation between WMHBV and HBV is considerable, with the most significant divergence of 32% located within the pre-S1 domain. The pre-S1 domain overlaps with the spacer domain of the polymerase, a region that can tolerate sequence divergence (27). The evolutionary gap between HBV and WMHBV spans host species that represent Old World primates and New World primates. The pre-S1 domain is believed to dictate the species specificity observed by primate hepadnaviruses and has been implicated in receptor binding and host range (7).
An array of proteins have been described as putative receptors for HBV; however, the biological significance of these molecules has not been confirmed. Most of our knowledge of hepadnaviral entry has been obtained from studies on duck hepatitis B virus (DHBV) and its cellular receptor carboxypeptidase D (5, 12, 14, 15, 31), while glycine decarboxylase (p120) has been implicated as a coreceptor in this process (23). Carboxypeptidase D is widely expressed in nonhepatic tissues and in different avian species that are nonpermissive for DHBV. This observation is suggestive of the existence of a tissue- and species-specific coreceptor for DHBV, a hypothesis that has not been excluded for HBV but has yet to be proven. The human homologue for carboxypeptidase D has not been shown to be involved in HBV attachment and entry. It is entirely possible that the evolutionary distance between the avian and primate hepadnaviruses resulted in the selection of an unrelated receptor molecule. The availability of two closely related but biologically distinct primate hepadnaviruses provides the opportunity to determine the basis for the biological differences. The distinct host range phenotypes displayed by HBV and WMHBV in vivo provided an opportunity to examine the determinants of primate hepadnaviral infectivity and host range. There are limited resources to utilize for models of HBV infection. Only primary hepatocytes are susceptible to HBV infection, and optimizing this model to achieve maximal results has been technically problematic. Recently, Gripon et al. reported a hepatoma cell line susceptible to infection with HBV (10), and this in vitro model may prove useful in deciphering the steps of HBV infection.
In the present study, the hepatitis D virus (HDV) in vitro model for HBV infection initially reported by Sureau et al. (28-30) was utilized to examine determinants of host range. HDV is a defective virus that cannot replicate autonomously; it requires the helper functions of HBV. The role of HBV is limited to supplying the viral envelope, allowing the HDV RNA to be packaged and released as a viral particle, thereby providing a mode of transmission. The composition of the HDV envelope includes the HBV S, M, and L proteins, although the relative amount of each protein differs from that of HBV particles (3, 4). Previous studies using this system have demonstrated the absolute requirement for the L protein in infectivity despite the fact that HDV particles comprised of only the S protein can be assembled and secreted (28, 30).
In this report, we describe the in vitro production of recombinant HDV particles containing the primate hepadnaviral envelopes from HBV and WMHBV. The experimental design presented us with the opportunity to use HDV RNA replication as a uniform indicator of infection with HDV particles containing different envelopes. By using this system, we observed the influence of each envelope on host range without introducing different viral genomes into the assay. Primary cultures of human and spider monkey hepatocytes were infected with recombinant HDV particles, and the infectivity was assessed by analysis of HDV replication. The results demonstrated the preferential infectivity of HBV-HDV and WM-HDV on human and spider monkey hepatocytes, respectively. Based upon the unanticipated results with HDV particles containing chimeric L proteins, we discuss the possibility that the L protein is comprised of two domains that affect infectivity and host range.
MATERIALS AND METHODS
Plasmid constructions.
For the production of HBV envelope proteins in Huh7 cells, four expression vectors were constructed. All HBV constructs were derived from an infectious, greater-than-genome-length construct of genotype ayw3 (2). A BglII-to-BglII (nucleotides [nt] 1987 to 2425) fragment was cloned into the BamHI site of plasmid pZErO-1 (Invitrogen). The resulting recombinant plasmid, pZHB2.7, contains the HBV pre-S1-pre-S2-S gene and can direct the expression of S, M, and L HBV envelope proteins. It includes the HBV promoter upstream of the pre-S1 region for expression of the mRNA for the L protein, the HBV promoter within the pre-S1 region for expression of the M and S mRNAs, and the HBV polyadenylation signal. For the construction of plasmid pZWM2.7, a greater-than-genome-length construct of WMHBV DNA (17) was digested to release a KpnI-to-SspI (nt 1980 to 2669) fragment that was blunted at the SspI site. The KpnI-SspI fragment was inserted between the KpnI and EcoRV sites of the pZErO-1 plasmid. This recombinant plasmid contains the WMHBV pre-S1-pre-S2-S gene and can direct the expression of S, M, and L WMHBV envelope proteins, as described for HBV above. Two plasmids were constructed to analyze the function of the first 40 amino acids of the pre-S1 region in primate hepadnaviral infection. pZHu40WM2.7 was derived from pZWM2.7 by using overlapping PCR to amplify HBV nt 2839 (BglII site) to 2975 and WMHBV nt 2949 to 3182 (EcoRI). The resulting chimeric fragment was inserted between the BglII and EcoRI sites in pZWM2.7. This recombinant plasmid encodes HBV pre-S1 aa 1 to 40 and WMHBV pre-S1 aa 41 to 108, followed by the WMHBV pre-S2-S gene. pZWM40HB2.7 is derived from pZHB2.7 by using overlapping PCR to amplify WMHBV nt 2839 (BglII site) to 2972 and HBV nt 2955 to 3182 (EcoRI site). The resulting chimeric fragment was inserted between the BglII and EcoRI sites in pZHB2.7. This recombinant plasmid encodes WMHBV pre-S1 aa 1 to 40 fused to HBV pre-S1 aa 41 to 108, followed by the HBV pre-S2-S gene.
The recombinant plasmid pSVLD3, a gift from John Taylor, contains a head-to-tail trimer of full-length HDV cDNA for expression of HDV genomic RNA under the control of the simian virus 40 late promoter (13).
Hepatocytes.
Primary human hepatocytes were purchased from BD Gentest (BD Biosciences Discovery Labware, Woburn, Mass.) and were obtained with the appropriate informed consent of the donor or donor's next of kin, in compliance with the Uniform Anatomical Gift Act and all other local, state, and federal laws and regulations governing the recovery and distribution of human tissue. Spider monkey hepatocytes were isolated from a liver wedge from a spider monkey in accordance with protocols approved by the Institutional Animal Care and Use Committee. Black-handed spider monkeys (Ateles geoffroyi) were housed at the Southwest National Primate Research Center at the Southwest Foundation for Biomedical Research and were cared for by members of the Department of Comparative Medicine in accordance with the Guide for the Care and Use of Laboratory Animals. Primary spider monkey hepatocytes were prepared and cultivated as described for the cultivation of primary primate hepatocytes (1, 16, 20). Hepatocytes were isolated by a two-step perfusion method using Liberase CI collagenase (Roche Applied Science, Indianapolis, Ind.), seeded at a density of approximately 3 × 106 cells per well in six-well plates (BD Falcon Primaria, BD Biosciences Discovery Labware, Bedford, Mass.), and incubated at 37°C and 10% CO2. Both human and spider monkey primary hepatocytes were maintained in a simplified formulation of our original serum-free medium as described previously (19), and the medium was changed every 72 h. Primary hepatocytes were used 3 to 6 days postplating for HDV infection studies.
Preparation of recombinant HDV particles.
The procedure for the production of recombinant HDV particles has been previously described (1, 30). The human hepatoma cell line Huh7 was maintained in Dulbecco's modified Eagle’s medium-Ham's F-12 medium (Mediatech, Inc., Herndon, Va.) containing 10% fetal bovine serum (HyClone, Logan, Utah), 2 mM l-glutamine (Mediatech, Inc.), and 50 μg of gentamicin sulfate per ml. For production of recombinant HDV particles, Huh7 cells were transfected with a mixture of cloned HDV cDNA and one of the HBV or WMHBV envelope protein expression plasmids. Cells were seeded at 8.4 × 106 per 100-mm-diameter tissue culture dish and transfected with 27 μg of the envelope protein expression plasmid, 3 μg of pSVLD3 (HDV cDNA), and 60 μg of TransIT-LT1 (Mirus Corp.) at 24 h after being seeded. Cells were exposed to the lipid-DNA transfection mixture for 6 h, flooded with fresh culture medium, and then incubated at 37°C and 10% CO2. To remove residual DNA, transfected cultures were washed with phosphate-buffered saline extensively on three separate occasions, days 0, 3, and 6 posttransfection, and the medium was changed every 3 days. Culture medium was harvested on days 9, 12, and 15 posttransfection, pooled, and clarified by centrifugation at 3,300 × g at 4°C for 30 min.
In vitro infections.
Cells were used for in vitro infections 3 to 6 days postplating and were exposed to HDV and 5% polyethylene glycol (PEG) 8000 for 16 h. The concentration of HDV genomes in the inoculum was estimated by quantitative reverse transcription (RT)-PCR (see below). Inocula were adjusted to contain 6.4 × 107 genome equivalents (ge) of HDV RNA/ml by adding serum-free medium. After exposure to the inoculum, cells were washed and incubated in 2 ml of fresh serum-free medium. Cells were harvested on different days thereafter for detection of intracellular HDV RNA by Northern hybridization or quantitative RT-PCR.
Analysis of HDV RNA.
For analysis by Northern hybridization, portions (500 μl) of the inocula were clarified by centrifugation at 13,000 × g at 4°C for 30 min. Recombinant HDV particles were precipitated from the culture medium by incubation on ice with 9% PEG 8000. Recombinant HDV particles were then pelleted by centrifugation at 13,000 × g at 4°C for 20 min. The pellet was suspended in 1 ml of TRIzol reagent (Invitrogen Corp., Carlsbad, Calif.), and RNA from the recombinant HDV particles was prepared per the manufacturer's specifications. Infected cells were harvested on different days postinoculation with 1 ml of TRIzol reagent, total cellular RNA was isolated as described by the manufacturer, and 5 μg of total cellular RNA was analyzed by Northern hybridization for the presence of HDV RNA. RNA was heated for 10 min at 70°C in RNA sample buffer containing 63% formamide and 2.1 M formaldehyde. RNA samples were subjected to electrophoresis through a 1% agarose-2.2 M formaldehyde gel and then transferred to a GeneScreen Plus hybridization transfer membrane (Perkin Elmer Life Sciences Inc.) for hybridization to an HDV-specific antigenomic Riboprobe (Promega) as described previously (1). After hybridization, the membrane was washed, dried, and autoradiographed at −80°C with intensifying screens. To quantify the data, the hybridized membrane was digitally scanned with a PhosphorImager 445 SI from Molecular Dynamics (Amersham Bioscences Corp.), and the data were analyzed by using the IPLab gel analysis software (Scanalytics, Inc., Fairfax, Va.).
HDV RNA was also quantified by a real-time, 5′ exonuclease RT-PCR (TaqMan) assay by using the ABI 7700 sequence detector (PE Biosystems, Foster City, Calif.). The primers and probe were selected by using the Primer Express software designed for this purpose (PE Biosystems). The forward primer consisted of nt 1007 to 1030 (5′-TATCCTATGGAAATCCCTGGTTTTC-3′), the reverse primer consisted of nt 1077 to 1061 (5′-CCCGGAGTCCCCCTTCT-3′), and the probe consisted of nt 1037 to 1057 (5′-TGTCCAGCCCCTCCCCG-3′). The fluorogenic probe was labeled with 6-carboxyfluorescein and 6-carboxytetramethylrhodamine and was obtained from Synthegen (Houston, Tex.). The primers and probe were used at 10 pmol/50-μl reaction mixture. To remove any residual HDV plasmid DNA that would interfere with RNA estimations, RNA was treated with DNase prior to RT-PCR. After isolation of RNA, 4 U of DNase I RNase-Free (Ambion, Inc., Austin, Tex.) per 1 μg of total cellular RNA or 25 U of DNase I RNase-Free per 100 μl of inoculum was added, and samples were incubated at 37°C for 30 min. RNA samples were then heated at 95°C for 10 min to denature the HDV RNA secondary structure. The ability to accurately determine the HDV RNA concentration was dependent on the latter denaturation step. The reactions were performed using a Brilliant Plus single-step RT-PCR kit (Stratagene, La Jolla, Calif.) and included a 30-min, 48°C reverse transcription step, followed by 10 min at 95°C, and then 40 cycles of amplification using the universal TaqMan standardized conditions: 15 s at 95°C for denaturation and 1 min at 60°C for annealing and extension. The standards used to establish genome equivalents were synthetic RNAs transcribed from a cDNA clone of HDV RNA. Synthetic RNA was prepared by using a MEGAscript T7 kit (Ambion, Inc.) and was purified by DNase treatment, RNazol extraction, and ethanol precipitation. RNA was quantified by determining optical density, and 10-fold serial dilutions were prepared from 106 to 10 molecules by using tRNA as a carrier. These standards were run in all TaqMan assays in order to calculate genome equivalents in the experimental samples.
RESULTS
Effect of PEG on in vitro infection of hepatocytes with the HDV model for HBV infections.
The use of PEG to enhance HBV infections in vitro initially met with some skepticism (8, 24). PEG is a highly hydrated polymer that can bring vesicle membranes to near molecular contact by making water between them thermodynamically unfavorable (21). In this study, we explored the effects of PEG prior to utilizing this methodology in studies on the species specificity of recombinant HDV particles.
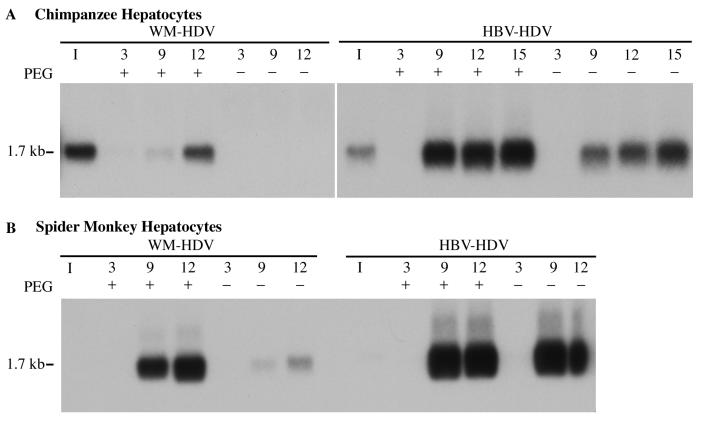
Initially, we confirmed the enhancing effects of PEG on infections of primary hepatocytes with recombinant HDV particles. Primary chimpanzee (Fig. 1A) and spider monkey hepatocytes (Fig. 1B) were inoculated with HBV-HDV or WM-HDV in the presence or absence of PEG. Cells were exposed to the inocula for 16 h at 37°C and then washed to remove excess PEG and virus. The cells were harvested at different days postinoculation and analyzed for HDV RNA by Northern hybridization. An increased infectivity for both HBV-HDV and WM-HDV particles in chimpanzee hepatocytes was observed in the presence of PEG (Fig. 1A). HBV-HDV particles were capable of efficiently infecting chimpanzee hepatocytes in the absence of PEG as previously described (29), yet PEG significantly increased the efficiency of infection (Fig. 1A). No infectivity was observed for WM-HDV in chimpanzee hepatocytes in the absence of PEG. WM-HDV infection was much less efficient in the presence of PEG than that observed for HBV-HDV, even though analysis of the WM-HDV inoculum indicated that cultures were exposed to a viral titer of WM-HDV that was higher than that of HBV-HDV (Fig. 1A). This host range difference was anticipated, since WMHBV does not efficiently infect chimpanzees in vivo (18). In spider monkey hepatocytes, WM-HDV particles showed a greater infectivity in the presence of PEG than they did in chimpanzee cells, and some infection was observed in the absence of PEG (Fig. 1B). Surprisingly, HBV-HDV also efficiently infected spider monkey hepatocytes with little apparent difference in the presence or absence of PEG. Absolute quantification of infections was performed in later experiments.
FIG. 1.
WM-HDV and HBV-HDV infection of primary hepatocytes in the presence or absence of PEG. Chimpanzee (A) and spider monkey (B) primary hepatocytes were inoculated with WM-HDV and HBV-HDV particles in the presence or absence of 5% PEG 8000. Cultures were harvested on days 3, 9, and 12 postinoculation (day 15 included for cells infected with HBV-HDV in panel A), and 5 μg of total cell RNA (approximately 15% of RNA from a 35-mm dish) was analyzed by Northern blot hybridization using a Riboprobe for HDV genomic RNA. RNA extracted from the equivalent of 5% of the inocula was analyzed under the same conditions (lane I). The size of HDV genomic RNA (in kilobases) is indicated.
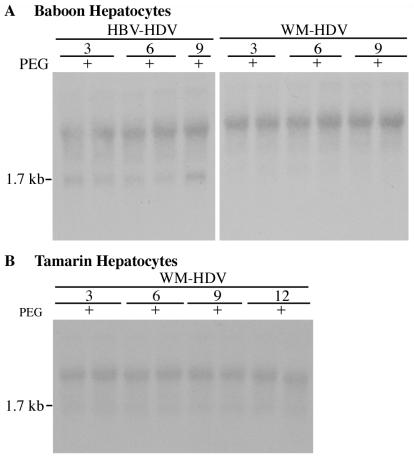
To confirm the specificity of the in vitro HDV infection system in the presence of PEG, hepatocytes from Old World and New World monkeys that are not susceptible to HBV infection were examined. Hepatocytes from baboons (Fig. 2A) and tamarins (Fig. 2B) were inoculated with recombinant HDV particles. Baboon hepatocytes infected with HBV-HDV and WM-HDV in the presence of PEG were harvested in duplicate on days 3, 6, and 9 postinoculation (Fig. 2A). No HDV replication was detected by Northern analysis, pointing to the absence of HDV infection in baboon hepatocytes (Fig. 2A). For reference, compare the HBV-HDV infection illustrated in Fig. 1A (2.5-h exposure) with that shown in Fig. 2A (7-day exposure). Even after extensive overexposure of the autoradiograph (7-day exposure with intensifying screens), no infection of tamarin hepatocytes with WM-HDV could be detected (Fig. 2B). We further demonstrated specificity by showing that recombinant HDV particles containing only the S protein of HBV were not infectious on human hepatocytes in the presence of PEG (see the discussion of chimeric L proteins below).
FIG. 2.
Lack of infection of baboon and tamarin hepatocytes with HBV-HDV and WM-HDV. Cultures of baboon (A) and tamarin (B) hepatocytes were inoculated in duplicate with HBV-HDV (A) and WM-HDV (A and B) in the presence of 5% PEG. Cultures were harvested on days 3, 6, and 9 postinoculation, and total cellular RNA (5 μg) was analyzed by Northern blot hybridization using a Riboprobe for HDV genomic RNA. RNA extracted from 10% of the inocula was analyzed under the same conditions (lane I). The size of HDV genomic RNA (in kilobases) is indicated.
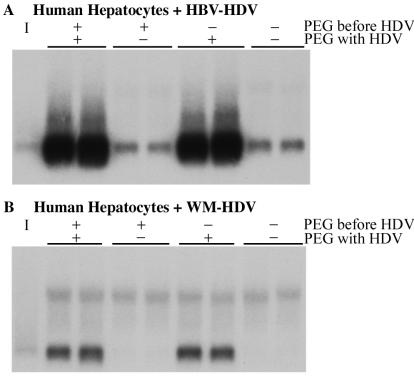
To determine if PEG was altering the cellular membranes of primary hepatocytes in such a manner as to enhance susceptibility to infection with HDV particles, primary human hepatocytes were incubated with serum-free medium containing 5% PEG for 6 h at 37°C, washed to remove the PEG, and then inoculated with HBV-HDV (Fig. 3A) or WM-HDV (Fig. 3B) particles in the presence or absence of 5% PEG overnight. Hepatocytes were harvested on day 9 postinoculation and analyzed for the presence of HDV RNA. Hepatocytes incubated with inocula and PEG simultaneously were susceptible to infection (Fig. 3), while pretreatment of the cultures with PEG appeared to have no effect, indicating that the presence of PEG with the viral inocula is necessary for the enhanced phenotype.
FIG. 3.
Requirement for simultaneous exposure of hepatocytes to PEG and virus for enhanced infection. Human hepatocytes were infected in duplicate with HBV-HDV (A) and WM-HDV (B) with or without prior exposure to PEG and with or without exposure to PEG during virus incubation. Preexposure to PEG (PEG before HDV, +/−) consisted of incubation in the presence of 5% PEG for 6 h at 37°C and then the removal of PEG by washing. Exposure to PEG plus virus (PEG with HDV, +/−) followed the standard protocol for exposure to virus in the presence or absence of 5% PEG. Cultures were harvested on day 9 postinoculation, and total cellular RNA (5 μg) was analyzed by Northern blot hybridization using a Riboprobe for HDV genomic RNA. RNA extracted from 10% of the inocula was analyzed under the same conditions (lane I).
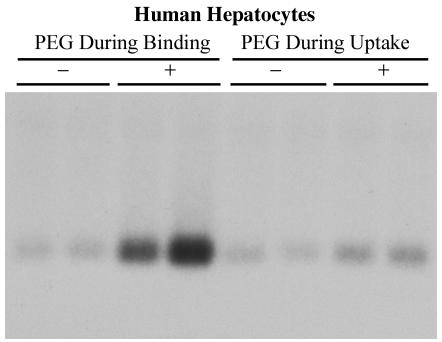
To identify the mechanism by which PEG enhances infectivity, we looked at whether PEG facilitates binding of HDV particles to the cell or uptake of bound HDV into the cell, perhaps by a fusogenic mechanism. HBV-HDV particles were allowed to bind to primary human hepatocytes for 2 h at 4°C in the presence or absence of PEG. After the 2-h incubation, cells were washed vigorously to remove unbound viral particles and incubated overnight at 37°C in the presence or absence of PEG. Hepatocytes were harvested on day 12 postinoculation and analyzed for the presence of HDV RNA. An obvious increase in efficiency of infection was observed in hepatocytes that were exposed to PEG during viral binding, while no significant enhancement was observed when particles were bound in the absence of PEG and then exposed to PEG after binding (Fig. 4). These results indicate that PEG facilitates the binding of recombinant HDV particles to the cell rather than facilitating the uptake of bound particles into the cell. Due to the requirement of WM-HDV for PEG for detectable infections, subsequent experiments were conducted in the presence of PEG. All subsequent infections utilized inocula precisely balanced by quantitative RT-PCR, such that infectivity of HBV-HDV and WM-HDV particles could be accurately compared, and all experiments were conducted using a single preparation of each envelope construct.
FIG. 4.
PEG enhances virus binding but not postbinding events. PEG During Binding: HBV-HDV particles were allowed to adsorb onto human hepatocytes for 2 h at 4°C in the presence (+) or absence (−) of 5% PEG; the cultures were then washed and incubated overnight at 37°C in the absence of PEG. PEG During Uptake: virus was allowed to adsorb onto hepatocytes for 2 h at 4°C in the absence of PEG; the cultures were washed and then incubated overnight at 37°C in the presence (+) or absence (−) of PEG. Cultures were harvested on day 12 postinoculation, and total cellular RNA (5 μg) was analyzed by Northern blot hybridization using a Riboprobe for HDV genomic RNA. RNA extracted from 10% of the inocula was analyzed under the same conditions (lane I).
Infectivity of recombinant HDV particles in human hepatocytes.
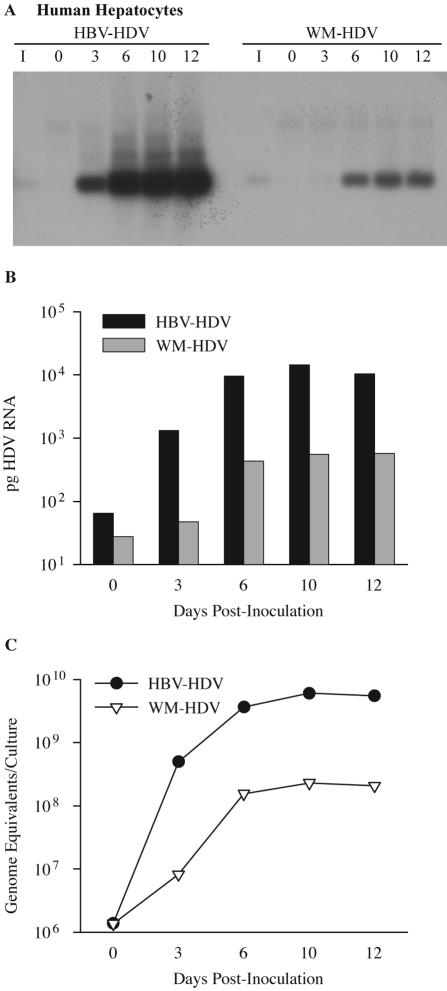
Human hepatocytes were inoculated with 6.4 × 107 HDV ge of HBV-HDV or WM-HDV and harvested on days 0, 3, 6, 10, and 12 postinoculation for detection of intracellular HDV RNA. HDV RNA from HBV-HDV particles was not detected on day 0 but was present at increasing levels starting on day 3 (Fig. 5). These results clearly demonstrated that HBV-HDV particles produced in Huh7 cells following transfection were infectious in human hepatocytes, as previously shown with chimpanzee hepatocytes (29). WM-HDV exhibited a low efficiency of infection in human hepatocytes (Fig. 5A), similar to that demonstrated in chimpanzee hepatocytes (Fig. 1A). These results established a preferential infectivity displayed by HBV-HDV particles over WM-HDV particles for human hepatocytes. Hybridization signals for the inocula were not detected in Fig. 5A, because the autoradiography exposure time was below that necessary to detect 10% of the inocula (6-h exposure to film).
FIG. 5.
Infectivity of HDV pseudotypes of HBV and WMHBV in human hepatocytes. (A) Human hepatocytes were infected with HBV-HDV or WM-HDV particles. Cultures were harvested on days 0, 3, 6, 10, and 12 postinoculation, and total cellular RNA (5 μg) was analyzed by Northern blot hybridization using a Riboprobe for HDV genomic RNA. RNA extracted from 10% of the inocula was analyzed under the same conditions (lane I). (B) Autoradiographs from panel A were scanned with a phosphorimager. The amount of HDV RNA is expressed in picograms per culture and was derived by linear regression analysis of HDV RNA standards loaded on the same gel. (C) Levels of HDV RNA from the same cultures analyzed in panels A and B were quantified by TaqMan RT-PCR and expressed as genomic equivalents per culture.
To quantify results from the infections, autoradiographs of Northern blots detecting HDV RNA replication were analyzed with a phosphorimager. Standards of known amounts of synthetic HDV RNA were included on the same gel and were plotted to arrive at a linear regression model of total picograms of HDV RNA per infected culture. The amount of HDV RNA at time zero was difficult to estimate by this method and was considerably lower than the amount of HDV RNA that was present in the inoculum. HDV RNA at time zero presumably represented virus particles attached to the culture dish or the cell surface in addition to the virus particles that were internalized by the cells following 16 h of incubation at 37°C with the inocula. From day 3 to day 10, HDV RNA increased from 1,300 to 14,000 pg per culture (Fig. 5B). In contrast, for WM-HDV infection of human hepatocytes, the peak level of HDV RNA was only 580 pg on day 12. Comparison of peak HDV RNA levels for HBV- and WM-HDV-infected human hepatocytes revealed a 25-fold increase in infection by HBV-HDV.
Since the estimation of low levels of HDV RNA at day 0 by Northern hybridization was difficult, we chose to use a quantitative RT-PCR assay (TaqMan) to better quantify and monitor HDV RNA during the course of in vitro infections. The growth curve of HDV from human hepatocytes that were infected with HBV-HDV displayed an exponential 360-fold increase between days 0 and 3 postinoculation to levels of 5.0 × 108 ge per culture (Fig. 5C and Table 1). Peak levels of intracellular viral RNA were observed (6.0 × 109 ge per culture) on day 10, with a 4,400-fold increase in intracellular HDV RNA from days 0 to 10. Only a twofold increase in HDV RNA levels occurred from days 6 to 10, suggesting that near-maximum replication occurs by day 6. In contrast, human hepatocytes inoculated with WM-HDV exhibited a delay in the exponential phase of the growth curve, with only a sixfold increase from days 0 to 3. However, a 110-fold increase occurred by day 6, and peak levels on day 10 (2.3 × 108 ge per culture) represented a 170-fold increase from day 0. At maximal levels, HDV RNA from an infection of human hepatocytes with HBV-HDV displayed a 26-fold increase when compared to an infection with WM-HDV. This approximated the results observed using phosphorimaging.
TABLE 1.
Infectivity levels of recombinant HDV particles in human and spider monkey hepatocytesa
| Cell type and day postinoculation | Infectivity levels for:
|
|||
|---|---|---|---|---|
| HBV-HDV | WM-HDV | WM40-HDV | Hu40-HDV | |
| Human hepatocytes | ||||
| 0 | 1.4 × 106 | 1.4 × 106 | 3.8 × 106 | 1.3 × 106 |
| 3 | 5.0 × 108 | 8.2 × 106 | 8.9 × 106 | 1.5 × 107 |
| 6 | 3.7 × 109 | 1.6 × 108 | 1.5 × 108 | 4.9 × 108 |
| 9 or 10 | 6.0 × 109 | 2.3 × 108 | 3.5 × 108 | 5.4 × 108 |
| 12 | 5.5 × 109 | 2.1 × 108 | ||
| Spider monkey hepatocytes | ||||
| 0 | 1.2 × 106 | 2.6 × 106 | 3.5 × 106 | 2.4 × 106 |
| 3 | 2.9 × 105 | 6.3 × 105 | 1.3 × 106 | 1.1 × 106 |
| 6 | 1.4 × 107 | 9.3 × 107 | 3.9 × 106 | 4.7 × 108 |
| 9 | 2.9 × 108 | 5.5 × 108 | 4.6 × 107 | 3.0 × 109 |
| 12 | 8.3 × 108 | 1.2 × 109 | 3.0 × 108 | 8.2 × 109 |
Values were determined by TaqMan RT-PCR and are expressed as genome equivalents per infected culture.
Infectivity of recombinant HDV particles in spider monkey hepatocytes.
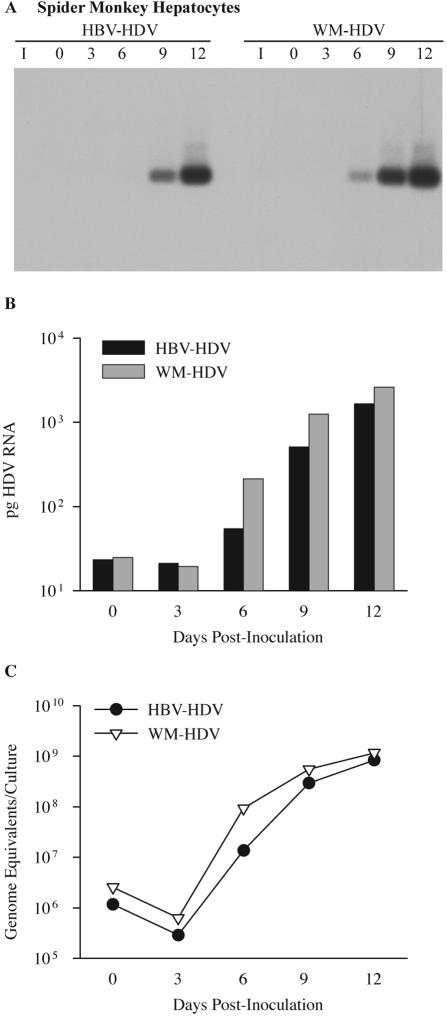
The infectivity of recombinant HDV particles was also determined in primary cultures of spider monkey hepatocytes. Spider monkey hepatocytes were inoculated with 6.4 × 107 ge of HBV-HDV or WM-HDV, and the cultures were harvested on days 0, 3, 6, 9, and 12 postinoculation for detection of intracellular HDV RNA. Unlike the infections in human hepatocytes, HDV RNA from HBV-HDV-infected cells was not detected until day 9 postinoculation, while HDV RNA from WM-HDV-infected cells was detected on day 6 postinoculation (Fig. 6A). Thus, the efficiency of infection by HBV-HDV particles was lower in spider monkey hepatocytes than in human hepatocytes, and the efficiency of WM-HDV infection, although greater than in human cells, was only marginally better than HBV-HDV in spider monkey cells.
FIG. 6.
Infectivity of HDV pseudotypes of HBV and WMHBV in spider monkey hepatocytes. (A) Spider monkey hepatocytes were infected with HBV-HDV and WM-HDV particles. Cultures were harvested on days 0, 3, 6, 9, and 12 postinoculation, and total cellular RNA was analyzed as described in the legend for Fig. 5. (B) Autoradiographs from panel A were scanned with a phosphorimager. The amount of HDV RNA is expressed in picograms per culture and was derived by linear regression analysis of HDV RNA standards loaded on the same gel. (C) Levels of HDV RNA from the same cultures analyzed in panels A and B were quantified by TaqMan RT-PCR and expressed as genomic equivalents per culture.
Analysis by phosphorimaging revealed that infection of spider monkey hepatocytes was nearly equivalent for WM-HDV and HBV-HDV (Fig. 6B). On day 12 at peak levels, HBV-HDV-infected cultures contained 1,700 pg of HDV RNA, while WM-HDV-infected cultures contained 2,600 pg, reflecting a twofold difference.
To better quantify the HDV RNA levels at early time points, quantitative RT-PCR was performed. RNA levels actually declined from days 0 to 3, probably due to the loss of nonspecifically adsorbed particles during a time when RNA replication was still low. Increases in HDV RNA in HBV-HDV- and WM-HDV-infected spider monkey hepatocytes were nearly parallel from days 3 to 12, with WM-HDV RNA levels minimally greater than those of HBV-HDV. By day 12 postinoculation, peak levels of intracellular viral RNA were observed for HBV-HDV (8.3 × 108 ge per culture) and WM-HDV (1.2 × 109 ge per culture), with a 710- and 460-fold increase, respectively, in intracellular HDV RNA occurring from day 0 (Fig. 6C and Table 1). The severalfold increase from day 0 was greater for HBV-HDV even though WM-HDV had higher peak RNA levels, because the day 0 HBV-HDV baseline was lower. These data indicate that although a substantial host range difference is observed for the two viruses in human hepatocytes, HBV-HDV and WM-HDV particles infect and replicate to near equal levels in spider monkey hepatocytes. Nonetheless, for WM-HDV, this represents a sixfold increase in the efficiency of infection in spider monkey hepatocytes in comparison to that in human hepatocytes, while it represented a sevenfold decrease in efficiency for HBV-HDV.
Infectivity of HDV pseudotypes with HBV and WMHBV chimeric L proteins in human and spider monkey hepatocytes.
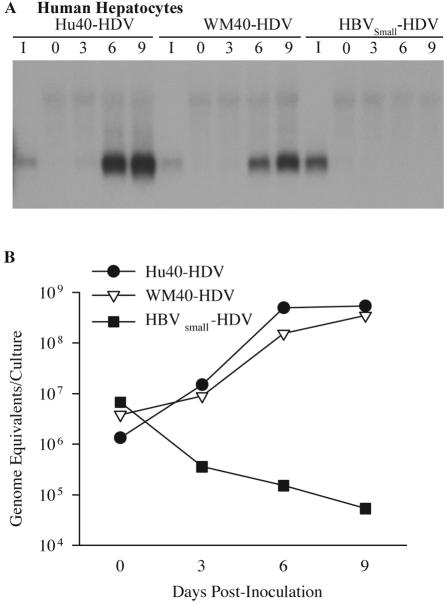
To test the role of the pre-S1 domain of the hepadnaviral L protein in host range, the infectivity of HDV pseudotypes containing L proteins with aa 1 to 40 exchanged from HBV (Hu40-HDV) or WMHBV (WM40-HDV) was determined in primary cultures of both human and spider monkey hepatocytes. As described in Materials and Methods, Hu40-HDV and WM40-HDV particles are similar to WM-HDV and HBV-HDV particles, respectively, except for the first 40 aa of the pre-S1 domain of the L protein; thus, the nomenclature emphasizes the origin of the amino terminus of the L protein. Primary hepatocytes were inoculated with 6.4 × 107 ge of Hu40-HDV or WM40-HDV, and the cultures were harvested on days 0, 3, 6, and 9 postinoculation. In human hepatocytes, HDV RNA was detected on day 6 postinoculation for both Hu40-HDV and WM40-HDV (Fig. 7A), with only a slightly more intense signal displayed by hepatocytes infected with Hu40-HDV. This suggests that the HBV sequence from aa 1 to 40 was not sufficient to dramatically enhance infection of WM-HDV in human hepatocytes (compare Fig. 5A and 7A).
FIG. 7.
Infectivity of HDV pseudotypes with chimeric HBV and WMHBV L proteins in human hepatocytes. (A) Human hepatocytes were infected with Hu40-HDV and WM40-HDV particles. Cultures were also inoculated with HDV pseudotypes containing only the HBV S protein (HBVSmall-HDV) in the presence of 5% PEG. Cultures were harvested on days 0, 3, 6, and 9 postinoculation, and total cellular RNA was analyzed as described in the legend for Fig. 5. (B) Levels of HDV RNA from the same cultures analyzed in panel A were quantified by TaqMan RT-PCR and expressed as genomic equivalents per culture.
The growth curve of HDV from human hepatocytes that were infected with Hu40-HDV displayed a 400-fold increase between days 0 and 9 postinoculation to levels of 5.4 × 108 ge per culture (Fig. 7B and Table 1). A similar increase was observed in human hepatocytes infected by WM40-HDV, with the peak viral level on day 9 designated as 3.5 × 108 ge per culture. These data demonstrate that the HBV sequence from aa 1 to 40 failed to convey to the WMHBV envelope the ability to infect human hepatocytes at an efficiency equivalent to that of HBV-HDV. Peak viral RNA levels for Hu40-HDV were 1 log lower than those of HBV-HDV (Table 1). In addition, WMHBV aa 1 to 40 reduced the infectivity of HBV-HDV in human hepatocytes approximately 30-fold, from 6.0 × 109 to 3.5 × 108 ge per culture at peak levels. Thus, although critical for efficient infection of human hepatocytes, the HBV sequence from aa 1 to 40 does not appear to comprise the entire host range determinant.
The absolute requirement for the L protein was demonstrated by producing particles that contained only the S protein of HBV (HBVSmall-HDV). Previous studies demonstrated that HDV particles containing only the S protein can be assembled but are not infectious in vitro on chimpanzee hepatocytes (28). Human hepatocytes were exposed to 1.9 × 108 ge of HBVSmall-HDV for 16 h at 37°C, and the cells were harvested at different days postinoculation. In agreement with the previous observation (28), no HDV infection of human hepatocytes with HBVSmall-HDV particles was detected, even in the presence of PEG (Fig. 7A). The lack of infectivity is especially evident in the RT-PCR data, where the level of bound virions continues to decline after day 0 (Fig. 7B).
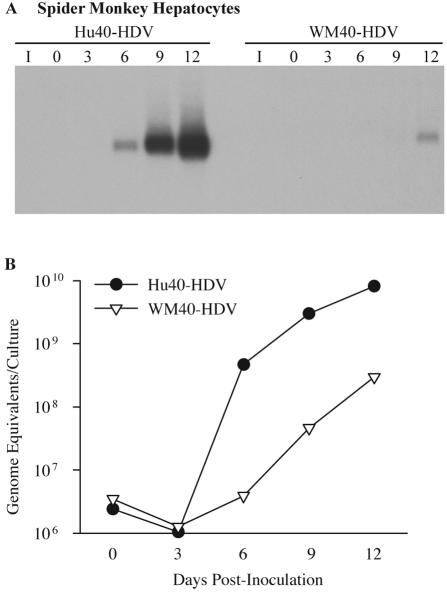
In spider monkey hepatocytes, HDV RNA was easily detected on day 6 postinoculation for the Hu40-HDV infection, whereas HDV RNA from WM40-HDV-infected cells was not detectable until day 12 (Fig. 8A). Similar to what was observed for Hu40-HDV in human hepatocytes, the WM40 region did not convey a high infectivity to the HBV envelope for spider monkey hepatocytes. Surprisingly, the Hu40 region dramatically increased the infectivity of the WMHBV envelope for spider monkey hepatocytes.
FIG. 8.
Infectivity of HDV pseudotypes with chimeric HBV and WMHBV L proteins in spider monkey hepatocytes. (A) Spider monkey hepatocytes were infected with Hu40-HDV and WM40-HDV particles. Cultures were harvested on days 0, 3, 6, 9, and 12 postinoculation, and total cellular RNA was analyzed as described in the legend for Fig. 5. (B) Levels of HDV RNA from the same cultures analyzed in panel A were quantified by TaqMan RT-PCR and expressed as genomic equivalents per culture.
Quantitative RT-PCR analysis of infected spider monkey hepatocytes revealed a 200-fold increase for Hu40-HDV RNA levels between days 0 and 6 postinoculation to 4.7 × 108 ge per culture and no detectable increase for WM40-HDV RNA levels during the same time period (Fig. 8B). HDV RNA from WM40-HDV-infected spider monkey hepatocytes increased 85-fold on day 12 to peak at only 3.0 × 108 ge per culture, while Hu40-HDV-infected cells increased 3,400-fold to 8.2 × 109 ge per culture. A 27-fold difference was observed in the infectivities of the two chimeric viruses on spider monkey hepatocytes, while the infectivities were nearly identical on human hepatocytes. These data demonstrated that aa 1 to 40 of the WMHBV L protein failed to convey to the HBV envelope the ability to infect spider monkey hepatocytes at the same efficiency as WMHBV and that, instead of reducing the infectivity, aa 1 to 40 of the HBV L protein actually increased the infectivity of the WMHBV envelope for spider monkey hepatocytes to a level greater than that of HBV-HDV on human hepatocytes (Table 1).
DISCUSSION
In this study, we demonstrate that recombinant HDV particles produced in vitro containing the primate hepadnaviral envelopes from HBV and WMHBV are infectious for human and spider monkey primary hepatocytes, with various levels of efficiency. The pattern of infectivity displayed by these viruses appeared to follow the natural host ranges for human HBV and WMHBV, since HBV-HDV and WM-HDV preferentially infected human and spider monkey hepatocytes, respectively.
Although the in vitro HDV infection system we employed utilizes PEG for enhancement of viral infection, the specificity of the system was not compromised. This was demonstrated by the lack of susceptibility of baboon and tamarin primary hepatocytes to recombinant HDV infection in the presence of PEG. In addition, HBVSmall-HDV particles were not infectious, corroborating previous work that demonstrated the requirement of the L protein for infectivity (28). In the absence of PEG, HBV-HDV exhibited detectable infectivity for both human and spider monkey hepatocytes, although a quantitative evaluation of the efficiency of infection in each species was not performed in those studies. In contrast, WM-HDV failed to detectably infect human hepatocytes and exhibited limited infectivity for spider monkey hepatocytes in the absence of PEG. PEG enhanced the infectivity of both viruses, and studies on the mechanism indicated that the enhancement occurred at the level of receptor binding and was not from a fusogenic event. Similar observations have been previously made with PEG by using HBV particles and human hepatocytes (8). Whether PEG induced subtle alterations in the host range of the viruses or induced differential effects on the infectivity of the two viruses could not be determined due to the low infectivity of WM-HDV in the absence of PEG.
Several factors should be considered in the interpretation of the data in this study. The results could be influenced by factors other than the efficiency of interaction of the various L proteins with the hepadnaviral receptor(s). The use of RNA copy number to balance the inocula could affect the comparison of the different viruses if viral preparations from different envelope constructs contain different ratios of infectious to noninfectious particles. HDV particles can be prepared that contain only the S envelope protein, and these particles are noninfectious (28; this study). There is no evidence that S-only particles are produced when abundant L protein is available for particle production. Indeed, this seems unlikely considering that S, M, and L proteins form complexes on the endoplasmic reticulum membrane prior to particle production. However, if S-only particles were produced, and the different constructs produced S-only particles at substantially different ratios, the outcome of the infections and interpretation of the data would be affected. Another factor to consider is the influence of PEG on the infections. The enhancing effect of PEG may not be identical for different envelope constructs. PEG may influence the conformation of the receptor or envelope in such a manner as to increase the affinity of the receptor-envelope interaction and thereby increase the number of productive interactions. Alternatively, PEG may cause the aggregation of particles and increase the number of genomes delivered to the cell per productive receptor interaction. This mechanism would still be dependent on the L protein-receptor interaction but could be potentially influenced by the ratio of infectious to noninfectious particles in different preparations if this allowed noninfectious particles containing a replication-competent genome to gain entry into the cell. The level of PEG required to induce precipitation of viral particles under the conditions used for infection is much greater than what was used to enhance infection; nonetheless, aggregation of particles may be involved. Considering that there is no evidence for the production of S-only particles in the presence of L protein and that this phenomenon would not address the differential infectivity of the various preparations on different species of hepatocytes, we believe that our interpretations of the data are valid.
An understanding of the biology of WMHBV is required to fully appreciate the data from these studies. HBV-HDV efficiently infects human hepatocytes and has a greatly reduced efficiency for infection of spider monkey hepatocytes. This is consistent with observations in vivo on the host range of this virus. HBV does not infect spider monkeys in vivo. WM-HDV does not infect human hepatocytes in the absence of PEG and, in comparison to HBV-HDV, has a greatly decreased efficiency for infection of human hepatocytes in the presence of PEG. This is also consistent with the observation that WMHBV does not infect chimpanzees. WM-HDV has approximately the same efficiency for infection of spider monkey hepatocytes as does HBV-HDV, and for WM-HDV, this represents an increase in the efficiency of infection in comparison to that for human hepatocytes. The question arises as to why WM-HDV does not infect spider monkey hepatocytes much more efficiently than does HBV-HDV. One possibility is that infection of spider monkey hepatocytes represents a jump in host range for both WM-HDV and HBV-HDV. This is consistent with in vivo observations. WMHBV induces a high-level viremia in woolly monkeys (∼109 ge/ml) but induces only a moderate viremia in spider monkeys (∼105 ge/ml). Thus, despite the permissiveness of spider monkeys for WMHBV infection, the level of viremia is restricted, and the present data can be interpreted to imply that the restriction occurs at the level of receptor interaction.
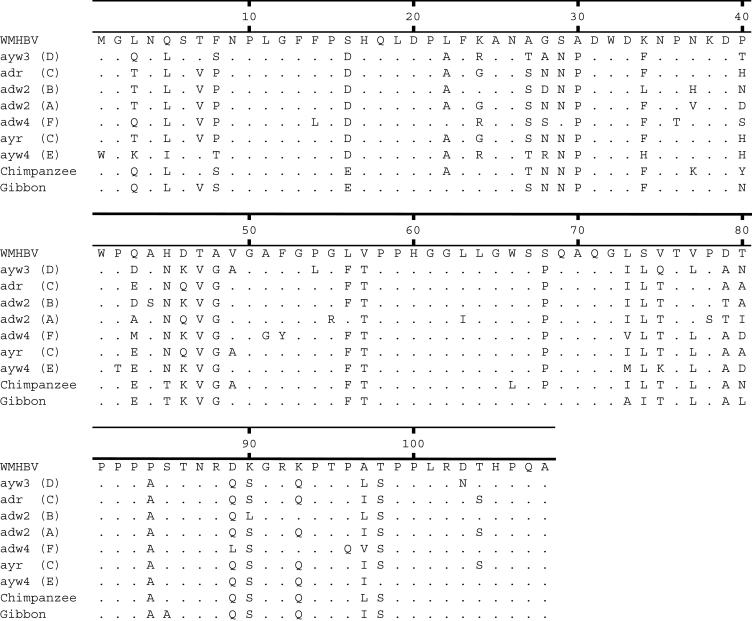
The host range differences of the two viruses were further explored by producing chimeric L proteins that exchanged the amino-terminal 40 aa of HBV and WMHBV. Both chimeras generated HDV particles infectious for both human and spider monkey hepatocytes. The WMHBV amino-terminal 40 aa of L protein dramatically reduced the infectivity of HBV-HDV on human hepatocytes, suggesting that critical residues for receptor interaction reside within this domain. However, the HBV sequence from the same region failed to increase the infectivity of WM-HDV on human hepatocytes. We interpret these data to suggest that, although necessary, this region is not sufficient for a high-affinity interaction with the human receptor. One interpretation of the data would imply that sequences downstream of residue 40 of the L protein might influence host range. This region differs by 12 aa between WMHBV and the ayw3 strain of HBV used in these studies, but only two of these residues are conserved across different HBV strains (residue 16, S → D; and residue 30, A → P) (Fig. 9). Most surprising in the analysis of the chimeric envelope proteins was the increased infectivity of Hu40-HDV on spider monkey hepatocytes. These data could be reconciled by assuming that the pre-S1 region has at least two critical domains for receptor interaction and infectivity: one that is located at the amino terminus of the L protein and one that is downstream of this region. Data from the DHBV model support this hypothesis. Although peptides from residues 2 to 41 block infection (32), the gp180 binding domain mapped to residues 43 to 108 (12). One possible interpretation of the data from the chimeric viruses is that the amino terminus of the HBV L protein exhibits the highest affinity for the spider monkey receptor(s), while the downstream region of the WMHBV L protein has the greatest affinity for the spider monkey receptor(s). Thus, neither wild-type virus exhibits maximum infectivity for spider monkey hepatocytes, while the Hu40 construct combines the best domains from both. The spider monkey receptor(s) may have by chance acquired a polymorphism that results in poor interaction with the amino-terminal domain of WMHBV and increased interaction with the same domain in the HBV L protein.
FIG. 9.
Alignment of HBV pre-S1 domains. The amino acid sequences of the pre-S1 domains for HBV genotypes A to F are displayed along with the sequences for WMHBV and HBV isolates from chimpanzees and gibbons. The GenBank numbers for the isolates are AY226578 (WMHBV), V01460 and J02203 (ayw3-D), L08805 (adr-C), M54923 (adw2-B), X02763 (adw2-A), X75663 (adw4-F), X04615 (ayr-C), X75664 (ayw4-E), D00220 (chimpanzee), and U46935 (gibbon).
Chimeric hepadnaviral envelopes have been used previously to define host range determinants. In the DHBV system, residues 22 to 90 of the L protein conveyed to heron HBV the ability to efficiently infect duck hepatocytes (11). Chimeric proteins containing WMHBV and HBV sequences have been used to examine the infectivity of hepadnaviral particles on human hepatocytes. In those studies, HBV residues 1 to 30 of the L protein restored infectivity of human hepatocytes for hepadnaviral particles containing a chimeric WMHBV envelope (7). In contrast, our findings with the HDV system suggest that HBV residues 1 to 40 are insufficient to provide the WMHBV envelope with the capacity to infect human hepatocytes at an efficiency equivalent to that of HBV-HDV. The reason for this discrepancy is not apparent but may be due to the differences in the HDV and HBV experimental systems. Due to the very high level of replication of HDV RNA, the HDV system should be a more sensitive determinant of infection. However, HBV and HDV particles may display subtle differences in receptor interactions, as well. Our results can be interpreted to imply that sequences downstream of residue 40 are important in receptor interaction. The same question has been approached in other studies. The effect on infectivity exhibited by five amino acid deletions throughout the pre-S1 domain of HBV suggested that residues 3 to 77 were needed for proper conformation and receptor binding (22). Studies have been conducted using peptides to inhibit infection, as well. As mentioned above, studies with DHBV demonstrated that peptides spanning residues 2 to 41 of the L protein inhibit infection of primary duck hepatocytes (32). This was surprising, since previous studies had suggested that a region between residues 43 and 108 was required for binding to the receptor candidate gp180 (12). Collectively, these studies can be used to build a model in which the amino terminus of the L protein interacts with one receptor component, while a downstream region interacts with gp180. Recently, a human hepatoma cell line that is permissive for HBV infection was isolated. In these studies, a myristylated peptide spanning residues 2 to 78 of the L protein inhibited HBV infection (10). We have analyzed peptides that span residues 2 to 45 with and without myristylation. Only the myristylated peptide inhibited HBV-HDV and WM-HDV infections, and a shorter peptide spanning residues 2 to 35 did not inhibit infection (unpublished observations). Further studies are needed to precisely map the receptor binding and host range determinants of primate hepadnaviruses.
In the future, the HBV-HDV and WM-HDV in vitro system should provide a mechanism to explore the authenticity of receptor candidates. Any valid receptor for HBV would presumably display a differential binding affinity for HBV and WMHBV L proteins. The sequence of the receptor candidate should differ across species of primates. It is difficult to predict the level of sequence variation that would account for the host range differences of HBV and WMHBV in humans, apes, woolly monkeys, and spider monkeys or the variations that would account for the lack of permissiveness of most primate species for either virus.
Acknowledgments
We thank Camille Sureau for his role in initially developing the HDV in vitro model and for his scientific contributions that prefaced our study.
This work was supported by grants RO1 AI46609 and P51 RR13986 from the National Institutes of Health. Azeneth Barrera is the recipient of an NIH graduate student fellowship.
REFERENCES
- 1.Barrera, A., and R. E. Lanford. 2004. Infection of primary chimpanzee hepatocytes with recombinant hepatitis D virus particles: a surrogate model for hepatitis B virus. Methods Mol. Med. 95:131-142. [DOI] [PubMed] [Google Scholar]
- 2.Beames, B., and R. E. Lanford. 1993. Carboxy-terminal truncations of the HBV core protein affect capsid formation and size of the encapsidated HBV RNA. Virology 194:597-607. [DOI] [PubMed] [Google Scholar]
- 3.Bonino, F., K. H. Heermann, M. Rizzetto, and W. H. Gerlich. 1986. Hepatitis delta virus: protein composition of delta antigen and its hepatitis B virus-derived envelope. J. Virol. 58:945-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonino, F., B. Hoyer, J. W.-K. Shih, M. Rizzetto, R. H. Purcell, and J. L. Gerin. 1984. Delta hepatitis agent: structural and antigenic properties of the delta-associated particle. Infect. Immun. 43:1000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breiner, K. M., S. Urban, and H. Schaller. 1998. Carboxypeptidase D (gp180), a Golgi-resident protein, functions in the attachment and entry of avian hepatitis B viruses. J. Virol. 72:8098-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruss, V., J. Hagelsten, E. Gerhardt, and P. R. Galle. 1996. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology 218:396-399. [DOI] [PubMed] [Google Scholar]
- 7.Chouteau, P., J. Le Seyec, I. Cannie, M. Nassal, C. Guguen-Guillouzo, and P. Gripon. 2001. A short N-proximal region in the large envelope protein harbors a determinant that contributes to the species specificity of human hepatitis B virus. J. Virol. 75:11565-11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gripon, P., C. Diot, and C. Guguen-Guillouzo. 1993. Reproducible high level infection of cultured adult human hepatocytes by hepatitis B virus: effect of polyethylene glycol on adsorption and penetration. Virology 192:534-540. [DOI] [PubMed] [Google Scholar]
- 9.Gripon, P., J. Le Seyec, S. Rumin, and C. Guguen-Guillouzo. 1995. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology 213:292-299. [DOI] [PubMed] [Google Scholar]
- 10.Gripon, P., S. Rumin, S. Urban, J. Le Seyec, D. Glaise, I. Cannie, C. Guyomard, J. Lucas, C. Trepo, and C. Guguen-Guillouzo. 2002. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. USA 99:15655-15660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa, T., and D. Ganem. 1995. The pre-S domain of the large viral envelope protein determines host range in avian hepatitis B viruses. Proc. Natl. Acad. Sci. USA 92:6259-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa, T., K. Kuroki, R. Lenhoff, J. Summers, and D. Ganem. 1994. Analysis of the binding of a host cell surface glycoprotein to the preS protein of duck hepatitis B virus. Virology 202:1061-1064. [DOI] [PubMed] [Google Scholar]
- 13.Kuo, M. Y.-P., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuroki, K., R. Cheung, P. L. Marion, and D. Ganem. 1994. A cell surface protein that binds avian hepatitis B virus particles. J. Virol. 68:2091-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroki, K., F. Eng, T. Ishikawa, C. Turck, F. Harada, and D. Ganem. 1995. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J. Biol. Chem. 270:15022-15028. [DOI] [PubMed] [Google Scholar]
- 16.Lanford, R. E., K. D. Carey, L. E. Estlack, G. C. Smith, and R. V. Hay. 1989. Analysis of plasma protein and lipoprotein synthesis in long-term primary cultures of baboon hepatocytes maintained in serum-free medium. In Vitro Cell. Dev. Biol. 25:174-182. [DOI] [PubMed] [Google Scholar]
- 17.Lanford, R. E., D. Chavez, A. Barrera, and K. M. Brasky. 2003. An infectious clone of woolly monkey hepatitis B virus. J. Virol. 77:7814-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanford, R. E., D. Chavez, K. M. Brasky, R. B. Burns III, and R. Rico-Hesse. 1998. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc. Natl. Acad. Sci. USA 95:5757-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanford, R. E., D. Chavez, L. Notvall, and K. M. Brasky. 2003. Comparison of tamarins and marmosets as hosts for GBV-B infections and the effect of immunosuppression on duration of viremia. Virology 311:72-80. [DOI] [PubMed] [Google Scholar]
- 20.Lanford, R. E., and L. E. Estlack. 1998. A cultivation method for highly differentiated primary chimpanzee hepatocytes permissive for hepatitis C virus replication. Methods Mol. Med. 19:501-516. [DOI] [PubMed] [Google Scholar]
- 21.Lentz, B. R., and J. K. Lee. 1999. Poly(ethylene glycol) (PEG)-mediated fusion between pure lipid bilayers: a mechanism in common with viral fusion and secretory vesicle release? Mol. Membr. Biol. 16:279-296. [DOI] [PubMed] [Google Scholar]
- 22.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1999. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J. Virol. 73:2052-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, J. S., S. P. Tong, and J. R. Wands. 1999. Identification and expression of glycine decarboxylase (p120) as a duck hepatitis B virus pre-S envelope-binding protein. J. Biol. Chem. 274:27658-27665. [DOI] [PubMed] [Google Scholar]
- 24.Mabit, H., C. Vons, S. Dubanchet, F. Capel, D. Franco, and M. A. Petit. 1996. Primary cultured normal human hepatocytes for hepatitis B virus receptor studies. J. Hepatol. 24:403-412. [DOI] [PubMed] [Google Scholar]
- 25.Macrae, D. R., V. Bruss, and D. Ganem. 1991. Myristylation of a duck hepatitis B virus envelope protein is essential for infectivity but not for virus assembly. Virology 181:359-363. [DOI] [PubMed] [Google Scholar]
- 26.Persing, D. H., H. E. Varmus, and D. Ganem. 1987. The preS1 protein of hepatitis B virus is acylated at its amino terminus with myristic acid. J. Virol. 61:1672-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radziwill, G., W. Tucker, and H. Schaller. 1990. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J. Virol. 64:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sureau, C., B. Guerra, and R. E. Lanford. 1993. Role of the large hepatitis B virus surface envelope protein in infectivity of the hepatitis delta virion. J. Virol. 67:366-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sureau, C., J. R. Jacob, J. W. Eichberg, and R. E. Lanford. 1991. Tissue culture system for infection with human hepatitis delta virus. J. Virol. 65:3443-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sureau, C., A. M. Moriarty, G. B. Thornton, and R. E. Lanford. 1992. Production of infectious hepatitis delta virus in vitro and neutralization with antibodies directed against hepatitis B virus pre-S antigens. J. Virol. 66:1241-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong, S. P., J. S. Li, and J. R. Wands. 1995. Interaction between duck hepatitis B virus and a 170-kilodalton cellular protein is mediated through a neutralizing epitope of the pre-S region and occurs during viral infection. J. Virol. 69:7106-7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urban, S., and P. Gripon. 2002. Inhibition of duck hepatitis B virus infection by a myristoylated pre-S peptide of the large viral surface protein. J. Virol. 76:1986-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]